Abstract
AMP-activated protein kinase (AMPK) senses changes in the intracellular AMP/ATP ratio, switching off energy-consuming processes and switching on catabolic pathways in response to energy depletion. Here, we show that AMPK down-regulates rRNA synthesis under glucose restriction by phosphorylating the RNA polymerase I (Pol I)-associated transcription factor TIF-IA at a single serine residue (Ser-635). Phosphorylation by AMPK impairs the interaction of TIF-IA with the TBP-containing promoter selectivity factor SL1, thereby precluding the assembly of functional transcription initiation complexes. Mutation of Ser-635 compromises down-regulation of Pol I transcription in response to low energy supply, supporting that activation of AMPK adapts rRNA synthesis to nutrient availability and the cellular energy status.
Keywords: AMPK, glucose deprivation, RNA polymerase I, TIF-IA, transcription
The maintenance of cellular energy levels in response to changes in nutrient availability, exercise, or stress stimuli is pivotal for cellular homeostasis. Disruption of this balance is associated with a number of pathologies including diabetes, cancer and heart disease. AMP-activated protein kinase (AMPK) plays a crucial role in translating changes in energy levels into adaptive cellular responses (1, 2). Stimuli that increase the cellular AMP/ATP ratio lead to phosphorylation-dependent activation of AMPK by upstream protein kinases, such as LKB1 and/or the calcium/calmodulin-dependent protein kinase (CaMKK) (3–5). AMPK serves important functions in the nucleus, modulating the activity of specific transcriptions factors and coregulators, such as HNF4a (6), ChRBP (7), p53 (8), and p27 (9). Thus, AMPK-dependent phosphorylation provides a direct link between cellular energy metabolism and regulation of gene expression.
Conditions that harm cell growth, including stress, nutrient starvation or toxic lesions, down-regulate transcription of rRNA genes (rDNA), whereas agents that stimulate growth and proliferation upregulate rDNA transcription (10–12). TIF-IA, the Pol I-associated transcription factor that transmits external signals to the nucleolar transcription machinery, is targeted by a variety of protein kinases that phosphorylate TIF-IA at multiple sites. For example, increase in pre-rRNA synthesis upon mitogenic stimulation correlates with phosphorylation of TIF-IA at Ser-633 and Ser-649 by ERK and RSK (13). Moreover, mTOR-dependent phosphorylation at Ser-44 activates TIF-IA, whereas Thr-200 phosphorylation by JNK2 impairs TIF-IA activity (14, 15). Given the central function of AMPK in sensing the cellular energy status, we surmised that AMPK-dependent phosphorylation of the Pol I transcription apparatus would be an efficient means to down-regulate rDNA transcription and ribosome biogenesis at low ATP levels. We show that activation of AMPK by glucose deprivation or treatment with the AMP-mimetic drug AICAR (5-amino-4-imidazolecarboxamide ribonucleotide) (16) leads to inactivation of TIF-IA both in vivo and in vitro. AMPK phosphorylates TIF-IA at serine 635 (Ser-635), and this phosphorylation impairs the interaction of TIF-IA with the TBP-containing promoter selectivity factor SL1. Consequently, recruitment of Pol I to the rDNA promoter and transcription complex formation is impaired. The results decipher a unique connection between the AMPK pathway and the nucleolar transcription machinery, implicating a mechanism that adapts rRNA synthesis to changes in cellular energy supply.
Results
Glucose Deprivation Leads to AMPK-Dependent Repression of Pol I Transcription.
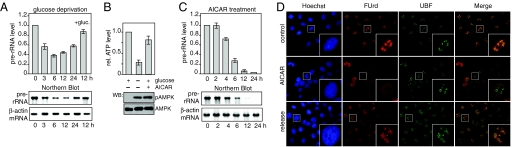
In mammalian cells, glucose deprivation leads to rapid down-regulation of rDNA transcription, the level of pre-rRNA decreasing by 50–70% within 6 h (Fig. 1A and Fig. S1). Transcription was restored after refeeding with glucose, indicating that inhibition of pre-rRNA synthesis is relieved when normocaloric conditions are reestablished. Consistent with glucose deprivation decreasing the cellular energy charge, ATP levels dropped by 70% (Fig. 1B Upper). The drop of cellular ATP and concomitant increase in AMP levels triggered phosphorylation-dependent activation of AMPK at Thr-172 (17), suggesting that AMPK down-regulates pre-rRNA synthesis by targeting component(s) of the Pol I transcription machinery (Fig. 1B Lower). Indeed, activation of AMPK by AICAR (5-amino-4-imidazolecarboxamide ribonucleotide) (16) treatment caused an even stronger decrease in pre-rRNA synthesis than glucose deprivation (Fig. 1 B and C). Although in low glucose Pol I transcription partially recovered, presumably due to AMPK-dependent activation of catabolic pathways that refill the ATP pool, Pol I transcription remained repressed after AICAR treatment, supporting the notion that AICAR activates AMPK independent of the cellular energy charge.
Fig. 1.
Glucose restriction or AICAR treatment inhibits Pol I transcription. (A) HEK293T cells were cultured for the indicated times (0–24 h) in glucose-free DMEM, or cultured for 12 h in glucose-rich medium after 12 h of glucose deprivation (+ gluc.). Pre-rRNA levels were determined by RT-qPCR and normalized to β-actin mRNA (Upper). Data are from three independent experiments, error bars represent the mean ± SD. (Lower) Northern blot of the same RNA samples. (B) ATP levels were determined in HEK293T cells cultured for 6 h in glucose-rich, in glucose-free, or in glucose-rich DMEM supplemented with 0.5 mM AICAR. Error bars, the mean ± SD from three independent experiments (Upper). pAMPK was detected on immunoblots with a phosphoThr172-specific antibody, the total level of AMPK with α-AMPK antibody (Lower). (C) HEK293T cells were incubated with 0.5 mM AICAR for the indicated times. (D) U2OS cells were either maintained in DMEM (control), were cultured for 12 h in the presence of 0.5 mM AICAR, or were first treated for 12 h with 0.5 mM AICAR and then released into AICAR-free medium for 12 h. Cells were pulse-labeled with FUrd, and incorporation into nascent RNA was detected by indirect immunofluorescence. Cells were costained with α-UBF antibodies and Hoechst 33342.
To demonstrate that AMPK interferes with Pol I transcription, we pulse-labeled nascent RNA with fluorouridine (FUrd) in the absence or presence of AICAR and visualized FUrd-labeled RNA by indirect immunofluorescence. Consistent with the high transcription rate of rRNA genes, FUrd staining was most pronounced in nucleoli, colocalizing with the basal Pol I transcription factor UBF (Fig. 1D Top). Nucleolar labeling was abolished after treatment with AICAR, indicating that AMPK abrogated pre-rRNA synthesis (Fig. 1D Middle). Noteworthy, AICAR treatment induced relocalization of UBF to the nucleolar periphery reminiscent of redistribution of UBF observed in actinomycin D-treated cells (18). Nucleolar transcription fully recovered after removal of AICAR, demonstrating that repression of Pol I transcription by AMPK is reversible (Fig. 1D Bottom).
TIF-IA Rescues Pol I Transcription in Extracts from Glucose-Deprived Cells.
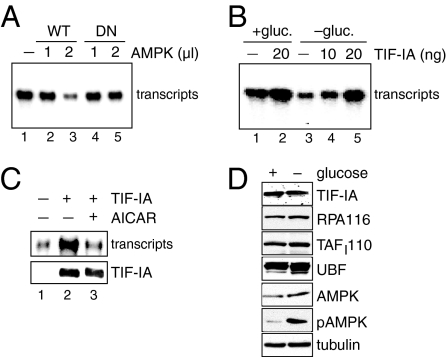
To explore the molecular mechanism underlying AMPK-dependent down-regulation of Pol I transcription, we assayed the effect of AMPK on rDNA transcription in vitro. Addition of immunopurified HA-tagged AMPK (Fig. S2) to a fractionated nuclear extract inhibited the synthesis of Pol I-specific runoff transcripts in a dose-dependent manner (Fig. 2A lanes 1–3), whereas the kinase-deficient mutant HA-AMPK/DN did not affect transcription (Fig. 2A lanes 4 and 5).
Fig. 2.
Activation of AMPK inhibits TIF-IA. (A) AMPK impairs Pol I transcription in vitro. Immunopurified HA-AMPK (WT) or HA-AMPK/DN (DN) were preincubated with a partially purified nuclear extract (NE) for 20 min at 30 °C in the presence of 10 μM ATPγS and 0.3 mM AMP before transcription was started by adding template DNA and nucleotides. Runoff transcripts were analyzed by PAGE and PhosphorImaging. (B) TIF-IA rescues Pol I transcription in extracts from glucose-deprived cells. Forty micrograms of nuclear extract from HEK293T cells cultured in glucose-rich (+ gluc.) or in glucose-free (− gluc.) medium were used in in vitro transcription assays supplemented with 10–20 ng of immunopurified TIF-IA. (C) TIF-IA from AICAR-treated cells is inactive. HEK293T cells expressing Fl-TIF-IA were cultured in the absence or presence of AICAR. TIF-IA was immunopurified and assayed for transcriptional activity using a nuclear extract from glucose-deprived HEK293T cells. A silver-stained PAA gel is shown (Lower). (D) The level of Pol I and basal Pol I-specific transcription factors is not altered during glucose deprivation. Extracts from HEK293T cells cultured in glucose-rich or -free DMEM were analyzed on immunoblots using the indicated antibodies.
We then compared the transcriptional activity of nuclear extracts from glucose-deprived and control HEK293T cells. Transcription of the rDNA template was markedly reduced in extracts from glucose-deprived cells (see Fig. 2B), indicating that AMPK-dependent repression of Pol I transcription can be mimicked in vitro. Importantly, transcriptional repression was overcome by addition of recombinant TIF-IA, demonstrating that glucose deprivation has inactivated TIF-IA (Fig. 2B lanes 3–5). In support of this conclusion, TIF-IA from control cells, but not from AICAR-treated cells, restored transcription in extracts from glucose-deprived cells (Fig. 2C). The levels of individual components of the Pol I transcription apparatus, including TIF-IA, Pol I (RPA116), SL1 (TAFI110), and UBF, were comparable in both extracts, indicating that neither the synthesis nor stability of these proteins were affected (Fig. 2D).
AMPK Phosphorylates TIF-IA at Serine 635.
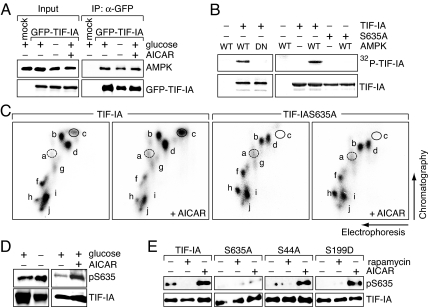
TIF-IA is phosphorylated at multiple sites by numerous protein kinases (13–15, 19). Group-based phosphosite prediction (http://gps.biocuckoo.org) revealed that Ser-635, a residue conserved within the C-terminal domain of vertebrate TIF-IA (Fig. S3A), is a putative AMPK target site. In support of AMPK targeting TIF-IA, coimmunoprecipitation experiments revealed that TIF-IA and AMPK interact with each other (Fig. 3A and Fig. S3B). To examine whether TIF-IA is phosphorylated by AMPK, we monitored phosphorylation of GST-TIF-IA by immunopurified AMPK in vitro. As shown in Fig. 3B, HA-AMPK, but not the kinase-deficient mutant HA-AMPK/DN, was capable to phosphorylate TIF-IA. Substituting Ser-635 by alanine (TIF-IAS635A) compromised phosphorylation, supporting that AMPK targets Ser-635 (Fig. 3B Right).
Fig. 3.
AMPK phosphorylates TIF-IA at Ser-635. (A) TIF-IA interacts with AMPK. GFP-TIF-IA from HEK293T cells cultured in glucose-rich or -free DMEM or treated with AICAR was immunoprecipitated and coprecipitated AMPK was detected on Western blots. (B) AMPK phosphorylates TIF-IA in vitro. GST-TIF-IA (TIF-IA) or GST-TIF-IAS635A (S635A) were incubated with HA-AMPK/WT or HA-AMPK/DN in the presence of [γ-32P]ATP and separated by SDS/PAGE. (Lower) The Coomassie-stained gel shows the amount of GST-TIF-IA and GST-TIF-IAS635A. (C) AMPK phosphorylates TIF-IA in vivo. HEK293T cells expressing Fl-TIF-IA or Fl-TIF-IAS635A were labeled for 4 h with 32P-orthophosphate in the absence or presence of 2 mM AICAR, and TIF-IA was subjected to tryptic phosphopeptide mapping. (D) Activation of AMPK increases Ser-635 phosphorylation. Fl-TIF-IA was immunoprecipitated from glucose-deprived (Left) or AICAR-treated (Right) HEK293T cells. Phosphorylation of Ser-635 was monitored with antibodies against phosphoSer635 (pS635); TIF-IA levels were determined with α-TIF-IA antibodies. (E) Ser-635 phosphorylation does not depend on mTOR signaling. Fl-TIF-IA or the indicated mutants (S635A, S44A, and S199) were immunopurified from untreated, rapamycin-treated (50 nM, 4 h) or AICAR-treated (1 mM, 12 h) HEK293T cells. Ser-635 phosphorylation was assayed by immunoblotting with α-pS635 antibodies (Upper); TIF-IA levels were determined with α-Flag antibodies (Lower).
To demonstrate AMPK-dependent phosphorylation of Ser-635 in vivo, we metabolically labeled TIF-IA with [32P]orthophosphate in the absence or presence of AICAR, and mapped the tryptic phosphopeptides by electrophoresis and chromatography. Upon stimulation by AICAR, labeling of spot c was markedly enhanced in wild-type but not mutant TIF-IAS635A (Fig. 3C). Spot c comprises the C-terminal tryptic peptide that harbors Ser-635, Ser-633, and Ser-649 (Fig. S3C). Ser-633 and Ser-649 are phosphorylated by ERK and RSK, phosphorylation at Ser-635 precluding phosphorylation of Ser-633 and Ser-649 (13). Therefore, single and multiple phosphorylations at these phosphoacceptor sites give rise to spots c, d, f, and g, peptide c being exclusively phosphorylated at Ser-635 (Fig. S3C). The different position of spot d in the maps of wild-type and mutant TIF-IA is due to the serine-alanine substitution, which increases the mobility of phosphopeptide d (Fig. S3D). Consistent with a functional cross-talk between AMPK and mTOR (20, 21), phosphorylation of Ser-199 (spot a), which decreases in response to mTOR signaling (14), was slightly increased after AICAR treatment (Fig. 3C). Unexpectedly, phosphorylation of Ser-44 (spot b), which is enhanced in response to mTOR signaling, was not decreased upon AMPK activation, indicating that mTOR and AMPK regulate TIF-IA independently. This result is supported by immunoblotting with phosphoSer635-specific antibodies, showing that Ser-635 phosphorylation was strongly enhanced in glucose-deprived and AICAR-treated but not in rapamycin-treated cells (Fig. 3 D and E). As expected, no phosphorylation was detected in TIF-IAS635A. Mutations of Ser-44 and Ser-199, residues that are targeted by mTOR signaling, did not affect AICAR-induced Ser-635 phosphorylation (Fig. 3E), consistent with AMPK inactivating TIF-IA directly by Ser-635 phosphorylation rather than indirectly by inhibition of mTOR.
AMPK-Dependent Phosphorylation Inactivates TIF-IA.
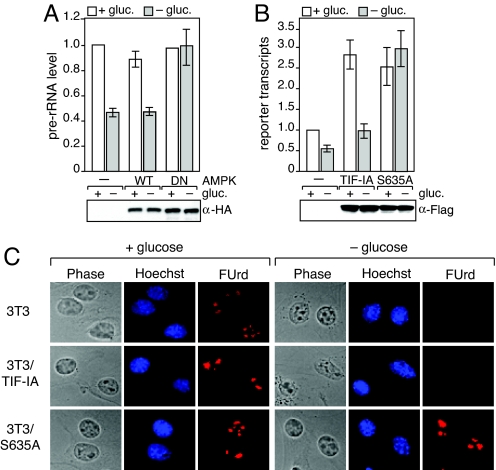
Given that phosphorylation by AMPK inhibits TIF-IA activity in conditions of low energy supply, inactivation of AMPK should prevent down-regulation of Pol I transcription upon glucose restriction. To test this assumption, we overexpressed HA-AMPK or the dominant-negative mutant HA-AMPK/DN and monitored pre-rRNA synthesis in normocaloric and glucose-deprived cells. Glucose deprivation reduced pre-rRNA levels by ≈50% in mock-transfected cells or in cells overexpressing the catalytic subunit of AMPK (HA-AMPK/WT) (Fig. 4A). However, upon overexpression of the dominant–negative mutant HA-AMPK/DN, pre-rRNA levels remained unaffected, regardless whether cells were cultured in the presence or absence of glucose, supporting that activation of AMPK is required for down-regulation of rDNA transcription upon depletion of the cellular ATP pool.
Fig. 4.
AMPK-dependent phosphorylation of TIF-IA at S635 down-regulates Pol I transcription. (A) Overexpression of a dominant-negative mutant of AMPK (DN) counteracts repression of Pol I transcription under glucose-restriction. (Upper) HEK293T cells expressing HA-AMPK/WT or HA-AMPK/DN were cultured for 6 h in the presence (white bars) or absence (gray bars) of glucose. Left bars represent values from mock-transfected cells. Pre-rRNA levels were determined by RT-qPCR and normalized to β-actin mRNA. (Lower) The immunoblot shows equal expression levels of HA-AMPK/WT and HA-AMPK/DN. (B) Overexpression of TIF-IAS635A prevents down-regulation of Pol I transcription upon glucose-deprivation. HEK293T cells were transfected with the rDNA reporter pHrP2-BH alone (−) or were cotransfected with expression plasmids encoding Fl-TIF-IA (TIF-IA) or Fl-TIF-IAS635A (S635A). Cells were cultured in glucose-rich medium (white bars) or glucose-free-medium for 6 h before harvesting (gray bars). The graph represents the level of reporter transcripts normalized to β-actin mRNA as determined by RT-qPCR in three independent experiments. (C) TIF-IAS635A rescues rRNA synthesis in glucose-deprived cells. Mock-infected NIH 3T3 cells and cells stably expressing Fl-TIF-IA or Fl-TIF-IAS635A were cultured for 12 h in the presence or absence of glucose. After pulse-labeling with FUrd, nascent RNA was visualized by indirect immunofluorescence with α-BrU antibodies.
To demonstrate that inhibition of pre-rRNA synthesis was due to phosphorylation of TIF-IA at Ser-635, we transfected HEK293T cells with expression plasmids encoding Flag-tagged TIF-IA or mutant TIF-IAS635A together with an rDNA minigene reporter plasmid (pHrP2-BH), and monitored reporter transcripts by RT-PCR. In control cells, both wildtype and mutant TIF-IA stimulated transcription approximately 3-fold (Fig. 4B). Removal of glucose, however, impaired transcription in cells expressing wild-type TIF-IA but not in cells expressing TIF-IAS635A. To analyze transcription of endogenous rDNA in the presence of wildtype or mutant TIF-IA, we monitored FUrd-incorporation into nascent RNA using NIH 3T3 cells that stably express TIF-IA or TIF-IAS635A (Fig. S4). Upon glucose deprivation, FUrd-incorporation was virtually abolished both in control cells and in cells harboring ectopic TIF-IA (Fig. 4C Top and Middle). In contrast, expression of TIF-IAS635A prevented down-regulation of pre-rRNA synthesis upon glucose withdrawal (see bright nucleolar FUrd-labeling in the presence and absence of glucose in Fig. 4C Bottom. Thus, mutation of S635 renders TIF-IA resistant to inactivation by AMPK, reinforcing that phosphorylation of Ser-635 by AMPK down-regulates Pol I transcription in response to energy limitation.
Phosphorylation of TIF-IA at S635 Impairs Transcription Complex Formation.
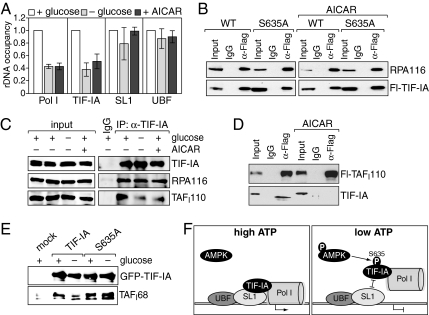
TIF-IA plays a key role in transcription initiation complex formation, recruiting Pol I to the transcription start site by association with the preinitiation complex comprising UBF and the TBP-TAFI complex SL1 (22, 23). We used ChIP to examine whether AMPK-dependent phosphorylation of TIF-IA affects the association of TIF-IA and/or Pol I with rDNA. After glucose deprivation or AICAR treatment, promoter occupancy of Pol I and TIF-IA, but not of UBF and SL1, decreased by ≈60%, suggesting that AMPK prevented TIF-IA-dependent recruitment of Pol I (Fig. 5A). Pol I occupancy at the rDNA coding region dropped accordingly, whereas no change in UBF occupancy was observed (Fig. S5 and Table S1).
Fig. 5.
Activation of AMPK inhibits transcription complex formation. (A) AMPK impairs rDNA occupancy of TIF-IA and Pol I. Cross-linked chromatin from HEK293T cells cultured in glucose-rich (white bars) or glucose-free DMEM (light gray bars), or treated with 1 mM AICAR for 6 h (dark gray bars) was immunoprecipitated with antibodies against TIF-IA, Pol I (RPA116), SL1 (TAFI110), and UBF. Precipitated DNA was assayed by qPCR. The bar diagram shows rDNA promoter occupancy upon AMPK activation (− glucose, + AICAR) relative to normocaloric conditions (+ glucose). Error bars, the mean ± SD from four independent experiments. (B) Phosphorylation of Ser-635 does not impair the association of TIF-IA with Pol I. Lysates from untreated or AICAR-treated HEK293T cells expressing Fl-TIF-IA (WT) or Fl-TIF-IAS635A (S635A) were incubated with α-Flag antibody (α-Flag) or mouse IgG, and the immunoprecipitates were assayed on Western blots with antibodies against Pol I (RPA116) or the Flag-epitope. The upper blot represents 80%, and the lower blot represents 10% of the immunocomplexes. (C) Activation of AMPK prevents the association of TIF-IA with SL1. TIF-IA was immunoprecipitated from HEK293T cells that were treated as in A and coprecipitation of Pol I and SL1 was monitored with α-RPA116 and α-TAFI110 antibodies. (D) AICAR treatment impairs the interaction of TIF-IA with SL1. HeLa cells stably expressing Flag-TAFI110 were treated with AICAR or left untreated, and subjected to immunoprecipitation with α-Flag antibodies as in B. Immunoprecipitates were analyzed on immunoblots using antibodies against TAFI110 (Upper) and TIF-IA (Lower). (E) Pull-down assay. Immobilized GFP-TIF-IA and GFP-TIF-IAS635A from HEK293T cells grown in the presence or absence of glucose were incubated with partially purified SL1, and bound SL1 was monitored with antibodies against TAFI68. (F) Model depicting the role of AMPK-dependent phosphorylation of TIF-IA in transcription complex formation. Under normocaloric conditions (high ATP) Ser-635 of TIF-IA is unphosphorylated, enabling TIF-IA to interact with SL1 and to recruit Pol I to the transcription start site (Left). Upon activation of AMPK (low ATP), TIF-IA is phosphorylated at Ser-635, which abrogates recruitment of the TIF-IA/Pol I complex to promoter-bound SL1 and prevents transcription complex assembly (Right).
To examine whether phosphorylation by AMPK affects the interaction of TIF-IA with Pol I and/or with SL1, we immunoprecipitated Flag-tagged TIF-IA or TIF-IAS635A from untreated, glucose-deprived or AICAR-treated cells, and monitored coprecipitated Pol I (RPA116) on immunoblots. Similar amounts of Pol I were coprecipitated with wildtype and mutant TIF-IA in untreated and treated cells, indicating that phosphorylation of Ser-635 does not impair the association of TIF-IA with Pol I (Fig. 5B). Likewise, the interaction between Pol I and endogenous TIF-IA was not affected upon glucose-deprivation or AICAR-treatment (Fig. 5C Upper). In contrast, the association between TIF-IA and SL1 was strongly reduced upon glucose deprivation or AICAR treatment (Fig. 5C Lower). To prove this observation, we precipitated SL1 from a HeLa cell line that stably expresses Flag-tagged TAFI110 (24) and monitored coprecipitation of TIF-IA. Again, AICAR treatment abolished the interaction between TIF-IA and SL1 (Fig. 5D), in accord with AMPK impairing the interaction of TIF-IA with SL1. Consistently, GFP-TIF-IA from glucose-deprived cells was less capable to pull-down partially purified TAFI68, the second largest subunit of mouse SL1, than GFP-TIF-IA from control cells. GFP-TIF-IAS635A, however, bound to TAFI68 with similar efficiency, regardless whether it was prepared from control or glucose-deprived cells (Fig. 5E). These results demonstrate that Ser-635 phosphorylation affects the association of the TIF-IA/Pol I complex with promoter-bound SL1, thereby abrogating an early and important step in the assembly of productive transcription initiation complexes.
Discussion
It has long been recognized that a given nutritional state affects the cellular equilibrium in which the synthesis of ATP is balanced by its use in protein synthesis and other energy-consuming processes. If energy levels are low and the intracellular AMP/ATP ratio is elevated, AMPK switches on energy-producing pathways and switches off energy-consuming pathways to restore cellular ATP levels. Therefore, under conditions of nutrient shortage, cells down-regulate energy-consuming processes, like ribosome biogenesis and protein synthesis. At low cellular ATP levels, transcription of rRNA genes decreases, whereas elevated ATP levels enhance rRNA synthesis (25). AMPK targets multiple proteins ranging from key metabolic enzymes to transcription and translation factors (26). Here, we show activation of AMPK either by glucose-deprivation or by the AMP-mimetic drug AICAR triggers phosphorylation of TIF-IA at Ser-635, leading to inactivation of TIF-IA, disruption of transcription initiation complexes and inhibition of rRNA synthesis (Fig. 5F). These results uncover a yet unrecognized mechanism mammalian cells use to couple the cellular energy status to ribosome biogenesis, cell growth, and proliferation.
TIF-IA, the mammalian homolog of yeast Rrn3p (27, 28), confers most of growth-dependent control of rDNA transcription. Conditions that support growth and proliferation, such as nutrients and growth factors, induce mTOR- and MAPK-dependent signaling pathways that activate TIF-IA by phosphorylating specific serine residues (Ser-44, Ser-633, and Ser-649) (13, 14). Conversely, conditions that harm cell proliferation and activate JNK2, inactivate TIF-IA by triggering phosphorylation at Thr-200 (15). JNK2-, MAPK- and mTOR-dependent phosphorylation of TIF-IA regulates the interaction of TIF-IA with Pol I, affecting recruitment of Pol I to the rDNA promoter and the formation of productive transcription complexes. In contrast, AMPK-mediated phosphorylation of TIF-IA at Ser-635 does not compromise binding of TIF-IA to Pol I but abrogates the interaction between promoter-bound SL1 and TIF-IA, which also impairs transcription complex assembly. These results uncover another level of Pol I transcriptional regulation, TIF-IA sensing not only external signals but also translating changes of intracellular energy supply into changes of rRNA synthesis. Likewise, in yeast Pol I recruitment to rDNA depends on Rrn3p, the homolog of TIF-IA (28), and is controlled by the TOR-pathway (29). Phosphorylation of Pol I, rather than Rrn3p, regulates formation of the Rrn3p/Pol I complex (30), indicating that yeast and mammals use different mechanisms to regulate Pol I transcription. It will be interesting to learn whether SNF1, the yeast homolog of AMPK that is not activated by AMP (26), serves a similar function as AMPK, adapting rDNA transcription to the cellular energy status.
AMPK is known to repress mTOR activity by activating TSC2, a negative regulator of mTOR, and by phosphorylation of the mTORC1 component Raptor (20, 21). Therefore, one would expect that down-regulation of mTOR signaling at low energy supply is sufficient to impair TIF-IA activity. Indeed, phosphopeptide mapping of metabolically labeled TIF-IA revealed that AICAR treatment caused hyperphosphorylation of Ser-199, a residue that is also phosphorylated upon rapamycin-mediated inhibition of mTOR (14). However, activation of AMPK did not affect phosphorylation of Ser-44, which is dephosphorylated by PP2A upon abrogation of mTOR-signaling (14). This observation may be explained by the fact that activation of AMPK will compromise PP2A activity in a FOXO3-dependent manner (31, 32), preventing PP2A-dependent dephosphorylation of Ser-44. Coregulation by mTOR-signaling and AMPK has also been reported for the elongation factor 2 kinase (eEF2K), a negative regulator of eE2F, which is inactivated by mTOR-dependent phosphorylation at Ser-366 (33) and activated by AMPK-mediated phosphorylation of Ser-398 (34). Thus, AMPK affects both eE2FK and TIF-IA, key factors required for translation and ribosome biogenesis, ensuring a fine tuned regulation of the cellular protein synthesis machinery in response to nutrient and energy availability.
Recently, Murayama et al. (35) have described another mechanism that couples the cellular energy status to repression of rDNA transcription. They have identified a protein complex, energy-dependent Nucleolar Silencing Complex (eNoSC), which contains the NAD+-dependent histone deacetylase SIRT1, the heterochromatic histone methyltransferase SUV39H1, and Nucleomethylin (NML), a nucleolar protein that specifically binds to the heterochromatic histone mark H3K9me2. eNoSC changes the ratio of active to silent rRNA genes in response to the cellular energy status. Upon prolonged glucose depletion, increased NAD+ levels activate SIRT1 and SUV39H1, which in turn trigger heterochromatin formation and transcriptional silencing. Thus, cells use at least two mechanisms to down-regulate nucleolar activity according to the overall level of energy supply, a fast response involving AMPK-dependent inactivation of TIF-IA and a slow response involving SIRT1- and SUV39H1-dependent epigenetic regulation.
Materials and Methods
Cell Culture, Transfection, Retroviral Infection, and RNA Analysis.
Cells were cultured in DMEM containing 25 mM glucose and 10% FCS. For glucose deprivation, cells were transferred to glucose-free DMEM. Transfection was performed using the calcium phosphate-DNA coprecipitation method. Infection of NIH 3T3 cells was carried out with pBabe based retroviruses encoding Flag-TIF-IA and TIF-IAS635A. Cellular pre-rRNA and β-actin mRNA was analyzed on Northern blots using specific riboprobes. Alternatively, pre-rRNA levels or reporter gene transcripts were measured by reverse transcription using random hexamer primers and quantitative real-time PCR. Data were normalized to the level of β-actin-mRNA. Primer sequences are given in Table S2.
FUrd Labeling of Nascent RNAs and Immunofluorescence.
Cells grown on coverslips were labeled with 2 mM fluorouridine (FUrd) for 15 min. Cells were fixed in 2% para-formaldehyde, permeabilized with methanol at −20 °C, and incubated with α-BrU and α-UBF antibodies, followed by incubation with the appropriate secondary antibodies coupled to Cy3 or FITC.
Extract Preparation and In Vitro Transcription.
Nuclear extracts were prepared from HEK293T cells grown in glucose-rich or -free DMEM. In vitro transcription assays contained 40 μg of nuclear extract or 15 μg of nuclear extract fractionated on a DEAE-column (280 mM KCl elution step) and 40 ng of template pHrP2 linearized with NdeI. To assay TIF-IA activity, 10–50 ng of immunopurified Flag-TIF-IA expressed in HEK293T cells were added. The amount and purity of Flag-TIF-IA was monitored on silver-stained polyacrylamide gels.
ChIP, Immunoprecipitation, and Pull-Down.
ChIP assays were performed as described in ref. 36. For immunoprecipitation, cells were lysed in buffer AM-200 (20 mM Tris·HCl, pH 7.9, 200 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol) containing 0.2% Nonidet P-40, PhosphoSTOP (Roche) and protease inhibitors (Roche), tagged proteins were precipitated with α-Flag or α-HA antibodies and eluted with buffer E (20 mM Tris·HCl, pH 7.9, 300 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, and 0.1% Nonidet P-40) containing 200 μg/mL epitope peptide. HeLa cells expressing Flag-TAFI110 have been described (24). For pull-down assays, immobilized GFP-TIF-IA was incubated with partially purified SL1. After washing and elution, proteins were analyzed by immunoblotting.
In Vitro Kinase Assays, Metabolic Labeling, and Tryptic Phosphopeptide Analysis.
For in vitro phosphorylation, GST-TIF-IA and immunopurified HA-AMPK were incubated for 30 min at 30 °C in the presence of 10 μCi [γ-32P]ATP (5,000 Ci/mmol). 32P-labeled GST-TIF-IA was separated by SDS/PAGE and visualized with a PhosphorImager. Tryptic phosphopeptide mapping of metabolically labeled Flag-TIF-IA was performed as described in ref. 13. For AICAR treatment, cells were preincubated for 2 h with 2 mM AICAR before metabolic labeling.
ATP Assay.
Cellular ATP levels were measured using the ATP Bioluminescence Assay Kit (Roche) according to the manufacturer's instructions.
Additional Details.
For additional details of plasmids, antibodies, expression and purification of AMPK, and AMPK assay, see SI Text.
Supplementary Material
Acknowledgments.
We thank K. L. Guan (University of California, San Diego, CA) for providing HA-AMPK expression plasmids. This work was supported by the Deutsche Forschungsgemeinschaft, the European Union Network “Epigenome,” and the Fonds der Chemischen Industrie.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909873106/DCSupplemental.
References
- 1.Hardie DG, Carling D. The AMP-activated protein kinase—fuel gauge of the mammalian cell? Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 2.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Shaw RJ, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Hawley SA, et al. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Woods A, et al. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Leclerc I, et al. Hepatocyte nuclear factor-4alpha involved in type 1 maturity-onset diabetes of the young is a novel target of AMP-activated protein kinase. Diabetes. 2001;50:1515–1521. doi: 10.2337/diabetes.50.7.1515. [DOI] [PubMed] [Google Scholar]
- 7.Kawaguchi T, et al. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: Regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J Biol Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- 8.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Liang J, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 10.Moss T. At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 14.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19:933–941. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-aminoimidazole-4-carboxamide ribonucleoside. Eur J Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 17.Hawley SA, et al. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 18.Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bierhoff H, Dundr M, Michels AA, Grummt I. Phosphorylation by casein kinase 2 facilitates rRNA gene transcription by promoting dissociation of TIF-IA from elongating RNA polymerase I. Mol Cell Biol. 2008;28:4988–4998. doi: 10.1128/MCB.00492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 21.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan X, et al. Multiple interactions between RNA polymerase I, TIF-IA and TAF(I) subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 2002;3:1082–1087. doi: 10.1093/embo-reports/kvf212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller G, et al. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heix J, et al. Mitotic silencing of human rRNA synthesis: Inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grummt I, Grummt F. Control of nucleolar RNA synthesis by the intracellular pool sizes of ATP and GTP. Cell. 1976;7:447–453. doi: 10.1016/0092-8674(76)90175-6. [DOI] [PubMed] [Google Scholar]
- 26.Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 27.Bodem J, et al. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 2000;1:171–175. doi: 10.1093/embo-reports/kvd032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moorefield B, Greene EA, Reeder RH. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc Natl Acad Sci USA. 2000;97:4724–4729. doi: 10.1073/pnas.080063997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claypool JA, et al. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fath S, et al. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc Natl Acad Sci USA. 2001;98:14334–14339. doi: 10.1073/pnas.231181398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni YG, et al. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc Natl Acad Sci USA. 2007;104:20517–20522. doi: 10.1073/pnas.0610290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greer EL, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, et al. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 35.Murayama A, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Ford E, et al. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.