Abstract
The use of plasmon coupling in metal nanoparticles has shown great potential for the optical characterization of many biological processes. Recently, we have demonstrated the use of “plasmon rulers” to observe conformational changes of single biomolecules in vitro. Plasmon rulers provide robust signals without photobleaching or blinking. Here, we show the first application of plasmon rulers to in vivo studies to observe very long trajectories of single biomolecules in live cells. We present a unique type of plasmon ruler comprised of peptide-linked gold nanoparticle satellites around a core particle, which was used as a probe to optically follow cell-signaling pathways in vivo at the single-molecule level. These “crown nanoparticle plasmon rulers” allowed us to continuously monitor trajectories of caspase-3 activity in live cells for over 2 h, providing sufficient time to observe early-stage caspase-3 activation, which was not possible by conventional ensemble analyses.
Keywords: caspase, live cell imaging, plasmonic nanoparticles, protease sensor, single-molecule imaging
Single molecule imaging has enabled the exploration of biomolecular dynamics and has revealed processes at work that are lost by extrapolation of ensemble assays (1–17). During the past few decades, optical single-molecule techniques such as single-fluorophore tracking (4, 5) and single-molecule fluorescence resonance energy transfer (1, 6–8) have proven to be effective tools for such purposes and have provided new biological information related to enzyme activity (6); transcription (7); protein dynamics (8); identification of rare intermediates and kinetic heterogeneity during RNA folding (5, 9); and RNA-protein interactions (10). Despite recent advances, however, most studies rely on biomolecules either immobilized on substrates or confined in a matrix, and observation of single-molecule behaviors inside live cells remains challenging (11–15). The inherent limitations of molecular dyes for single-molecule imaging, including low signal intensities, complex blinking phenomena, and photobleaching, are even more pronounced in live cell studies (16). Specifically, the fast photobleaching (less than a minute) of fluorescent dyes (17) hinders the ability to continuously monitor signaling pathways that transpire on the time scale of hours. New fluorophores such as green fluorescent proteins (GFPs) and quantum dots (QDs) have led to significant improvements (18, 19). For example, GFPs can be expressed as chimeras with a protein of interest to study subcellular localization in live cells, but are still prone to photobleaching (18). QDs show less bleaching, but blinking can make the interpretation of single-molecule data obtained with QDs ambiguous (20). Recent advances suggest that blinking can be controlled, but there are still remaining issues, such as a broad photoluminescence spectrum and biocompatibility (21–23).
We recently reported an alternative imaging technique using gold nanoparticle plasmon rulers for the optical readout of in vitro single-DNA hybridization and cleavage kinetics (24, 25). Because gold nanoparticles effectively scatter visible light and do not suffer from blinking or photobleaching (26), they have great potential to provide the highly enhanced signal intensity required for single-molecule detection and greatly extend the observation time for monitoring biological processes in living cells (27). To test this hypothesis, we chose caspase-dependent apoptotic signaling as a case study because apoptosis is an important biological process for maintaining homeostasis and function of the immune system and is highly related to various autoimmune diseases and cancer (28). Previous studies with ensemble techniques have shown that the activation of caspase-3 through the apoptotic signaling pathway can take from several minutes to hours (29). The long time-scale of these signaling events has made them difficult to measure continuously at the single-molecule level with established techniques. Here, we show an in vivo application of plasmon rulers in a study of caspase-3 signaling at the single-molecule level in living cells. The signal provided by these plasmon rulers allowed continuous observation of caspase-3 activation for over 2 h, unambiguously identifying early-stage activation of caspase-3 in apoptotic cells.
Results and Discussion
Design and Synthesis of Crown Nanoparticle Plasmon Rulers.
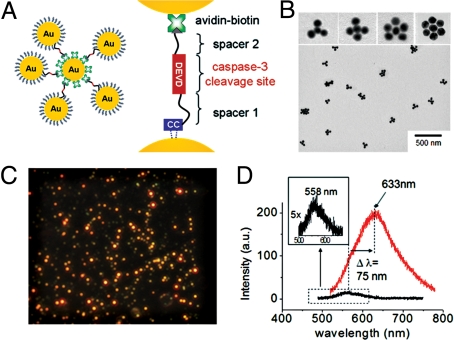
To apply the plasmon rulers to in vivo cell signaling, it is necessary to construct a nanoparticle assembly that provides a very clear signal at the single-molecule level in the highly heterogeneous and high-background scattering environment of live cells. For the design of such an assembly, it is important to recall that the polarizability, and hence the light-scattering intensity, for a pair of particles in close proximity (within one diameter) is significantly greater than that of 2 separate particles (24, 25). In fact, this applies to assemblies where the particle is surrounded by several others. Here we find, for instance, that a crown nanoparticle assembly with a core 40-nm particle surrounded by 5 others (Fig. 1 A and B) scatters light ≈44× more intensely than a single particle, along with a substantial spectral red shift (Δλ = 75 nm) (Fig. 1 C and D; supporting information (SI) Fig. S1). For this reason, we developed a series of crown nanoparticle plasmon rulers for this in vivo study of caspase-3 activation (Fig. 1B). These assemblies are linked together by peptides containing the DEVD sequence via NeutrAvidin-biotin and Au-thiol chemistries (SI Methods). It is well known that caspase-3 cleaves the DEVD sequence most efficiently; therefore, caspase-3 cleavage of one of the DEVD peptides (30) in the crown nanoparticle is expected to result in an immediate reduction in light-scattering intensity. Recent reports also suggest that other caspases can exhibit some activity for DEVD (31). Although the cleavage activities of caspase-6, 8, and 10 to this substrate are negligible (25 −103 times less active) compared with caspase-3, caspase-7 cleaves DEVD with similar efficiency. To minimize such cross-reactivity, we used a cell line in which caspase-7 is not expressed at the mRNA level. In addition, we inserted a glycine residue at the P1′ site, which is known to be optimal for caspase-3 cleavage activity. The use of a substrate containing a peptide bond avoids selectivity issues that have been observed with substrates containing activated amide bonds, such as aminocoumarin and para-nitroanilide (32, 33).
Fig. 1.
Crown nanoparticle plasmon rulers. (A) Schematic of crown nanoparticle probes, which are composed of a NeutrAvidin-coated gold-core nanoparticle with multiple biotinylated gold satellite nanoparticles. Peptides with the caspase-3 cleavage sequence DEVD crosslink the core and satellite nanoparticles via avidin-biotin interactions. (B) Transmission electron microscopy (TEM) of crown nanoparticles. Approximately 3–6 satellite nanoparticles are linked to the core nanoparticle. (C) A representative scattering image of crown nanoparticles. Intense and bright red-colored spots correspond to a single-crown nanoparticle. (D) Representative scattering spectra of a single-crown nanoparticle (red) and a single gold nanoparticle (black). A single-crown nanoparticle probe exhibited a significant red shift (Δλ = 75 nm) and highly increased scattering intensity (≈44 times) compared with a single gold nanoparticle.
In Vitro Studies and Analyses of Enzymatic Kinetics.
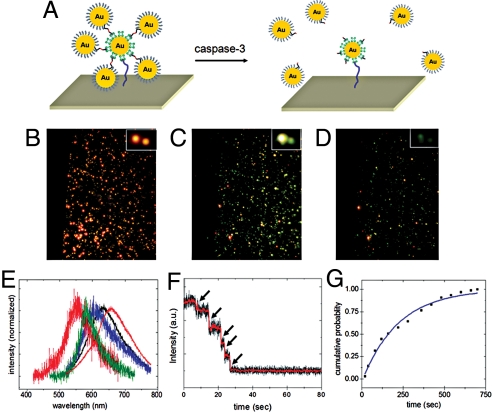
We first investigated the capabilities of the crown nanoparticles to detect caspase-3 activity under in vitro conditions. The NeutrAvidin-functionalized crown nanoparticles were immobilized on the biotinylated surface of a glass flow chamber (Fig. 2A). The scattering color and intensity were monitored under a dark-field microscope with a 100-watt tungsten lamp for illumination. Upon addition of caspase-3 (250 ng/mL), initial intense red-colored spots (Fig. 2B) gradually turned into yellow and then dim green spots as time elapsed (Fig. 2 C and D). Consistent with these observations, stepwise spectral blue-shifts in the scattering spectrum of a single-crown nanoparticle were detected due to sequential loss of satellite nanoparticles by caspase-3-mediated peptide cleavage (Fig. 2E). Single-particle trajectories of the scattering intensity recorded by electron-multiplying charge-coupled device (EMCCD; temporal frequency: 120 Hz) also show a stepwise decrease in the scattering intensity corresponding to each individual proteolytic event (Fig. 2F and Movie S1).
Fig. 2.
In vitro caspase-3-mediated cleavage of crown nanoparticle plasmon rulers. (A) Schematic of experimental design. Crown nanoparticles were immobilized on a glass flow chamber via avidin-biotin chemistry. After equilibrating with buffer, caspase-3 molecules were introduced, and the color and intensity changes of crown nanoparticles were monitored under dark-field microscopy. (B–D) Scattering color changes upon caspase-3 treatment of crown nanoparticles. Intense red-colored spots (B) progressed to yellow spots (C) and then dim green spots (D). (E) A representative spectral shift of a single-crown nanoparticle upon caspase-3 cleavage. Initially, the crown nanoparticle exhibited a scattering peak maximum at 654 nm (red). Gradually, blue shifts were observed as time elapses, corresponding to the sequential cleavage of satellite nanoparticles. (F) Single-crown nanoparticle intensity trace as a function of time. The trajectory was recorded at a temporal resolution of 100 Hz by using an intensified CCD detector. Stepwise-intensity drops were observed, which correspond to the sequential satellite nanoparticle cleavages. (G) Determination of rate constant using crown nanoparticle rulers as a substrate. The cumulative probability of cutting is shown as a function of time. Fitting the curve yielded first-order rate constant k. Using k and literature-reported Km, kcat was calculated to be 6.17 s−1.
To analyze the kinetics of cleavage by caspase-3, each proteolytic event was counted (n = 300) and plotted as a function of time (Fig. S2). The cumulative probability was calculated by dividing the number of cleavage events up to a given time, by the total number of cleavage events observed during the experimental time-course (Fig. 2G). The findings of this in vitro experiment fit well to a first-order kinetic model, with a kinetic rate constant (k) of 0.0046 s−1. Using literature values for Km and the rate constant derived from this work, the catalytic kinetic constant (kcat) was calculated to 6.17 s−1 (34). This value falls within the range of previously reported kcat values (2.4−8.2 s−1) obtained in ensemble studies (34). Control experiments either in the absence of caspase-3 or with crown nanoparticle probes constructed from a peptide lacking the DEVD sequence showed only minimal cleavage (Fig. S3).
Intracellular Delivery and Distribution of Plasmon Rulers.
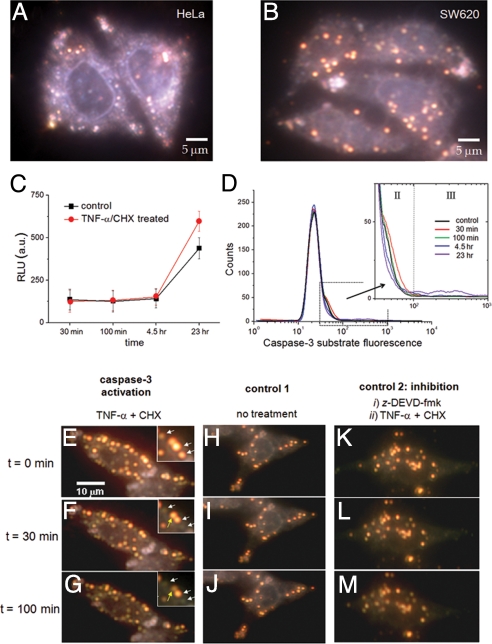
The in vitro experiments showed that the crown nanoparticles could be used to monitor caspase-3 activity and were modified for live cell experiments. To deliver the NeutrAvidin-functionalized crown nanoparticle probes inside of cells, they were conjugated with a biotinylated form of the cell penetration peptide, TAT. Either HeLa or SW620 cells were incubated with the TAT-modified crown nanoparticle plasmon rulers for 12 h to allow sufficient time for loading into cells, and then imaged with a dark-field microscope. The background scattering from the cell details its boundary, and the bright red spots inside suggest successful delivery of the plasmon rulers (Fig. 3 A and B). Without TAT modification, there was no indication of the nanoparticle delivery inside cells (Fig. S4); however, some of the nonspecifically bound plasmon rulers were seen on the culture plate at a high dose of nanoparticles (Fig. S5).
Fig. 3.
Cellular delivery of crown nanoparticles in live cells and their utilization for single-molecule imaging of caspase-3 activation in apoptotic cells. (A and B) Crown nanoparticles were first conjugated with the cell penetration peptide TAT and delivered into HeLa cells and colon cancer cells SW620. Bright red-colored spots correspond to individual crown nanoparticles. (C and D) Ensemble caspase-3 activity was measured by either a luminescence assay (Caspase-Glo 3/7; Promega) or flow cytometry 30 min, 100 min, 4.5 h, and 23 h after the addition of TNF-α/CHX. (C) The luminescence assay shows minimal caspase-3 activation at early stages. TNF-α/CHX induced caspase-3 activation is evident only at 23 h. (D) Flow cytometry findings are consistent with the luminescence assay. Shoulder peaks that might be indicative of caspase-3 activity appear at early time points. (E–G) Caspase-3 activation was induced by the addition of the apoptotic inducers TNF-α and CHX. As time elapses, some red spots turn into either dim red or green dots. (H–J) Vehicle-treated cells show almost no signal change during the entire observation time. (K–M) Cells were pretreated with the caspase-3 inhibitor z-DEVD-fmk, and subsequently treated with TNF-α/CHX. There are no indications of proteolysis throughout the time course.
Though definitive proof of cytosolic localization of the gold nanoparticles awaits further experimentation, their subcellular distribution can be inferred from the following: TAT peptides release a wide range of cargos, such as gold nanoparticles, from endosomal compartments following endocytosis (35). SW620 cells were incubated with the TAT-conjugated crown nanoparticles for 12 h to provide time for TAT-mediated endosomal escape. The nanoparticles that remain endosomally trapped will eventually fuse with lysosomes, which are not accessible to caspase-3 (36). The apoptotic inducer used in this study has been shown to permeabilize lysosomal membranes, facilitating the release of proapoptotic cathepsins (37). Therefore, the efficiency of our cytosolic delivery might be a function both of TAT-induced endosomal escape and lysosomal permeabilization during apoptosis. The caspase-3-mediated cleavage events described subsequently suggest that most crown nanoparticles are cytosolic.
The scattering signals from the plasmon rulers are highly intense compared with the background scattering from the cell, allowing continuous imaging at rates up to 250 Hz with an EMCCD. In addition, the scattering signal lasts more than ≈8 h under continuous illumination of visible light, with no signal decrease in healthy cells. The highly intense and stable scattering properties of crown nanoparticles should allow the detection of single-particle trajectories with high temporal resolution during caspase-3 activation.
Ensemble Assay of Caspase-3 Activity in Live Cells.
Before investigating caspase-3 activity in live cells with the crown nanoparticle plasmon rulers, we first measured caspase-3 activity in cells via conventional noncontinuous fluorescence-based ensemble techniques. SW620 cells were treated with the apoptotic inducers, tumor necrosis factor-α (TNF-α), and cycloheximide (CHX). SW620 cells were chosen because caspase-7, the major competitor for the DEVD sequence, is absent in SW620 cells at the mRNA level (http://biogps.gnf.org/#goto = welcome). In addition, it is well documented that SW620 cells are resistant to death receptor-induced apoptosis and exhibit low levels of caspase-3 activity upon addition of death receptor agonists (38). We reasoned that low levels of caspase-3 activity would be an excellent test of the single-molecule sensitivity of the crown nanoparticles. To confirm the resistance of SW620 cells to death receptor-mediated apoptosis, we used ensemble analyses with peptide substrates containing the DEVD sequence. Analysis of caspase activity with a luminescent assay (Caspase-Glo 3/7; Promega) showed almost no caspase-3 activity in cell lysates 4.5 h after the addition of TNF-α/CHX. In fact, death receptor-induced caspase-3 activity was detectable only after 23 h (Fig. 3C). To investigate caspase-3 activity on a cell-by-cell basis, we also performed flow cytometry with a cell-permeable fluorescent DEVD substrate. Consistent with the in vitro data, only a small percentage of cells show high levels of caspase-3 activity (labeled III in Fig. 3; fluorescence intensity between 102 and 103) 23 h after addition of TNF-α/CHX (Fig. 3D, III, and Figs. S6 and S7). Interestingly, at early time points, an unknown shoulder peak also appears (labeled II) and then decreases gradually (Fig. 3D, II).
In Vivo Plasmon Rulers for Imaging of Caspase-3 Activation.
Use of the crown nanoparticle plasmon rulers allows continuous observation of caspase-3 activation at the single-molecule level, and provides a clearer understanding of the origin of the shoulder peaks in the flow cytometry and the early-stage caspase-3 activation. Some crown nanoparticles exhibited sudden intensity drops after a short period (30 min and 100 min) following the addition of TNF-α/CHX (Fig. 3 E–G and Movie S2), which correspond to the cleavage of satellite nanoparticles by activated caspase-3. Because caspase-3 is present predominantly in the cytosol, the observed cleavage of crown nanoparticles during apoptosis is consistent with cytosolic localization. However, some nanoparticles (≈15%) do not show any signal reduction after addition of an apoptotic inducer. This is possibly due to residual endosomal trapping of a portion of the nanoparticle plasmon rulers. The peptide in our crown nanoparticle is a selective caspase-3 cleavage sequence and is unlikely to be cleaved by other endosomal proteases, such as papain-like proteases, because these proteases have been shown to be unable to degrade peptide substrates with P1 Asp (39).
To verify that the sudden intensity changes of the crown nanoparticle probes were due to the caspase-3 activity, a number of control experiments were performed. In the absence of TNF-α/CHX, virtually no changes in the scattering signals of the plasmon rulers were observed over the entire 2-h measurement period (Fig. 3 H–J). As another control, we pretreated the cells with a cell-permeable caspase-3 inhibitor, z-DEVD-fmk, and activated apoptotic signaling by the addition of TNF-α/CHX. No sign of cleavage was observed over the course of the experiment (Fig. 3 K–M). In a separate experiment, we measured the behavior of the crown plasmon nanoparticle rulers that are nonspecifically bound on the cell culture plate. The scattering signal of these rulers remained unchanged upon addition of TNF-α/CHX to cells (Fig. S5). These findings suggest that caspase-3 is activated at early time points during death receptor-induced apoptosis, and this activation is barely detectable by currently available ensemble analyses.
Interestingly, unlike small molecular probes, the movement of crown plasmon rulers is minimal during the time course. It is well known that the presence of networks of cytoskeleton filaments restricts the diffusion of large particles in cells (39, 40). Although there should be regional and cell-dependent differences, the size of void spaces (50–250 nm) is similar to that of plasmon rulers (≈100 nm) (40, 41). Their movement in the cytosol is hindered by such cytoskeletal sieving, and therefore they diffuse slowly. However, the cleaved satellite nanoparticles are smaller than the void size. Thus, upon cleavage, the freed individual particles can readily escape from the region of interest, which is defined as the diffraction limit (Fig. S8). Such limited diffusion of our plasmon rulers in the cytosol enables facile interpretation of single-particle trajectories with minimal signal perturbation by the probe movement.
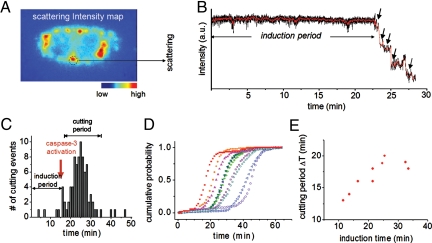
Single-particle trajectories of the crown nanoparticle probes were obtained with an EMCCD enabling kinetic analysis of the enzymatic reaction in vivo and providing more details on early-stage caspase-3 activation. Fig. 4 A and B show the scattering intensity map of a whole cell labeled with crown nanoparticles and a representative time trace of a single crown nanoparticle probe upon addition of the apoptotic inducers TNF-α/CHX, respectively (see also Figs. S9 and S10). Each cleavage event, represented by a sudden intensity drop in the time traces (Fig. 4B, arrows), was counted independently for each particle in all cells (Fig. 4C). Only minimal cleavage events were observed directly after the addition of TNF-α/CHX, but after several minutes, the number of cutting events increased dramatically (Fig. 4 B and C). The time lag (≈16 min) between the addition of the TNF-α/CHX and the majority of cleavage events is indicative of the induction time required for signaling events upstream of caspase-3 activation, such as initiator caspase-8 activation.
Fig. 4.
Single-particle traces in live cells as a function of time. (A) Scattering intensity map of a single cell having internalized multiple-crown nanoparticles plasmon rulers. (B) Trajectory of a single-crown nanoparticle upon addition of TNF-α/CHX. Stepwise decreases in signal intensity were observed, which correspond to the sequential cleavage of satellite nanoparticles by caspase-3. (C and D) Statistical analyses of crown nanoparticle cleavages in a single cell as a function of time (C). Only minimal proteolysis was observed from 0 min to 16 min. Then, a rapid increase in cleavages occurs in the period from 16 to 35 min, indicative of caspase-3 activation. Caspase-3-mediated proteolysis of 10 different cells in a group (D). Differences in induction times across cells in the same population reflect cell-by-cell heterogeneity against caspase-3 activation. (E) A plot of the induction time (i.e., time from addition of apoptotic inducer until 5% of cleavages have occurred) vs. the cutting period (i.e., time following the induction period and ending when 95% of cleavages have occurred) of crown nanoparticle plasmon rulers. A slower cleavage rate was observed from cells exhibiting a longer induction time for initial cleavage.
To further understand early-stage caspase-3 activation, we also analyzed the cleavage statistics of nanoparticles located inside 10 individual cells from the same group (Fig. S11). Interestingly, we observed significant variations in the induction times between different cells in the same population, ranging from 15 to 50 min (Fig. 4D). In addition, we found that differences in the induction time (labeled in Fig. 4C) correlate with the time required for the majority of cleavages to occur (defined as cutting period and labeled in Fig. 4C) (Fig. 4E). A longer cutting period was observed for cells exhibiting a longer induction time. Though the cutting period and induction time were correlated within a cell, variability in both measurements across cells in the same population reflect the heterogeneity in the resistance of cells to caspase-3 activation. Such findings are consistent with recent single-cell studies describing how cells in a group can exhibit different caspase-3 activation behaviors in response to the same apoptotic stimulus, and that such differences determine whether cells die or survive after a certain period (29, 42). This variability is believed to be dependent on each cell's individual resistance to death receptor-induced apoptosis (29, 42).
The crown nanoparticle plasmon ruler technique provides a new way of probing single-molecule activities in live cells. In this work, we have shown that crown nanoparticle plasmon rulers can be used for continuous observation of caspase-3 activity, establishing plasmon rulers as robust tools for single-molecule imaging in live cells. This would not be possible with conventional single-molecule imaging techniques, which are limited by much shorter continuous observation windows or discontinuous snapshot imaging. Caspase-3 activation kinetics were successfully analyzed at the single-molecule level. Although the present study probes only enzyme activity, the design of crown nanoparticle plasmon rulers with alternate peptides of interest will allow other dynamics such as conformational changes to be studied in live cells. Furthermore, by using different types of plasmon rulers in terms of nanoparticle composition, size, shape, and number of particles, it should be possible to realize multicolor imaging of different signaling molecules in live cells. This will allow the activity of different signaling effectors to be correlated within the same cell at the single-molecule level.
Methods
Reagents.
Gold nanoparticles (40 nm) were purchased from Ted Pella Inc. and were ligand exchanged with P,P′-(bis-p-sulfonatephenyl)-P′′-phenylphosphine (BSPP) according to a literature protocol (25) and then concentrated to 5 nM. Either biotin-GSEGGSESEDEVDGGSNSGGRLCC or biotin-CH2CH2(OCH2CH2)6DEVDG-GCH2CH2(OCH2CH2)12CC peptides were used as caspase-3 substrates and were purchased from New England Peptide Inc. HOOCCH2(OCH2CH2)6S-S(OCH2CH2)6-CH2COOH and biotin-CH2(OCH2CH2)6S-S(OCH2CH2)6-CH2-biotin were purchased from PolyPure Inc. and reduced by treatment with BSPP before use.
Optical Setup.
Scattering images were taken using an Axioplan 2 upright microscope (Zeiss), with a dark-field immersion oil condenser. The microscope was modified by the addition of an iXon EMCCD detector (512 × 512 pixel chip; Andor) on the imaging port of the SpectraPro 2300i spectrometer (Acton).
Synthesis of Crown Nanoparticles.
Synthesis of NeutrAvidin (Ntv)-coated gold nanoparticles and peptide-conjugated gold nanoparticles are described in SI Methods. Fifteen microliters of Ntv-coated gold nanoparticles (1 nM) was added to 300 μL of peptide-conjugated gold nanoparticles (5 nM). After an overnight reaction at 4 °C, crown nanoparticles were separated from highly ordered aggregates and excess reactant nanoparticles via a sucrose-density gradient separation protocol with detection by UV-Vis absorption spectrometry.
In Vitro Single-Molecule Experiments.
The glass flow chamber was modified with BSA-biotin (Roche) as described in the literature (23). A dilute solution of crown nanoparticle plasmon rulers was flushed into the chamber. The chamber was equilibrated with T30 buffer (10 mM Tris, 30 mM NaCl) and then flushed with 200 μL of 250 ng/mL caspase-3 in 10 mM phosphate buffer containing 50 mM NaCl. The scattering color images and intensities of the crown nanoparticle plasmon rulers in the field of view were continuously monitored by either a color camera (CoolSNAP cf; Roper Scientific) or an EMCCD. Also, we recorded the scattering spectra of individual crown nanoparticle plasmon rulers during caspase-3-mediated cleavage. For comparison, we used biotin-GSEGGSESEIETDGGSNSGGRLCC peptide-conjugated crown nanoparticles.
Caspase-Glo 3/7 Assay.
SW620 cells were maintained in DMEM, 10% FBS, 1× glutamax, and 1× penicillin/streptomycin. Four 96-well clear plates were seeded with 2 × 104 SW620 cells/well for a seeding density of 6.25 × 104 cells/cm2. Cells were allowed to attach overnight. On average, cells were at 30% confluency before addition of apoptotic inducer. Before induction, media was removed and replaced with the following reagents in SW620 growth media: 1 μM staurosporine (0.1% final DMSO; Sigma, catalog no. S6942); 0.2 μg/mL TNF-α (Invitrogen, catalog no. PHC3016) and 2 μg/mL cycloheximide (CHX) (1% final DMSO; Sigma, catalog no. C4859); or vehicle controls with matching concentrations of DMSO. A total of 12 replicates were performed. At 23 h, 4.5 h, 100 min, and 30 min after addition of apoptotic inducer, Caspase-Glo 3/7 Reagent (Promega) was added to all wells in a 1:1 ratio following manufacturer's instructions. After 10 min on a plate shaker at room temperature, 90% of the lysate volume was transferred to a 96-well solid-white plate (E&K Scientific, catalog no. EK-25075). Cell lysates were analyzed with an Analyst HT microplate reader (Molecular Devices), and data points were blank subtracted.
Flow Cytometry.
A 6-well plate was seeded with 5.7 × 105 SW620 cells/well for a seeding density of 6 × 104 cells/cm2. Cells were allowed to attach overnight. On average, cells were at 30% confluency before addition of apoptotic inducer. Addition of apoptotic inducers was performed as described previously. At 23 h, 4.5 h, 100 min, and 30 min after addition of apoptotic inducer, media was removed from all wells. To recover caspase-3-positive cells that might have detached during the time course, cells from the media were harvested by centrifugation at 1,000 RCF for 5 min and combined with cells recovered from the plate. The cells on the plate were detached using TrypLE Express (Gibco, catalog no. 12604–013) following manufacturer's instructions. After harvesting by centrifugation, supernatant was removed and cells were resuspended in 75 μL of PhiPhiLux G1D2 (OncoImmunin, catalog no. A304R1G). This reagent contains a peptide with the sequence DEVD-GI that is homodoubly labeled with a fluorophore (λex = 505 nm; λem = 530 nm). Cells were incubated at 37 °C, 5% CO2 for 1 h. Following incubation, 1 mL of flow cytometry dilution buffer (OncoImmunin) was added to each tube, and cells were harvested by centrifugation. Following supernatant removal, cell pellets were loosened by gently flicking, resuspended in 1 mL of flow cytometry dilution buffer, and transferred to 12 × 75-mm polystyrene round-bottom test tubes (BD Biosciences, catalog no. 352054). 2 × 104 cells were analyzed with a Beckton Dickinson FACSCalibur cytometer. Unstained cells (not loaded with substrate) were adjusted to the first decade of the FL1 channel, and data analysis was performed with FlowJo version 7.2.4. To facilitate analysis of the shoulder peaks, the major peak of each histogram (located between fluorescent intensity of 101 to 102) were overlaid and plotted in Fig. 3D.
Single-Molecule Imaging of Caspase-3 Catalytic Reaction in SW620 Cell Lines.
Crown nanoparticles (≈0.15 pmol) were treated with 4.5 pmol of biotinylated TAT peptides for 1 h. Then, the TAT-conjugated nanoparticles were mixed with 2 mL of culture media. SW620 cells were incubated with the nanoparticle solution for 12 h. The cells were washed with 2 mL of SW620 growth media 3 times. The cells were treated with 0.2 μg/mL of TNF-α and 2 μg/mL of CHX and continuously monitored by either color camera (CoolSNAP cf; Roper Scientific) or an EMCCD (scan speed: 100 images per min) for 2 h.
Supplementary Material
Acknowledgments.
We thank Prof. J. Wells (University of California, San Francisco), Prof. M. Shuman (University of California, San Francisco), Prof. C. Larabell (Lawrence Berkeley National Laboratory), Dr. W. Gu, Dr. A. Mastroianni, Dr. S. Claridge, and C. Choi for helpful discussion. We also thank Dr. Dennis Wolan and Julie Zorn for advice on caspase-3 activity assays and for providing us with purified recombinant caspase-3, and A. Fischer and M. Yasukawa for cell culture. This work was supported by the National Institutes of Health Grant R01-GM77856, NOT-OD-09–056 and the US Air Force Office of Scientific Research-Korea Ministry of Education-Science and Technology Nano-Bio-Information Technology Program K20716000001-07A0400-00100; Lawrence Berkeley National Laboratory LB08003826, Leukemia and Lymphoma Society Fellowship 5552-06 (to D.R.H.), National Institute of General Medical Sciences Grant 1 R25 GM56847 (to C.T.), and National Institutes of Health Grants CA72006 and CA128765 (to C.S.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907367106/DCSupplemental.
References
- 1.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1627–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 2.Giepmans BNG, Adams SR, Elissman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Mehta AD, Rief M, Spudich JA, Smith DA, Simmons RM. Single-molecule biomechanics with optical methods. Science. 1999;283:1689–2695. doi: 10.1126/science.283.5408.1689. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang X, et al. A single molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 6.Lu H, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 7.Harada Y, et al. K. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 8.Talaga DS, et al. Dynamics and folding of single two-stranded coiled-coil peptides studied by fluorescent energy transfer confocal microscopy. Proc Natl Acad Sci USA. 2000;97:13021–13026. doi: 10.1073/pnas.97.24.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Z, Srividya N, Sosnick T, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc Natl Acad Sci USA. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha T, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 11.Sako Y, Minoguchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 12.Choi PJ, Cai L, Frieda K, Xie XS. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322:442–445. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 14.Teramura Y, et al. Single-molecule analysis of epidermal growth factor binding on the surface of living cells. EMBO J. 2006;25:4215–4222. doi: 10.1038/sj.emboj.7601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois A, Canva M, Brun A, Chaput F, Boilot JP. Photostability of dye molecules trapped in solid matrices. Appl Opt. 1996;35:3193–3199. doi: 10.1364/AO.35.003193. [DOI] [PubMed] [Google Scholar]
- 17.Rasnik I, McKinney SA, Ha T. Nonblinking and longlasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 18.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 19.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 20.Nirmal M, et al. Fluorescence intermittency in single cadmium selenide nanocrystals. Nature. 1996;383:802–804. [Google Scholar]
- 21.Hohng S, Ha T. Near-complete suppression of quantum dot blinking in ambient conditions. J Am Chem Soc. 2004;126:1324–1325. doi: 10.1021/ja039686w. [DOI] [PubMed] [Google Scholar]
- 22.Fomenko V, Nesbitt DJ. Solution control of radiative and nonradiative lifetimes: A novel contribution to quantum dot blinking suppression. Nano Lett. 2008;8:287–293. doi: 10.1021/nl0726609. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Non-blinking semiconductor nanocrystals. Nature. 2009;459:686–689. doi: 10.1038/nature08072. [DOI] [PubMed] [Google Scholar]
- 24.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nat Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 25.Reinhard BM, Sheikoleslami S, Mastorianni A, Alvisatos AP, Liphardt J. Use of plasmon coupling to reveal the dynamics of DNA bending and cleavage by single EcoRV restriction enzymes. Proc Natl Acad Sci USA. 2007;104:2667–2672. doi: 10.1073/pnas.0607826104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yguerabide J, Yguerabide EE. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications—II. Experimental characterization. Anal Biochem. 1998;262:157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 27.Huang X, El-Sayed IH, Qian W, El-Sayed MA. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 28.Igney FH, Krammer PH. Death and anti-death: Tumour resistance to apoptosis. Nat Rev Cancer. 2002;2:277–287. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 29.Albeck JG, et al. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornberry NA, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. J Biol Chem. 1997;272:17909–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 31.McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: Implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 32.Harris JL, Peterson EP, Hudig D, Thornberry NA, Craik CS. Definition and redesign of the extended substrate specificity of granzyme B. J Biol Chem. 1998;273:27364–27373. doi: 10.1074/jbc.273.42.27364. [DOI] [PubMed] [Google Scholar]
- 33.Mahrus S, Kisiel W, Craik CS. Granzyme M is a regulatory protease that inactivates proteinase inhibitor 9, an endogenous inhibitor of granzyme B. J Biol Chem. 2004;279:54275–54282. doi: 10.1074/jbc.M411482200. [DOI] [PubMed] [Google Scholar]
- 34.Talanian RV, et al. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 35.Fuente JM, Berry CC. TAT peptide as an efficient molecule to translocate gold nanoparticles into the cell nucleus. Bioconjugate Chem. 2005;16:1176–1180. doi: 10.1021/bc050033+. [DOI] [PubMed] [Google Scholar]
- 36.Werneburg NW, Guicciardi ME, Bronk SF, Kaufmann SH, Gores JG. Tumor necrosis factor-related apoptosis-inducing ligand activates a lysosomal pathway of apoptosis that is regulated by Bcl-2 proteins. J Biol Chem. 2007;282:28960–28970. doi: 10.1074/jbc.M705671200. [DOI] [PubMed] [Google Scholar]
- 37.Claus V, et al. Lysosomal enzyme trafficking between phagosomes, endosomes, and lysosomes in J774 macrophages. Enrichment of cathepsin H in early endosomes. J Biol Chem. 1998;273:9842–9851. doi: 10.1074/jbc.273.16.9842. [DOI] [PubMed] [Google Scholar]
- 38.Ndozangue-Touriguine O, et al. A mitochondrial block and expression of XIAP lead to resistance to TRAIL-induced apoptosis during progression to metastasis of a colon carcinoma. Oncogene. 2008;27:6012–6022. doi: 10.1038/onc.2008.197. [DOI] [PubMed] [Google Scholar]
- 39.Rozman-Pungercar J, et al. Inhibition of papain-like cysteine proteases and legumain by caspase-specific inhibitors: When reaction mechanism is more important than specificity. Cell Death Differ. 2003;10:881–888. doi: 10.1038/sj.cdd.4401247. [DOI] [PubMed] [Google Scholar]
- 40.Provance DW, MacDowall A, Marko M, Luby-Phelps K. Cytoarchitecture of size-excluding compartments in living cells. J Cell Sci. 1993;106:565–578. doi: 10.1242/jcs.106.2.565. [DOI] [PubMed] [Google Scholar]
- 41.Luby-Phelps K, Castle PE, Taylor DL, Lanni F. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc Natl Acad Sci USA. 1987;84:4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan MJ, Thorburn A. Measurement of caspase activity in individual cells reveals differences in the kinetics of caspase activation between cells. Cell Death Diff. 2001;8:38–43. doi: 10.1038/sj.cdd.4400800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.