Abstract
Rab GTPases and their effectors mediate docking, the initial contact of intracellular membranes preceding bilayer fusion. However, it has been unclear whether Rab proteins and effectors are sufficient for intermembrane interactions. We have recently reported reconstituted membrane fusion that requires yeast vacuolar SNAREs, lipids, and the homotypic fusion and vacuole protein sorting (HOPS)/class C Vps complex, an effector and guanine nucleotide exchange factor for the yeast vacuolar Rab GTPase Ypt7p. We now report reconstitution of lysis-free membrane fusion that requires purified GTP-bound Ypt7p, HOPS complex, vacuolar SNAREs, ATP hydrolysis, and the SNARE disassembly catalysts Sec17p and Sec18p. We use this reconstituted system to show that SNAREs and Sec17p/Sec18p, and Ypt7p and the HOPS complex, are required for stable intermembrane interactions and that the three vacuolar Q-SNAREs are sufficient for these interactions.
Keywords: biochemical reconstitution, Rab effector
Rab GTPases are central regulators of intracellular membrane trafficking (1), mediating vesicle formation (2, 3) and transport (4), and membrane docking (5). Like other small GTPases, Rab proteins cycle between GTP-bound and GDP-bound forms (6); this cycling is regulated by guanine nucleotide exchange factors (GEFs) (7) and GTPase-activating proteins (GAPs) (8). In their GTP-bound forms, Rab GTPases interact with a diverse set of proteins and protein complexes termed effectors (9). Despite the wealth of data regarding the physical interactions in which Rab GTPases participate, Rab function is poorly understood, for want of systems for studying Rab proteins and their effectors in the context of chemically defined membrane fusion.
Membrane fusion reactions have been reconstituted from purified components in vitro, including Ca2+- and polyethylene glycol-stimulated fusion (10, 11), viral fusion (12), and SNARE-mediated fusion (13). Regulation of SNARE-dependent membrane fusion by the SNARE-interacting protein Sec1/Munc18 (SM) (14, 15) and the SNARE-binding proteins synaptotagmin and complexin (16–25) has also been reconstituted. We have recently reported reconstituted membrane fusion that requires yeast vacuolar SNAREs, regulatory lipids, and the homotypic vacuole fusion and protein sorting (HOPS)/class C vacuole protein sorting (class C Vps) complex, a six-subunit effector and GEF for the yeast vacuolar Rab GTPase Ypt7p (26–28). The Vps33p subunit of the HOPS complex is a SM protein (27), whereas the Vps39p subunit contains the Ypt7p GEF activity (26).
We now report reconstitution of membrane fusion that requires purified Ypt7p, HOPS complex, vacuolar SNAREs (Vam3p, Vti1p, Vam7p, and Nyv1p), ATP hydrolysis, and the SNARE chaperones Sec17p and Sec18p [the yeast homologs of soluble N-ethylmaleimide sensitive factor attachment proteins (SNAPs) and N-ethylmaleimide sensitive factor (NSF), respectively]. Membrane fusion is not accompanied by lysis, and only Ypt7p that is in its GTP-bound state can support fusion. Previous studies of vacuole fusion have used assays of content mixing (29, 30). In this study, we use an assay of proteoliposome lipid mixing under conditions in which the proteoliposome membranes remain intact, shielding lumenally oriented lipids from external aqueous probes. This fusion also preserves the curvature of the initial liposome population, an independent measure of lysis-free fusion. The lipid mixing that we measure therefore represents authentic membrane fusion. Biochemical reconstitution of Rab5-dependent membrane fusion using purified components has also recently been reported (31).
We have used our in vitro system to investigate the molecular mechanisms of membrane docking, a key intermediate in membrane fusion. Docking is the close association of two membranes before fusion and has been proposed to consist of two steps: tethering, a reversible, Rab GTPase-dependent and SNARE association-independent association, followed by assembly of membrane-bridging “transSNARE” complexes (32–34). Both Ypt7p and the HOPS complex are required for vacuole docking in vitro (5, 35), and in general Rab GTPases and their effectors play a key role in bringing membranes in proximity before fusion (9). The HOPS complex might link apposed membranes, because it has a myriad of binding partners in addition to Ypt7p: SNARE complexes (36), the SNAREs Vam3p (37) and Vam7p (35) in their monomeric states, and phosphoinositides (35). However, it remains unclear whether Ypt7p–HOPS complex interactions or, more broadly, Rab–effector interactions mediate tethering by forming a direct physical link between membranes. We show in this study that Ypt7p and the HOPS complex are required for clustering of reconstituted proteoliposomes, but we also find that they are insufficient for this clustering; Sec17p, Sec18p, and vacuolar SNAREs are required as well, and the three vacuolar Q-SNAREs (38) are sufficient.
Results
We have recently reported proteoliposome fusion that requires SNAREs and the HOPS complex, but not Ypt7p (28). These proteoliposomes were made using the “standard” method, in which dried lipids are dissolved in a detergent solution of proteins, followed by removal of detergent (39). We reasoned that a different method of proteoliposome preparation might result in proteoliposomes that exhibit the physiological requirement for Ypt7p for membrane fusion. We therefore made proteoliposomes by using “direct” addition, incubation of protein/detergent solutions with preformed protein-free liposomes at a low detergent concentration, followed by detergent removal (39), to incorporate Ypt7p (40) and the vacuolar SNARE proteins Vam3p, Vti1p, Vam7p, and Nyv1p into liposomes that had been extruded from lipid mixtures similar in composition to vacuole membranes (see Methods). We also made proteoliposomes bearing SNAREs but lacking Ypt7p. The presence or absence of Ypt7p has no effect on the efficiency of incorporation of SNAREs (Fig. 1A Inset). The resulting proteoliposomes are 114 ± 3 nm in diameter, as estimated by thin-section electron microscopy, and most are unilamellar (Fig. 2A).
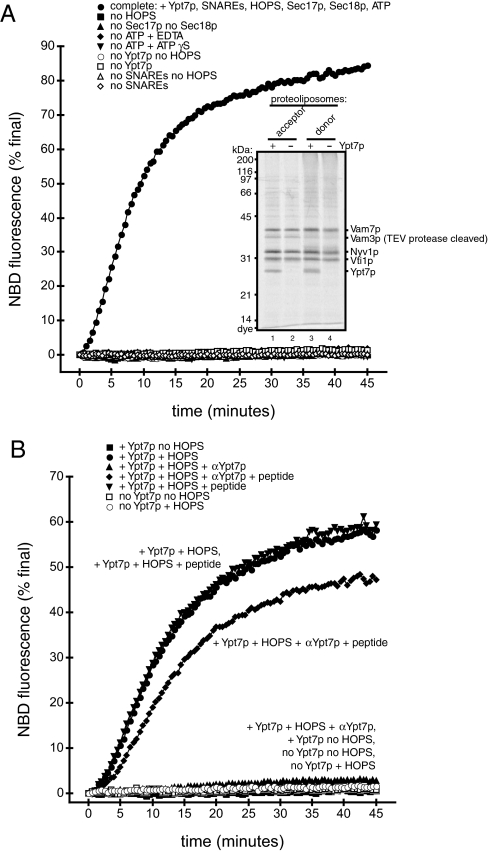
Fig. 1.
Ypt7p-dependent lipid mixing. (A) Lipid mixing of direct-method proteoliposomes requires Ypt7p, HOPS, Sec17p/Sec18p, ATP hydrolysis, and SNAREs. Fusion reactions (see Methods) used proteoliposomes of indicated composition and lacked the indicated soluble factors; any omitted components were replaced by their buffers. (Inset) Acceptor and donor proteoliposomes with SNAREs, with or without Ypt7p, were analyzed by SDS/PAGE and Sypro Ruby staining (5 nmol of total lipids per lane). (B) Anti-Ypt7p antibodies block lipid mixing. Anti-Ypt7p (1.2 M final; ▴), a mixture of anti-Ypt7p peptide (12 μM final; ⧫), Ypt7p peptide alone (12 M final; ▾), or RB150 (all others) were preincubated with the indicated proteoliposomes for 10 min at 27 °C. MgCl2, ATP, Sec17p, Sec18p, and HOPS complex or HOPS buffer, as indicated, were then added and reactions were carried out as described in Methods.
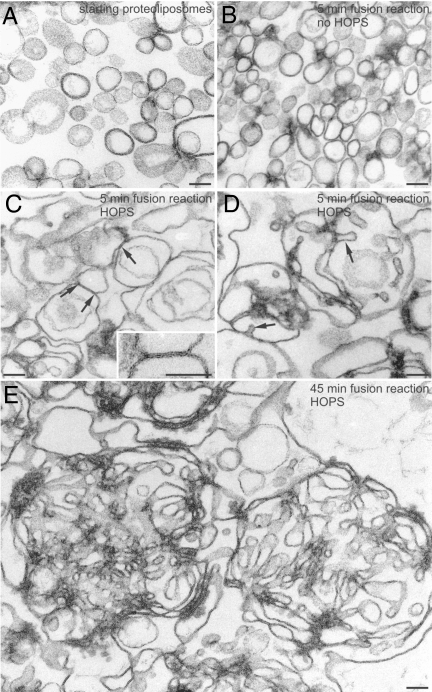
Fig. 2.
Electron microscopy analysis of proteoliposomes and fusion reactions. Donor proteoliposomes or donor-only fusion reactions were prepared as described. After 5 or 45 min at 27 °C for the reactions, and without incubation for the proteoliposomes alone, glutaraldehyde was added to a final concentration of 0.1% from a 2% stock in 0.1 M sodium phosphate, pH 7.3. Reactions were incubated at room temperature for 30 min, then centrifuged for 15 min at 14,000 rpm in an Eppendorf (Hamburg, Germany) 5415C microcentrifuge at 4 °C. Pellets were covered with 450 μL of 1% low melting point agarose, then processed for transmission electron microscopy as described (79). (A) Starting proteoliposomes not incubated under fusion conditions (mean size = 114 ± 3 nm). (B) Proteoliposomes incubated for 5 min in a fusion reaction without HOPS complex. (C and D) Proteoliposomes incubated for 5 min in a fusion reaction with HOPS. Liposomes are larger and pleomorphic, possibly as a result of incipient fusion. In C, the arrows point to regions where juxtaposed liposomes have established a close contact. The area delimited by two arrows is shown at higher magnification in the Inset. Note that the external leaflets of the proteoliposome membranes in this region have apparently merged into a single osmiophilic line resulting in the formation of a pentalaminar structure suggestive of a fusion event. (D) Large liposomes are frequently endowed with membrane infoldings, resulting in the formation of tubular structures (arrows). (E) Proteoliposomes incubated for 45 min in the fusion reaction with HOPS. Fused liposomes have generated tangled skeins of tubular membranes. (Scale bars, 100 nm.)
To assay membrane fusion, each combination of proteins was incorporated into two types of liposomes: “donor” liposomes, which contain quenching concentrations (1.5 mole percent each) of rhodamine- and nitrobenzoxadiazole (NBD)-conjugated phosphatidylethanolamines, and “acceptor” liposomes lacking these fluorophores (41). In donor liposomes, NBD fluorescence is quenched by Förster resonance energy transfer (FRET) to rhodamine. Upon fusion of donor and acceptor liposomes, the surface concentration of NBD- and rhodamine-conjugated lipids is reduced, FRET is abrogated, and NBD fluorescence increases (41).
Fusion of the membranes of these proteoliposomes requires Ypt7p and SNAREs, purified HOPS complex, and the SNARE chaperones Sec17p and Sec18p (Fig. 1A and Fig. S1). ATP hydrolysis is also required, because EDTA or the poorly hydrolyzable ATP analog ATPγS blocks fusion. Neither ATPγS nor EDTA inhibits HOPS function. HOPS-stimulated fusion of standard-method proteoliposomes bearing only Nyv1p with proteoliposomes bearing Vam3p, Vam7p, and Vti1p, which does not require ATP or Sec17p/Sec18p (28), is not inhibited by ATPγS or EDTA (Fig. S2). As an additional test of the requirement for Ypt7p for fusion, an anti-Ypt7p antibody (42) reduces fusion nearly to background levels (Fig. 1B). Preincubation of this antibody with the peptide against which it had been raised (42) relieves inhibition, whereas the peptide has no stimulatory activity (Fig. 1B); thus, antibody inhibition is specific for Ypt7p.
Lysis of vacuolar membranes has been observed when vacuolar SNAREs are overexpressed (43). Neuronal SNAREs can also cause proteoliposome lysis at high concentrations (44). We therefore used electron microscopy to examine whether lipid mixing (Fig. 1A) is accompanied by an increase in proteoliposome size. We observed little change in size when Ypt7p- and SNARE-bearing proteoliposomes were incubated under fusion conditions but without HOPS complex (Fig. 2B). However, HOPS induces a marked increase in proteoliposome size within only 5 min (Fig. 2C), accompanied by invaginations of the larger proteoliposomes (Fig. 2D). We also observed pentalaminar structures that could be fusion intermediates (Fig. 2C Inset). After 45 min of incubation with HOPS, the large proteoliposomes are extensively folded (Fig. 2E and Fig. S3). These invaginations and folds are consistent with a fusion reaction that preserves the high curvature of the small starting proteoliposomes (Fig. 2 A and B), that is, fusion without lysis.
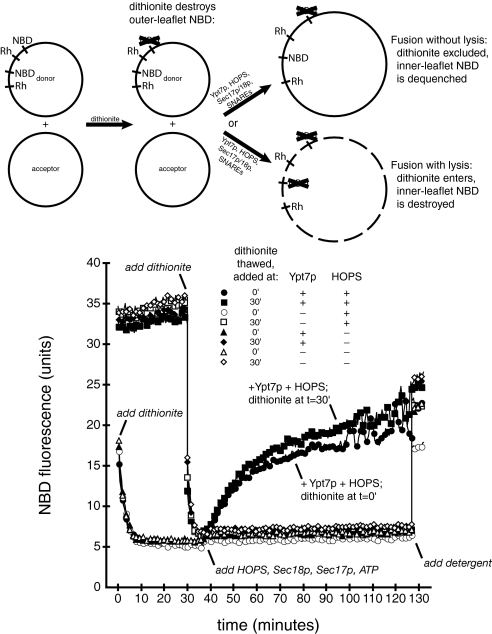
We also used sodium dithionite to test whether Ypt7p-dependent proteoliposome lipid mixing is caused by lysis followed by coalescence of membrane fragments (28). This reducing agent destroys only accessible NBD fluorescence because it crosses intact lipid bilayers very slowly (45). In aqueous solution, the reducing activity of sodium dithionite decays fully within 30 min (28). Sodium dithionite decreases the fluorescence of a mixture of donor and acceptor proteoliposomes (Fig. 3). If this mixture is incubated at 27 °C until the sodium dithionite has decayed, and then HOPS complex, Sec17p, Sec18p, and ATP are added to trigger lipid mixing (Fig. 3), NBD fluorescence increases at the same rate as in a reaction containing fresh sodium dithionite (Fig. 3). Thus, lipid mixing does not cause proteoliposome lysis and access of sodium dithionite to the NBD on the interior leaflet of the proteoliposome membranes. This assay therefore demonstrates authentic fusion of the inner membrane monolayer without interruption of the membrane permeability barrier.
Fig. 3.
Lipid mixing is not accompanied by lysis. Fusion reactions (see Methods) used proteoliposomes with SNAREs, with Ypt7p (filled symbols) or without Ypt7p (open symbols). Sodium dithionite (40 mM; Sigma) was prepared by addition of solid sodium dithionite to ice-cold RB150+, frozen immediately in aliquots, stored at −80 °C, and thawed just before use. At t = 0, one set of proteoliposomes in RB150+ (13.2 μL; circles and triangles) received freshly thawed sodium dithionite (2 μL) and was incubated at 27 °C. A second set of proteoliposomes (13.2 μL; squares and diamonds) was incubated at 27 °C without sodium dithionite. At t = 30 min, freshly thawed sodium dithionite (2 μL) was added, at room temperature, to this second set of proteoliposomes, and reactions were returned to 27 °C. At t = 37 min, MgCl2, ATP, Sec17p, Sec18p, and HOPS complex (circles and squares) or HOPS buffer (triangles and diamonds) were added, in a total volume of 4.8 μL, at room temperature. Reactions were incubated at 27 °C for 60 min, followed by addition of 2 μL of 1% Thesit and 5-min incubation at 27 °C. Raw fluorescence units are shown.
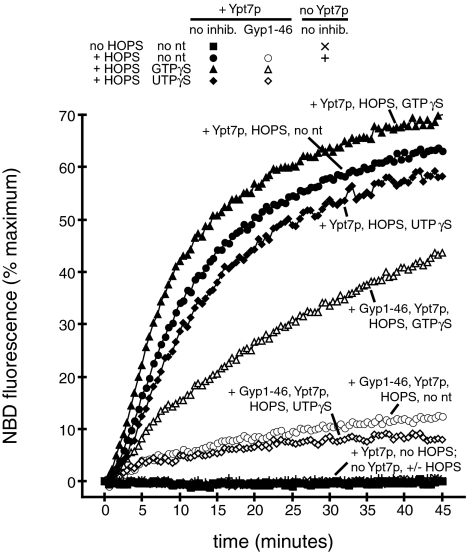
We next investigated whether the nucleotide-binding state of Ypt7p regulates membrane fusion. Added GTP is not necessary for proteoliposome fusion (Fig. 1A), nor for fusion of purified vacuoles (46). Is purified Ypt7p already in its GTP-bound form, and is this GTP-bound Ypt7p required for proteoliposome fusion? To test these questions, we used Gyp1–46 (47), a catalytic fragment of a GAP that stimulates GTP hydrolysis by Ypt7p (8). Ypt7p- and HOPS complex-dependent fusion (Fig. 4) is inhibited by Gyp1–46, suggesting that functional Ypt7p is GTP-bound. To show that this inhibition is caused by modulation of Ypt7p-bound nucleotide we used GTPγS, a slowly hydrolyzable GTP analog that prevents inhibition of vacuole fusion by Gyp1–46 (46) but does not block fusion (30). GTPγS relieves inhibition of proteoliposome fusion by Gyp1–46 but has little effect on fusion in the absence of Gyp1–46 (Fig. 4). As a control for nonspecific effects of GTPγS we added UTPγS, which does not relieve inhibition by Gyp1–46 (Fig. 4). Thus, GTP-bound Ypt7p is required for proteoliposome fusion.
Fig. 4.
Lipid mixing requires GTP-bound Ypt7p. Fusion reactions (see Methods) used proteolipsomes with SNAREs, with or without Ypt7p as indicated. During the preincubation, reactions received RB150 (squares and circles), GTPγS (100 μM final; triangles), or UTPγS (100 μM final; diamonds), and RB150 (filled symbols) or Gyp1–46 (2 μM final; open symbols). The symbols for reactions without Ypt7p are behind the filled squares and show no detectable increase in NBD fluorescence.
Requirements for Intermembrane Interactions.
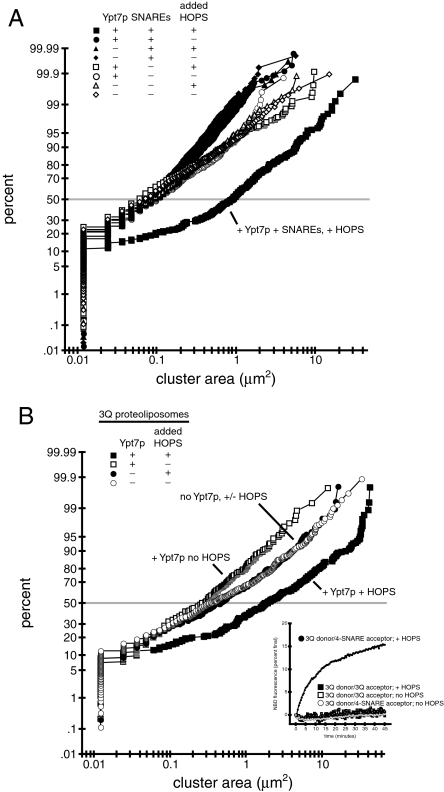
Ypt7p and the HOPS complex are required for vacuole docking (5, 35), but it is unclear whether they suffice. We therefore used microscopy to find the minimal set of factors required for clustering of proteoliposomes. Fusion reactions using proteoliposomes with or without Ypt7p, and with or without SNAREs, each in the presence or absence of HOPS complex, were imaged and the area occupied by each cluster of proteoliposomes in a field was measured with ImageJ (National Institutes of Health, Bethesda). Cumulative distribution plots for each treatment are shown in Fig. 5A, and representative images are shown in Fig. S4A; histogram analysis of selected distributions is presented in Fig. S4B. Individual proteoliposomes are too small to allow measurement of their size by this method. It is likely that some fusion occurs in the clustering assay using proteoliposomes bearing SNAREs. However, fusion cannot occur without clustering (see below). Thus, if an increase in cluster size is detected, then clustering must have taken place, regardless of whether the cluster consists of small unfused proteoliposomes, larger fused proteoliposomes, or a combination of the two.
Fig. 5.
Proteoliposome clustering requires Ypt7p, HOPS complex, Sec17p/Sec18p, and SNAREs. Proteoliposome fusion reactions, including Sec17p, Sec18p, and ATP, were prepared as in Methods, except that only donor proteoliposomes were used. After 20 min at 27 °C, 3 μL of each reaction was mixed on a microscope slide (Gold Seal no. 3051) with 5 μL of a mock reaction without proteoliposomes or HOPS. These mixtures were covered with 22-mm cover slips (Corning no. 2870-22) and randomized, and random fields were imaged with an Olympus BX51 microscope with a 100-W mercury arc lamp (Olympus), 3% U-RSL6 UV/IR filter (Olympus), TRITC/DiI filter set (Chroma Technologies), 1.4 NA Plan Aprochromat ×60 objective (Olympus), Sensicam QE CCD camera (Cooke), and IPLab software (Scanalytics). Measurement of cluster sizes was done with ImageJ using an intensity threshold of 50. At least two images from each reaction were used for measurement. Representative images used for A are depicted in Fig. S4A. (A) Ypt7p, HOPS complex, and SNAREs all are required for proteoliposome clustering. A cumulative distribution plot showing proteoliposome cluster sizes is shown. Proteoliposomes were with Ypt7p (squares and circles) or without Ypt7p (triangles and diamonds) and with SNAREs (filled symbols) or without SNAREs (open symbols). Reactions received HOPS complex (squares and triangles) or HOPS buffer (circles and diamonds) as indicated. The distribution for the reaction with + Ypt7p + SNARE proteoliposomes with added HOPS complex is significantly different (P < 0.0001) from all other distributions by the Wilcoxon-Mann–Whitney test (Kaleidagraph). (B) The three vacuolar Q-SNAREs suffice for Ypt7p- and HOPS complex-dependent proteoliposome clustering. A cumulative distribution plot showing proteoliposome cluster sizes is shown. Reactions contained proteoliposomes with Vti1p, Vam7p, and Vam3p, with Ypt7p (squares) or without Ypt7p (circles), and with added HOPS complex (filled symbols) or HOPS buffer (open symbols). The distribution for the reaction using Ypt7p-bearing proteoliposomes, with HOPS complex, is significantly different (P < 0.0001) from the distribution for the reaction containing Ypt7p-bearing proteoliposomes, but lacking HOPS complex, by the Wilcoxon-Mann–Whitney test (Kaleidagraph). The distributions for the two reactions using proteoliposomes lacking Ypt7p are not significantly different (P = 0.4645) by the same test. The larger clusters in these distributions therefore derive from intrinsic aggregation and are not HOPS complex-dependent. (Inset) Nyv1p is required for proteoliposome fusion. Donor proteoliposomes with Ypt7p and the 3 Q-SNAREs were mixed with acceptor proteoliposomes with Ypt7p and with the three Q-SNAREs (squares) or all four SNAREs (circles), with HOPS (filled symbols) or without HOPS (open symbols), under fusion conditions (see Methods).
In the presence of ATP, Sec17p, and Sec18p, proteoliposomes containing SNAREs form large clusters in a Ypt7p- and HOPS complex-dependent manner (Fig. 5A). Clustering of proteoliposomes without SNAREs, however, is not stimulated by Ypt7p and HOPS complex (Fig. 5A). HOPS, along with Ypt7p and the SNAREs, also induces a significant increase in the distribution of mean fluorescence intensity in proteoliposome clusters (Fig. S4C), indicative of an increase in the number of proteoliposomes in these clusters. Thus, Ypt7p and the HOPS complex are insufficient for stable membrane-membrane interactions under the conditions of our assay; SNAREs are also required. In several cases, distributions of cluster areas for reactions using SNARE-free proteoliposomes are significantly different from distributions for reactions using SNARE-containing proteoliposomes without Ypt7p and/or HOPS complex (Fig. S4D). However, the median cluster sizes in these cases differ only slightly (14–33%), in contrast with the large differences (825–1,380%) in median size between reactions using SNARE- and Ypt7p-bearing proteoliposomes with HOPS complex and all other reactions (Fig. S4D).
Do these large clusters represent an on-pathway intermediate of membrane fusion? The lipid-mixing assay (Fig. 1A) shows that most of the proteoliposomes in a reaction undergo fusion. At the same time, the clustering assay (Fig. 5A) shows that most of the proteoliposomes in a reaction enter into larger clusters: the size distribution for the “complete” reaction diverges from the distributions for the reactions lacking Ypt7p and/or HOPS at roughly the 10th percentile. The small clusters that make up only 10% of the clusters in the complete reaction cannot account for the extent of lipid mixing shown in Fig. 1A. Thus, the proteoliposomes in the larger clusters must fuse to generate such a large extent of lipid mixing. The physiological nature of the clustering reaction is also strongly supported by the fact that it requires SNAREs, a Rab GTPase, and the HOPS complex, which are required for fusion in vivo and on isolated vacuoles (27, 48, 49).
How do the SNARE proteins mediate proteoliposome clustering? SNAREs can form “trans” complexes that bridge the space between membranes (34). Because SNARE-dependent proteoliposome fusion requires Sec17p and ATP hydrolysis by Sec18p (Fig. 1A), the SNAREs are likely to be in “cis” complexes, residing in the same membrane, at the beginning of a fusion reaction. Disassembly of these complexes by Sec17p and Sec18p (50) would be required for the formation of cis complexes containing only the three vacuolar Q-SNAREs (38) or of transSNARE complexes containing three Q-SNAREs and one R-SNARE (38, 51). We therefore performed clustering assays in the absence of Sec17p and Sec18p. Without these factors, Ypt7p-containing proteoliposomes do not exhibit HOPS complex-dependent clustering (Fig. S4E). Replacement of ATP with ATPγS, to block ATP hydrolysis by Sec18p, has the same effect (Fig. S4F).
To test whether “3Q” cis complexes or “3Q:1R” trans complexes mediate clustering, we made proteoliposomes bearing only the three vacuolar Q-SNAREs, Vam3p, Vti1p, and Vam7p (38). When Ypt7p is also present in these proteoliposomes, the HOPS complex induces a large increase in cluster size (Fig. 5B). No increase in size is induced by the HOPS complex when Ypt7p is not present (Fig. 5B). Therefore, the three vacuolar Q-SNAREs are sufficient for Ypt7p- and HOPS-dependent intermembrane interactions in the absence of membrane fusion (Fig. 5B Inset).
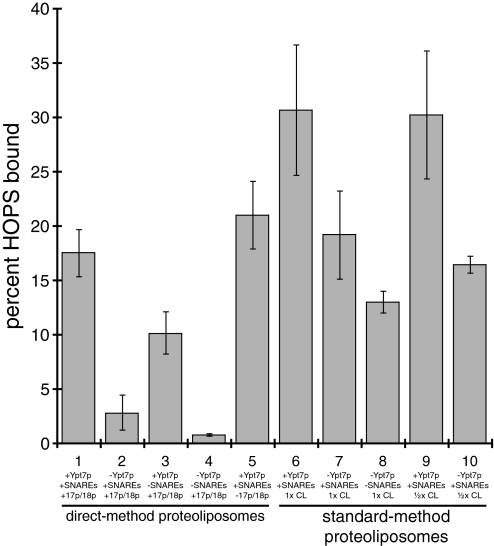
We next asked whether SNAREs promote proteoliposome clustering via recruitment of the HOPS complex to membranes by measuring HOPS binding to the direct-method proteoliposomes used in the clustering analysis. These proteoliposomes exhibit Ypt7p-dependent HOPS complex binding (Fig. 6, bars 1 and 2) that is stimulated by SNAREs (Fig. 6, bars 1–4). Without Sec17p and Sec18p, however, HOPS still binds efficiently to proteoliposomes (Fig. 6, bars 1 and 5), although clustering is abrogated in the absence of Sec17p and Sec18p (Fig. S4E). Thus, HOPS-proteoliposome binding is insufficient for proteoliposome clustering. We therefore conclude that the Ypt7p and the HOPS complex act together with a 3Q cis-SNARE complex of Vam3p, Vti1p, and Vam7p to mediate stable intermembrane interactions under the conditions of our assay.
Fig. 6.
HOPS complex binding to proteoliposomes. Donor-only proteoliposome fusion reactions (5× scale) containing the indicated components were incubated for 20 min at 27 °C then transferred to ice, mixed with 100 μL of 2 M sucrose in RB150+ in 5 × 41-mm ultracentrifuge tubes (Beckman no. 344090), and covered with 200 μL of 0.8 and 0.6 M sucrose in RB150+, then with 10 μL of RB150+. Gradients were centrifuged for 2 h and 30 min at 50,000 rpm at 4 °C in a SW-55 rotor (Beckman, Palo Alto, CA) using the appropriate inserts, and 20 μL of proteoliposomes was harvested from the top interface. Lipid yield was estimated by fluorescence (λex/λem 540/586 nm) and samples containing 4 nmol of lipids were analyzed by SDS/PAGE and Sypro Ruby staining. Bound HOPS complex was estimated by using a standard curve of purified HOPS.
Basis of Ypt7p Requirement for Fusion.
We next turned to the question of why the direct-method proteoliposomes presented here show the physiological requirement for Ypt7p for fusion whereas standard-method proteoliposomes do not (28). We found that differences in SNARE levels, or lipidic contaminants, have no measurable effect on Ypt7p dependence, whereas differences in cardiolipin levels and in the ability of proteoliposomes to bind HOPS complex are major factors in their dependence on Ypt7p for fusion.
SNARE levels or lipidic contaminants do not impact the extent of Ypt7p dependence for proteoliposome fusion. Direct-method proteoliposomes have lower levels of Vam3p than the other SNAREs (Fig. 1A Inset). However, standard-method proteoliposomes made with 25% of the usual level of Vam3p, which do not fuse efficiently, do not show enhanced dependence on Ypt7p for fusion (Fig. S5A). We therefore compared the lipid composition of standard-method and direct-method proteoliposomes by using mass spectrometry (Fig. S5 B and C). The only major lipid present in the standard-method proteoliposome analysis but not in the direct-method analysis (Fig. S5B) was identified as erucylamide, a fatty acid amide used in the manufacture of plastic films (52). However, erucylamide does not induce fusion of direct-method proteoliposomes lacking Ypt7p (Fig. S5D).
By contrast, both cardiolipin levels and HOPS complex-binding activity contribute to the extent of Ypt7p dependence for fusion. Cardiolipin levels are lower in direct-method proteoliposomes than in standard-method proteoliposomes (Fig. S5C). Cardiolipin forms water-insoluble nonlamellar structures in the presence of divalent cations (53); the MgCl2 present during preparation of direct-method proteoliposomes (see Methods) may reduce cardiolipin levels in the final proteoliposomes by causing precipitation of a fraction of the cardiolipin. Decreasing the amount of cardiolipin used to make standard-method proteoliposomes from 1.6% to 0.8% increases the degree of stimulation of fusion by Ypt7p (Fig. S5E). However, Ypt7p stimulates fusion of low-cardiolipin standard-method proteoliposomes far less than fusion of direct-method proteoliposomes (Fig. S5E and Fig. 1A). Thus, differences in cardiolipin content cannot fully account for the difference in Ypt7p dependence for fusion of direct-method and standard-method proteoliposomes.
These results led us to examine whether direct-method proteoliposomes and standard-method proteoliposomes have different HOPS complex-binding activities. As mentioned above, direct-method proteoliposomes bind HOPS complex in a Ypt7p-dependent manner (Fig. 6, bars 1 and 2); this binding is stimulated by SNAREs (Fig. 6, bars 1–4) but is unaffected by Sec17p and Sec18p (Fig. 6, bar 5). In contrast, standard-method proteoliposomes bind HOPS complex even in the absence of Ypt7p (Fig. 6, bars 7, 8, and 10), although HOPS binding to standard-method proteoliposomes is stimulated by Ypt7p (Fig. 6, bars 6 and 9). Standard-method liposomes without Ypt7p or SNAREs also bind HOPS complex (Fig. 6, bar 8), demonstrating a direct interaction between HOPS and liposome membranes. Standard-method proteoliposomes made with lowered cardiolipin levels bind HOPS to nearly the same extent as standard-method proteoliposomes made with normal amounts of cardiolipin, both with and without Ypt7p (Fig. 6, bars 6, 7, 9, and 10); thus, cardiolipin exerts its effect by modulating the propensity of membranes to fuse, not by altering HOPS complex-proteoliposome binding.
We conclude that the difference in requirement for Ypt7p for fusion between standard-method and direct-method proteoliposomes has two bases: the difference in requirement for Ypt7p for HOPS complex recruitment to membranes and the difference in cardiolipin content between standard-method and direct-method proteoliposomes.
Discussion
Reconstituted proteoliposomes can be prepared with more rigorous control of lipid and protein composition than purified organelles, which rely on intracellular transport for delivery of their constituents. Deletion or mutation of genes encoding trafficking proteins can affect delivery of other factors, complicating the interpretation of experiments using purified organelles derived from mutant sources. Reconstitution of Ypt7p-dependent membrane fusion and clustering therefore provides a chemically defined system for functionally dissecting the molecular interactions underlying docking.
Docking has been proposed to take place in two stages: tethering, a Rab GTPase-dependent, SNARE-independent, and reversible association, followed by trans-SNARE interactions (32, 34). Tethering may be mediated by “tethering factors” that interact simultaneously with binding partners, including Rab proteins, in apposed membranes (9, 54–57). However, no proposed Rab-dependent tethering factor has ever, to our knowledge, been shown to have direct membrane-bridging activity in a chemically defined membrane tethering reaction. In this study, proteoliposomes bearing Ypt7p alone cannot cluster in the presence of HOPS complex; SNAREs and Sec17p/Sec18p are also required, and the three vacuolar Q-SNAREs Vam3p, Vti1p, and Vam7p are sufficient), for intermembrane interactions (Fig. 5). We conclude that Ypt7p and the HOPS complex are insufficient for stable membrane-membrane associations in our assay, and that a Q-SNARE complex acts together with Ypt7p and the HOPS complex to bring membranes into proximity before fusion.
Many studies have suggested a role for SNARE proteins in prefusion membrane association. Antibodies against Sec18p and removal of ATP prevent vacuole docking; both of these treatments inhibit cis-SNARE complex disassembly (5, 50) and would be expected to block formation of 3Q cis-SNARE or 3Q:1R transSNARE complexes. In an in vitro assay for formation of the vesicular tubular cluster, an intermediate in transport from the endoplasmic reticulum (ER) to the Golgi apparatus, antibodies against Syntaxin 5, a homolog of Vam3p, inhibit vesicle “coisolation,” as does addition of a dominant negative mutant of α-SNAP that blocks NSF SNARE complex disassembly activity (58, 59). Docking of synaptic vesicles to the plasma membrane in Caenorhabditis elegans neuromuscular junctions also requires syntaxin (60). SNARE-dependent (21, 61) and syntaxin-, synaptotagmin I-, phosphatidylserine-, and Ca2+-dependent (19) proteoliposome clustering have also been reported, although in both cases this clustering was independent of Rab GTPases and effectors.
Other studies, however, have suggested that SNAREs are not required for intermembrane associations. Vacuoles lacking Vam3p are able to dock, and this docking is not inhibited by antibodies against Nyv1p (33). Also, vacuoles lacking Nyv1p can dock (62). However, these results do not preclude the action of other SNAREs in docking: Pep12p, a homolog of Vam3p, and Ykt6p, a homolog of Nyv1p, both can enter vacuolar SNARE complexes (36, 63). Other studies of the involvement of SNAREs in docking have used intact organelles (32, 64), which are likely to contain multiple sets of SNAREs that could also form “noncanonical” SNARE complexes (65) that mediate docking but not membrane fusion. Moreover, temperature-sensitive SNARE mutants that permit docking of ER-derived vesicles after incubation at a restrictive temperature that blocks membrane fusion (32) may be defective for fusion, but not for docking, at restrictive temperatures. Finally, a golgin protein acting in consort with an Arf GTPase (66), and several synaptotagmin isoforms (67, 68), can induce SNARE-independent, but also Rab- and Rab effector-independent, liposome clustering.
We have proposed that Ypt7p and the HOPS complex do not form a direct, stable bridge between membranes, but rather mediate docking by acting in consort with a 3Q complex consisting of Vam3p, Vti1p, and Vam7p. We cannot, however, eliminate the possibility that HOPS and Ypt7p mediate a transient intermembrane interaction that is too labile to be detected but that is an obligate intermediate before stable docking mediated by HOPS, Ypt7p, and a 3Q SNARE complex. Nor can we rule out the possibility that other vacuolar proteins and lipids, not included in this reconstitution, contribute to SNARE-independent membrane associations, although no other vacuolar tethering factors have yet been reported. Finally, care should be taken when interpreting results obtained with proteoliposomes that are smaller than vacuoles. However, the region of contact between docked vacuoles, the vertex ring (69), is a region of high curvature, and thus the proteoliposomes used in this study are likely to be an appropriate model for this site of intermembrane contact.
We have found that differences in cardiolipin content and HOPS complex binding activity underlie the difference in Ypt7p dependence for fusion of standard-method and direct-method proteoliposomes. Our standard-method proteoliposomes have higher cardiolipin content than our direct-method proteoliposomes (Fig. S5C). Cardiolipin stimulates Ypt7p-independent fusion of standard-method proteoliposomes (Fig. S5E), consistent with the finding that Mg2+-cardiolipin can mediate liposome fusion without leakage (70). Does cardiolipin play a role in vacuole fusion in vivo? Cardiolipin is synthesized exclusively in the inner membrane of mitochondria (71); its reported presence in vacuoles in lipid analysis of purified organelles (72) is therefore likely caused by mitochondrial contamination. Furthermore, vacuoles from yeast lacking cardiolipin synthase have normal morphology at 30 °C (73). At 37 °C, cardiolipin-deficient yeast have abnormal vacuole morphology, but this defect is suppressed by deletion of the gene encoding the sodium/proton exchanger Nhx1p or the gene encoding the mitochondrial signaling protein Rtg2p (73). Thus, cardiolipin is unlikely to be involved in vacuole fusion.
Differences in HOPS complex binding and fusion requirements between direct-method and standard-method proteoliposomes provide an opportunity to define Rab GTPase function. Ypt7p is required for HOPS binding to direct-method proteoliposomes, whereas standard-method proteoliposomes bind HOPS robustly even in the absence of Ypt7p (Fig. 6). Furthermore, direct-method proteoliposomes require Ypt7p for fusion (Fig. 1A) whereas standard-method proteoliposomes do not (28). SNAREs are also not required for HOPS binding to standard-method liposomes (Fig. 6). Thus, the HOPS complex binds standard-method proteoliposomes via direct interactions with the membrane. These interactions may be mediated by phosphoinositides, which bind the HOPS complex (35), or the interaction of highly curved membranes with the ArfGAP1 lipid packing sensor motif in residues 356–379 of the Vps41p subunit of the HOPS complex (74). Although more work will be required to learn the molecular basis for the difference in requirements for HOPS complex binding to direct-method and standard-method proteoliposomes, GTP-bound Ypt7p is required for HOPS complex association with the vacuole (46); thus, this requirement for direct-method proteoliposomes reflects a central physiological function of Ypt7p. We have recently found that that phosphorylation of the Vps41p subunit of the HOPS complex by the casein kinase I homolog Yck3p (75) abrogates HOPS-membrane interactions and causes Ypt7p dependence for fusion of standard-method proteoliposomes (40). This result is consistent with the finding, both by Mima et al. (28) and shown here, that Ypt7p-independent HOPS-proteoliposome interactions (Fig. 6) can support Ypt7p-independent membrane fusion (Fig. S5E). These results demonstrate that the primary function of Ypt7p is recruitment of the HOPS complex to membranes.
The studies presented here suggest a working model for vacuole tethering. Cis 3Q:1R SNARE complexes are disassembled by Sec17p/18p, allowing the assembly of cis 3Q SNARE complexes. The HOPS complex associates with membranes via its direct affinities for SNAREs, Ypt7p:GTP, and vacuolar lipids, but is optimally activated for tethering by associations with Ypt7p:GTP and the 3Q cis-SNARE complex. The vacuolar proteins and lipids that directly interact in trans during tethering are not known, but it is likely that tethering is needed for rapid formation of trans-SNARE complexes and subsequent fusion.
Methods
Reagents.
His6-Sec18p (28), his6-Sec17p (28), and anti-Ypt7p and Ypt7p peptide (42) have been described. Gyp1–46 (47) was the gift of Vincent Starai (University of Georgia, Athens). ATPγS and GTPγS were from Roche, and UTPγS was from Jena Bioscience. Nucleotides (as Mg2+ salts), Gyp1–46, GDI (guanosine nucleotide dissociation inhibitor), and anti-Ypt7p were in RB150 [20 mM NaHepes (pH 7.4), 150 mM NaCl, 10% (vol/vol) glycerol], Sec17p and Sec18p were in buffers as described (76, 77), and Ypt7p peptide was in 20 mM Pipes-KOH (pH 6.8) 200 mM sorbitol. Phosphoinositides were from Echelon Research, ergosterol was from Sigma, fluorescent lipids were from Invitrogen, and all other lipids were from Avanti Polar Lipids. Overexpression and purification of HOPS complex are described in SI Text Primers used for plasmid and strain construction are in Table S1.
Direct Incorporation of Proteins into Liposomes.
All lipids were dissolved in chloroform except for phosphoinositides, which were dissolved in 1:2:0.8 chloroform/methanol/water. Lipids were mixed in glass tubes at the following mole percentages (28, 72, 78): 43% 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC), 18% 1-palmitoyl-2-oleoyl phosphatidylethanolamine (POPE), 18% soy PI, 4.4% 1-palmitoyl-2-oleoyl phosphatidylserine (POPS, 2% 1-palmitoyl-2-oleoyl phosphatidic acid) (POPA), 1.6% heart cardiolipin, 8% ergosterol, 1% each PI (3)P, PI(4)P, PI(4,5)P2, dansyl-phosphatidylethanolamine (PE); for donor lipids, 41% POPC, 18% POPE, 18% soy PI, 4.4% POPS, 2% POPA, 1.6% heart cardiolipin, 8% ergosterol, 1% each PI (3)P, PI(4)P, PI(4,5)P2, 1.5% each NBD-PE, Rhodamine-PE. Lipids were dried under a stream of N2 gas followed by vacuum, then suspended to a final concentration of 10 mM in RB150+ (RB150 with 1 mM MgCl2) by incubation on ice for 1 h with occasional vortexing, followed by 10 freeze–thaw cycles. (Lipids were often stored at −80 °C under N2 gas after the last freeze.) Lipids were then passed 11 times through a 25-mm diameter, 1-μm pore filter (Nucleopore Track-Etch Membrane; Whatman) in an ER-1 extruder (Eastern Scientific) at room temperature. Dansyl-PE or NBD-PE and rhodamine-PE fluorescence were used to measure the concentration of extruded lipids (λex/λem 350/540 and 540/586 nm, respectively).
The molar ratios of proteins to lipids in incorporation reactions were 1:667 (SNAREs) and 1:1,333 (Ypt7p) for acceptor liposomes and 1:1,000 (SNAREs) and 1:2,000 (Ypt7p) for donor liposomes. Before protein incorporation, GST-Vam3p, Nyv1p, and Vti1p (28) were mixed and dialyzed for 4–6 h, using Fisherbrand dialysis tubing with a molecular mass cutoff of 6–8 kDa and a volume/cm of 1.67 mL (Thermo Fisher Scientific), into mock Ypt7p buffer. (Mock Ypt7p buffer and Vam7 buffer were used in place of SNAREs for SNARE-free liposomes.) This mixture was then supplemented with Vam7p (28), tobacco etch virus (TEV) protease (at a 1:1 molar ratio to GST-Vam3p) and either Ypt7p or mock Ypt7p buffer. Proteins were mixed with extruded liposomes that were diluted with RB150+ such that the incorporation reaction had an Reff, the ratio of the difference between the n-octyl-β-d-glucopyranoside concentration and its critical micelle concentration (CMC) to the lipid concentration (39), of 0.2 for donor liposomes and 0.3 for acceptor liposomes. (The CMC of n-octyl-β-d-glucopyranoside was considered to be 18.5 mM.) The volume of the liposome solution was calculated by using the following formula:
where Vprotein is the volume of the protein solution, [βOG]protein is the n-octyl-β-d-glucopyranoside concentration in the protein solution, and Mlipids is the number of moles of lipids in the liposome solution. In a typical preparation, 795 μL of proteins (1.5 nmol of each GST-Vam3p, Vti1p, Nyv1p,Vam7p, and TEV protease; 0.75 nmol of Ypt7p or an equivalent volume of mock Ypt7p buffer; 32.4 mM n-octyl-β-d-glucopyranoside) were mixed with 581.4 μL of acceptor lipids (1 μmol). Protein/lipid/detergent mixtures were incubated on ice for 1 h then dialyzed into RB150+ at 4 °C overnight. For small-scale incorporations, Slide-A-Lyzer Mini Dialysis units with a 10-kDa cutoff (Pierce) were used, whereas for large-scale incorporations Fisher dialysis tubing with a cutoff of 6–8 kDa and a volume/cm of 1.67 mL was used.
Dialyzed proteoliposomes were mixed with 80% Histodenz in RB150+ to a final concentration of 35% or 40% Histodenz and covered with 30% Histodenz in RB150+, then by RB150+; volumes and centrifuge tubes depended on the scale of the incorporation reaction. In the example above, the lipid/protein/detergent mixture (1,376.4 μL) was mixed with 1.1 mL of 80% Histodenz (35.5% Histodenz final) in an 11 × 60-mm ultracentrifuge tube (Beckman no. 328874); 0.6 mL of 30% Histodenz then 0.6 mL of RB150+ were layered over this mixture. Gradients were centrifuged in a Beckman (Palo Alto, CA) SW-60 rotor at 55,000 rpm at 4 °C for 3 h or in a TLS-55 rotor at 50,000 rpm at 4 °C for 2 h. Proteoliposomes were harvested from the top interface of each gradient and dialyzed against RB150+ overnight at 4 °C. Lipid concentrations were measured as described above. Efficiency of protein incorporation was assessed by SDS/PAGE and Sypro Ruby (Invitrogen) staining; in all cases, the presence or absence of Ypt7p in incorporation reactions made no detectable difference in the efficiency of SNARE incorporation (Fig. 1A Inset).
Standard method proteoliposomes were prepared as in Mima et al. (28) except that Ypt7p was added to the initial lipid/detergent/protein solution at a 1:2,000 Ypt7p/lipid molar ratio or the same volume of mock Ypt7p buffer was added to the initial lipid/detergent/protein solution.
Proteoliposome Fusion Reactions.
Fusion was performed in 384-well plates (Corning no. 3676) at 27 °C. Complete reactions were in RB150 with: acceptor proteoliposomes, 0.36 mM total lipids; donor proteoliposomes, 0.03 mM total lipids; 1 mM ATP-Mg2+; 5 mM free MgCl2; Sec18p, 150 nM hexamer; 50 nM Sec17p; 34 nM HOPS complex. Proteoliposomes, inhibitors, and inhibitor-reversal agents were mixed in a total volume of 15.2 μL on ice, then placed at 27 °C for 10 min. Reactions were moved to room temperature and MgCl2, ATP, Sec18p, Sec17p, and HOPS (or HOPS buffer) were added, premixed in a volume of 4.8 μL. Reactions were returned to 27 °C and fluorescence (λex/λem 460/538 nm) was measured for 60 min. Thesit (2 μL of a 1% solution) was added and fluorescence was measured after 5 min at 27 °C. Dequenching was calculated as described (28). Each graph represents data from one experiment representative of three or more experiments. For representation of fusion data as the normalized sum of dequenching values, each dequenching curve was first adjusted by subtracting the minimum dequenching value for that particular condition from every point in the curve. Adjusted dequenching values for 0–45 min were then added. Each sum was then normalized by dividing it by the average sum of all of the complete dequenching reactions by using proteoliposomes from the same preparation and multiplying by 100. Normalized values were averaged; means and standard deviations are presented in Fig. S2.
Supplementary Material
Acknowledgments.
We thank the Pole Facultaire de Microscopie Electronique (PFMU) at the University of Geneva Medical School for access to electron microscopy equipment, Vincent Starai for Gyp1–46, and Reza Kordestani and Christian Raetz (Duke University, Durham, NC) for lipid analysis. This work was supported by National Institutes of Health Grant GM23377.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903801106/DCSupplemental.
References
- 1.Pfeffer SR. Rab GTPases: Specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 2.Carroll KS, et al. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292:1373–1376. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- 3.Pagano A, Crottet P, Prescianotto-Baschong C, Spiess M. In vitro formation of recycling vesicles from endosomes requires adaptor protein-1/clathrin and is regulated by rab4 and the connector rabaptin-5. Mol Biol Cell. 2004;15:4990–5000. doi: 10.1091/mbc.E04-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahadoran P, et al. Rab27a: A key to melanosome transport in human melanocytes. J Cell Biol. 2001;152:843–850. doi: 10.1083/jcb.152.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer A, Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J Cell Biol. 1997;136:307–317. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabcenell AK, Goud B, Northup JK, Novick PJ. Binding and hydrolysis of guanine nucleotides by Sec4p, a yeast protein involved in the regulation of vesicular traffic. J Biol Chem. 1990;265:9366–9372. [PubMed] [Google Scholar]
- 7.Soldati T, Shapiro AD, Svejstrup AB, Pfeffer SR. Membrane targeting of the small GTPase Rab9 is accompanied by nucleotide exchange. Nature. 1994;369:76–78. doi: 10.1038/369076a0. [DOI] [PubMed] [Google Scholar]
- 8.Du LL, Collins RN, Novick PJ. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J Biol Chem. 1998;273:3253–3256. doi: 10.1074/jbc.273.6.3253. [DOI] [PubMed] [Google Scholar]
- 9.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parente RA, Lentz BR. Rate and extent of poly(ethylene glycol)-induced large vesicle fusion monitored by bilayer and internal contents mixing. Biochemistry. 1986;25:6678–6688. doi: 10.1021/bi00369a053. [DOI] [PubMed] [Google Scholar]
- 11.Papahadjopoulos D, Vail WJ, Pangborn WA, Poste G. Studies on membrane fusion. II. Induction of fusion in pure phospholipid membranes by calcium ions and other divalent metals. Biochim Biophys Acta. 1976;448:265–283. doi: 10.1016/0005-2736(76)90241-8. [DOI] [PubMed] [Google Scholar]
- 12.Harmsen MC, et al. Reconstitution and fusogenic properties of Sendai virus envelopes. Eur J Biochem. 1985;149:591–599. doi: 10.1111/j.1432-1033.1985.tb08966.x. [DOI] [PubMed] [Google Scholar]
- 13.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 14.Scott BL, et al. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J Cell Biol. 2004;167:75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J, et al. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 17.Schaub JR, et al. Hemifusion arrest by complexin is relieved by Ca2+-synaptotagmin I. Nat Struct Mol Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- 18.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Ca2+-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 20.Chicka MC, Hui E, Liu H, Chapman ER. Synaptotagmin arrests the SNARE complex before triggering fast, efficient membrane fusion in response to Ca2+ Nat Struct Mol Biol. 2008;15:827–835. doi: 10.1038/nsmb.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon TY, et al. Complexin and Ca2+ stimulate SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2008;15:707–713. doi: 10.1038/nsmb.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malsam J, et al. The carboxyl-terminal domain of complexin I stimulates liposome fusion. Proc Natl Acad Sci USA. 2009;106:2001–2006. doi: 10.1073/pnas.0812813106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maximov A, et al. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chicka MC, Chapman ER. Concurrent binding of complexin and synaptotagmin to liposome-embedded SNARE complexes. Biochemistry. 2009;48:657–659. doi: 10.1021/bi801962d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai H, Shen N, Arac D, Rizo J. A quaternary SNARE–synaptotagmin–Ca2+-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seals DF, et al. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mima J, et al. Reconstituted membrane fusion requires regulatory lipids, SNAREs, and synergistic SNARE chaperones. EMBO J. 2008;27:2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas A. A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci. 1995;17:283–294. [Google Scholar]
- 30.Jun Y, Wickner W. Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing. Proc Natl Acad Sci USA. 2007;104:13010–13015. doi: 10.1073/pnas.0700970104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohya T, et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 32.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ungermann C, Sato K, Wickner W. Defining the functions of trans-SNARE pairs. Nature. 1998;396:543–548. doi: 10.1038/25069. [DOI] [PubMed] [Google Scholar]
- 34.Collins KM, Wickner WT. Trans-SNARE complex assembly and yeast vacuole membrane fusion. Proc Natl Acad Sci USA. 2007;104:8755–8760. doi: 10.1073/pnas.0702290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins KM, Thorngren NL, Fratti RA, Wickner WT. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. EMBO J. 2005;24:1775–1786. doi: 10.1038/sj.emboj.7600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigaud JL, Levy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 40.Hickey CM, Stroupe C, Wickner W. The major role of the Rab Ypt7p in vacuole fusion is supporting HOPS membrane association. J Biol Chem. 2009;284:16118–16125. doi: 10.1074/jbc.M109.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 42.Eitzen G, Thorngren N, Wickner W. Rho1p and Cdc42p act after Ypt7p to regulate vacuole docking. EMBO J. 2001;20:5650–5656. doi: 10.1093/emboj/20.20.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starai VJ, Jun Y, Wickner W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc Natl Acad Sci USA. 2007;104:13551–13558. doi: 10.1073/pnas.0704741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dennison SM, Bowen ME, Brunger AT, Lentz BR. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McIntyre JC, Sleight RG. Fluorescence assay for phospholipid membrane asymmetry. Biochemistry. 1991;30:11819–11827. doi: 10.1021/bi00115a012. [DOI] [PubMed] [Google Scholar]
- 46.Eitzen G, et al. Sequential action of two GTPases to promote vacuole docking and fusion. EMBO J. 2000;19:6713–6720. doi: 10.1093/emboj/19.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickner W, Haas A. Yeast homotypic vacuole fusion: A window on organelle trafficking mechanisms. Annu Rev Biochem. 2000;69:247–275. doi: 10.1146/annurev.biochem.69.1.247. [DOI] [PubMed] [Google Scholar]
- 49.Seeley ES, et al. Genomic analysis of homotypic vacuole fusion. Mol Biol Cell. 2002;13:782–794. doi: 10.1091/mbc.01-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ungermann C, Nichols BJ, Pelham HR, Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol. 1998;140:61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parlati F, et al. Topological restriction of SNARE-dependent membrane fusion. Nature. 2000;407:194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- 52.Peloso CW, O'Connor MJ, Bigger SW, Scheirs J. Characterizing the degradation of the polymer slip additive erucamide in the presence of inorganic antiblock agents. Polym Degrad Stab. 1998;62:285–290. [Google Scholar]
- 53.Rand RP, Sengupta S. Cardiolipin forms hexagonal structures with divalent cations. Biochim Biophys Acta. 1972;255:484–492. doi: 10.1016/0005-2736(72)90152-6. [DOI] [PubMed] [Google Scholar]
- 54.Kümmel D, Heinemann U. Diversity in structure and function of tethering complexes: Evidence for different mechanisms in vesicular transport regulation. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- 55.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 56.Pfeffer S. Vesicle tethering factors united. Mol Cell. 2001;8:729–730. doi: 10.1016/s1097-2765(01)00371-9. [DOI] [PubMed] [Google Scholar]
- 57.Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am J Physiol. 2006;290:C11–C26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- 58.Bentley M, et al. SNARE status regulates tether recruitment and function in homotypic COPII vesicle fusion. J Biol Chem. 2006;281:38825–38833. doi: 10.1074/jbc.M606044200. [DOI] [PubMed] [Google Scholar]
- 59.Barnard RJ, Morgan A, Burgoyne RD. Stimulation of NSF ATPase activity by α-SNAP is required for SNARE complex disassembly and exocytosis. J Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hammarlund M, et al. Open syntaxin docks synaptic vesicles. PLoS Biol. 2007;5:e198. doi: 10.1371/journal.pbio.0050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tareste D, Shen J, Melia TJ, Rothman JE. SNAREpin/Munc18 promotes adhesion and fusion of large vesicles to giant membranes. Proc Natl Acad Sci USA. 2008;105:2380–2385. doi: 10.1073/pnas.0712125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Merz AJ, Collins KM, Wickner W. Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J Cell Biol. 2003;160:365–374. doi: 10.1083/jcb.200209095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerrard SR, Mecklem AB, Stevens TH. The yeast endosomal t-SNARE, Pep12p, functions in the absence of its transmembrane domain. Traffic. 2000;1:45–55. doi: 10.1034/j.1600-0854.2000.010108.x. [DOI] [PubMed] [Google Scholar]
- 64.Geumann U, et al. SNARE function is not involved in early endosome docking. Mol Biol Cell. 2008;19:5327–5337. doi: 10.1091/mbc.E08-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bethani I, et al. The specificity of SNARE pairing in biological membranes is mediated by both proofreading and spatial segregation. EMBO J. 2007;26:3981–3992. doi: 10.1038/sj.emboj.7601820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drin G, et al. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320:670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- 67.Araç D, et al. Close membrane-membrane proximity induced by Ca2+-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 68.Connell E, et al. Cross-linking of phospholipid membranes is a conserved property of calcium-sensitive synaptotagmins. J Mol Biol. 2008;380:42–50. doi: 10.1016/j.jmb.2008.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Seeley ES, Wickner W, Merz AJ. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 70.Ortiz A, Killian JA, Verkleij AJ, Wilschut J. Membrane fusion and the lamellar-to-inverted-hexagonal phase transition in cardiolipin vesicle systems induced by divalent cations. Biophys J. 1999;77:2003–2014. doi: 10.1016/S0006-3495(99)77041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlame M, Haldar D. Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J Biol Chem. 1993;268:74–79. [PubMed] [Google Scholar]
- 72.Zinser E, et al. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S, Tarsio M, Kane PM, Greenberg ML. Cardiolipin mediates cross-talk between mitochondria and the vacuole. Mol Biol Cell. 2008;19:5047–5058. doi: 10.1091/mbc.E08-05-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drin G, et al. A general amphipathic α-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 75.LaGrassa TJ, Ungermann C. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J Cell Biol. 2005;168:401–414. doi: 10.1083/jcb.200407141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thorngren N, et al. A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J. 2004;23:2765–2776. doi: 10.1038/sj.emboj.7600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haas A, Wickner W. Homotypic vacuole fusion requires Sec17p (yeast α-SNAP) and Sec18p (yeast NSF) EMBO J. 1996;15:3296–3305. [PMC free article] [PubMed] [Google Scholar]
- 78.Schneiter R, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orci L, et al. Budding from Golgi membranes requires the coatomer complex of nonclathrin coat proteins. Nature. 1993;362:648–652. doi: 10.1038/362648a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.