Lysander
Swift as a shadow, short as any dream, Brief as the lightning in the collied night, That, in a spleen, unfolds both heaven and earth; And ere a man hath power to say “Behold!” The jaws of darkness do devour it up: So quick bright things come to confusion.
William Shakespeare, A Midsummer Night’s Dream Act 1, scene 1, 141–149
Unlike T and B cells, NK cells lack variable, clonotypic receptors that recognize foreign antigens. Instead, NK cells depend on conserved receptors such as NKG2D. NKG2D recognizes a variety of inducible self-proteins that belong to the non-classical MHC class I family. They include ULBP (1–3), MIC (A & B) in human and H60 (a, b & c), Rae-1 (α-ε) and Mult1 in mice. These self-proteins are expressed due to pathological stimuli, share limited amino acid homology and form the molecular basis for NKG2D-mediated activation. Recent studies have vastly improved our understanding of NKG2D receptor-mediated activation, signaling and function. However, a detailed knowledge on the immunobiology of its ligands is lacking. How many is too many? Is NKG2D the only receptor for these ligands? Where are these ligands expressed? What are the molecular mechanisms that regulate their expression? Do normal cells express these ligands? Does the communication between NKG2D receptor and its ligands travel through a two way road? If so, what do the ‘target’ cells get in turn, only death? How efficient are these ligands as molecular targets for NK cell-mediated tumor immunotherapy?
Background
NK cells are the major effector lymphocytes of innate immune system that defend against many forms of viral infections and tumor growth(Karre, 2002b). NK cells are granular, bone marrow derived lymphocytes capable of executing ‘natural cytotoxicity’ of target cells without prior sensitizations(Moretta et al., 2001). NK cells also bridge innate and adoptive immune responses through the secretion of a variety of cytokines and chemokines(Biron et al., 1999),(Farag et al., 2003),(Moser and Loetscher, 2001). Effector phase of NK cells are regulated by inhibitory and activating receptors. Each NK cell is known to express one or more inhibitory receptors, which interact with specific MHC class I molecules on the target cells(Liu et al., 2000). Interaction of MHC class I with the inhibitory Ly49 receptors prevent the activation of NK cells and thereby the lysis of the target cell. Thus, NK cells utilize the Ly49 receptors to differentiate ‘self’ from ‘missing-self’(Karre et al., 1986;Karre, 2002a),. NK cells also use multiple activating receptors that are conserved and non-polymorphic. These include NKR-P1C, FcRγIII, Ly49D, Ly49H, NKG2C and NKG2D. These receptors associate with adaptor proteins that contain activation motifs for transducing signals.
NKG2D is expressed on all NK cells, some NKT, α/β-TCR+ and γ/δ-TCR+ T cells(Bauer et al., 1999;Wu et al., 1999;Vilarinho et al., 2007;Girardi et al., 2001;Groh et al., 2001). There are two known isoforms of NKG2D that could differentially associate with DAP10 or DAP12 adaptor proteins(Wu et al., 2000). NKG2D has the ability to recognize and bind to a variety of inducible self-proteins. Currently, nine murine and five human proteins have been defined as ligands for NKG2D receptor. These include ULBP (1–5)(Sutherland et al., 2001;Cosman et al., 2001), MIC (A & B)(Bauer et al., 1999) in human and H60 (a, b & c), Rae-1 (α, β, γ, δ & ε), Mult1 in mice(Diefenbach et al., 2000;Cerwenka et al., 2000;Carayannopoulos et al., 2002c). These ligands belong to non-classical MHC class I family and are inducible by multiple stress stimuli; therefore, defined as ‘induced-self’. Through our earlier studies, we have shown that ‘induced-self’ can be regulated by the inhibitory Ly49 receptors(Regunathan et al., 2005). Thus, ‘induced-self’ is a functional component of ‘missing-self’. NK cells can also mediate their effector functions if a non-compatible MHC is expressed on the target cells. NKG2D ligands may or may not be involved in this type of NK cell recognition, which is defined as ‘allo’.
Genomic organization of murine NKG2D ligands
Murine NKG2D ligands are located in chromosome 10 (Figure 1A). Based on their genomic and protein homology, we classify them into three families, H60, Rae-1 and Mult1. H60 family has three members (a, b and c) and the first identified member, Histocompatibility antigen 60a (H60a), is a minor histocompatibility antigen(Malarkannan et al., 1998;Takada et al., 2008). Retinoic acid induced early transcript (Rae-1) family was identified as responders to retinoic acid(Zou et al., 1996;Nomura et al., 1996) and have five members (α-ε). The third family, Murine ULBP like transcript (Mult1) has a single member and is distantly related to H60 and Rae-1(Carayannopoulos et al., 2002a;Diefenbach et al., 2003). The genomic locations of these genes have been assigned except for H60a and H60b which are approximately placed in the A3 band of chromosome 10(Takada et al., 2008) (Figure 1B). Currently, H60a is misassigned to Chromosome 5 in NCBI Build 37.1 (accession number: NW_001030795.1). H60b is in the contig Mm10_39532_37 (Accession number: NT_039492.7) and H60c is within the contig Mm10_39530_37 (Accession number: NT_39490.7). H60 genomic sequences vary between 8 to 15 kb and each gene contains 6 exons. In H60 genes, exon 1 encodes the leader sequence and exons 2 to 4 corresponds to the extracellular α1 and α2 MHC class I domains. In H60a and H60b, the exons 4 and 5 encode the transmembrane and cytoplasmic domains while in H60c they encode a GPI anchor. Rae-1 genes are located in a single cluster in the A2 band of Chromosome 10 and each gene spans approximately 10–12 kb (Figue 1A). Rae-1 family members have 4 exons and exon 1 encodes leader sequences. Exons 2 and 3 encode extracellular α1 and α2 MHC class I domains and exon 4 for GPI anchors. Interestingly, Mult1 gene is located between the exon 1 and 2 of H60b, which are about 120kb apart. Mult1 gene spans approximately 31 kb and it contains 7 exons. Exon 1 encodes the leader sequence, while the exons 2, 3 and part of exon 4 for the α1 and α2 extracellular domains. Remaining exons encode transmembrane and cytoplasmic domains. In human, NKG2D ligands are located in chromosome 6, which are grouped into two families, MIC and ULBP. MHC class I chain-related protein (MIC) has two members (A and B) and their genes are located within the MHC complex within 6q21.3. The UL16-binding protein (ULBP) family has 10 genes of which five are expressed. ULBP are located outside the MHC gene complex in 6q24.2–25.3 region, which is syntenic to a segment in murine chromosome 10 containing Rae-1 genes. No MIC equivalents present in the mouse and similarly H60 and Mult1 are not present in human.
Figure 1. Chromosomal localization and the genomic organization of NKG2D ligands.
(A) Comparative genomic map showing locations of murine NKG2D ligands in chromosome #10. NCBI Build 37.1 and contig sequences from Ensembl were used to generate the relative locations of these genes. Currently, the H60a gene is misassigned to Chromosome #5 in NCBI Build 37.1 (accession number: NW_001030795.1). H60b and H60c positions are estimated based on the information obtained from contigs Mm10_39532_37 (Accession number: NT_039492.7) and Mm10_39530_37 (Accession number: NT_39490.7). Other marker genes are included to indicate the relative mapping of NKG2D ligands. (B) Intron/exon organization of NKG2D ligands. All the nine members of the murine NKG2D ligands are presented. Exons (ex #) are represented as filled-in boxes with their length indicated as number of nucleotides.
A significant feature of murine NKG2D ligand genes is the presence of highly conserved Line-1 retrotransposon elements in the immediate upstream or within their first intronic sequences. Line-1 elements are the most abundant autonomous retrotransposons and constitute ~20% of the genomic mass in human or mouse(Boissinot and Furano, 2001). Recent studies have emphasized their biological roles as regulators of their host gene transcription and translation(Furano et al., 2004). Our analyses reveal that the Line-1 elements in the NKG2D ligand genomic sequences encode a protein, ORF2, which possesses endonuclease and reverse transcriptase activity and thereby 5′ UTR regulatory properties. H60b, all the Rae-1 and the Mult1 genes contain full-length ORF2 sequences within their first introns (data not shown). It was assumed these intronically inserted sequences are post-transcriptionally spliced out and do not affect the expression of the host genes. However, recent studies by Han et al. demonstrates that Line-1 elements can affect the RNA production of host genes through a combination of transcriptional elongation inhibition and premature polyadenylation(Han et al., 2004). Other studies indicate that micro RNAs (miRNAs) derived from Line-1 retrotransposons can effectively silence the translation of host genes(Aravin et al., 2007). Future studies should focus in delineating the role of intronic retrotransposons in regulating the expression and function of NKG2D ligands.
Evolution of murine NKG2D ligands
Organization and sequence homology of NKG2D ligand family indicate that they are generated by multiple rounds of gene duplication and divergence from an ancestral prototypic gene. To compare non-synonymous to synonymous substitutions in their genomic sequences, we generated internal sequence identity plots using PAML program(Yang, 2007). Since the genomic sequences of NKG2D ligands are distributed over multiple giga bases, we selected portions of contigs that represent the intron/exon organization of each gene, organized them into a single sequence and used it as our input for the PAML program. The order of arrangement and their sequence lengths are indicated in Figure 2A. The sequence identity plot demonstrates that the genes encoding Rae-1 isoforms possess the highest internal sequence identity within their family. Further, Rae-1 family genes also have significant regions of internal sequence identities to H60a and H60b genes. There are also major differences within H60 genes that indicate recent and possibly active diversifications in this family. This analysis also indicates that the Rae-1 family evolved by a series of localized tandem genomic duplications.
Figure 2. Internal sequence identity plot and phylogenetic tree analyses of NKG2D ligands.
(A) Non-synonymous to synonymous substitutions in the genomic sequences encoding for these ligands were compared in internal sequence identity plots generated by PAML program. Names and sequence numbers displayed in the X- and Y- axis denote individual genomic gene sequences used and the orientation. Since the genomic sequences of NKG2D ligands are distributed over multiple giga bases, we selected portions of contigs that represent the intron/exon organization of each gene, organized them into a single sequence and used it as our input into the PAML program. The center line represents identity; other lines indicate regions of internal sequence identity. Internal sequence identity that indicates possible gene duplication in Rae-1 gene family is shaded in yellow. Genomic similarities between H60a to H60 and Rae-1 (purple), H60b to H60c and Rae-1 (dark blue and green), Mult-1 to Rae-1 (light blue) are indicated. (B) Phylogenetic tree indicates the relationship of NKG2D ligands to select members of classical and non-classical MHC class I family. This tree was generated using ClustalW alignment program in MacVector software based on distance-matrix. We used amino acid sequences of the 9 members of the NKG2D ligand family along with select classical and non-classical MHC class I family members. Dendrogram was generated using UPGMA method with a ‘Systematic tie breaking’, which is based on the percent differences between amino acid sequences. Distances were calculated using an uncorrected ‘p’ and gaps were distributed proportionately among different amino acid sequences used.
Protein sequence and structure of NKG2D ligands are related to MHC class I family. To understand their evolutionary relationship with this family, we generated a phylogenetic tree with ClustalW alignment program in MacVector software based on distance-matrix. We compared amino acid sequences of all 9 murine NKG2D ligands with select classical and non-classical MHC class I family members. Dendrogram was generated using UPGMA method with a ‘Systematic tie breaking’, which is based on the percent differences between amino acid sequences. Distances were calculated using an uncorrected ‘p’ and gaps were distributed proportionately among different amino acid sequences. Dendrogram in Figure 2B suggests that the NKG2D ligand family evolved from an ancestral gene that has diverged from classical or non-classical MHC class I family. Dendrogram also supports our classification of NKG2D ligands into H60, Rae-1 and Mult1 families. Compared to Mult1, H60 and Rae-1 family are closely related suggesting that these two families diverged from a common ancestor. Among H60 family, H60a is closely related to H60b (75%) while H60a shares only 46% homology to H60c (Table 1). When we compared the ectodomains, which binds to NKG2D, H60b and H60c showed a higher homology to H60a (86 and 75%, respectively). Compared to H60, Rae-1 family members are more conserved with >88% amino acid homology and relatively smaller evolutionary distances. We also included an undefined gene sequence (dbj|BAE23092.1|) with 51% conserved homology over a stretch of 204 amino acids to Mult1 (New?) to indicate the presence of additional member(s) of Mult1 family in the genome. Evolutionary diversification of NKG2D ligands is also evident from their tissue specific expression pattern. For example, H60c is uniquely expressed in skin compared to H60a and H60b that are widely expressed in spleen, skeletal muscles and thymus. High level expression of Rae-1α isoform in neural tissues is another proof of evolutionary diversification of NKG2D ligands(Nomura et al., 1996).
Table 1. Amino acid homologies between NKG2D ligands.
Percent amino acid identities and similarities between each member of the NKG2D ligand family were determined using MacVector program. Highest homologies are seen among the Rae-1 family members. H60a and H60b have comparable homology between each other compared to H60c. However, similarities between inter subfamily members are very minimal. Mult1 has the least homology when compared to other members of the family. Thus, even with minimal amino acid homology NKG2D ligands conserved their functional ability of binding to NKG2D receptor.
| H60b | H60c | Rae1α | Rae1β | Rae1γ | Rae1δ | Rae1ε | Mult1 | |
|---|---|---|---|---|---|---|---|---|
| H60a | 75 (6) | 46 (6) | 24 (14) | 25 (14) | 23 (14) | 25 (38) | 24 (14) | 12 (11) |
| H60b | 49 (7) | 23 (16) | 24 (15) | 22 (16) | 23 (16) | 22 (16) | 10 (10) | |
| H60c | 19 (12) | 19 (12) | 18 (13) | 19 (12) | 19 (12) | 9 (8) | ||
| Rae1α | 94 (1) | 94 (1) | 92 (0) | 88 (2) | 12 (10) | |||
| Rae1β | 92 (2) | 94 (0) | 88 (2) | 11 (10) | ||||
| Raeγ | 91 (2) | 92 (1) | 12 (10) | |||||
| Rae1δ | 89 (1) | 11 (9) | ||||||
| Raeε | 12 (9) |
Protein structure and affinity of murine NKG2D ligands
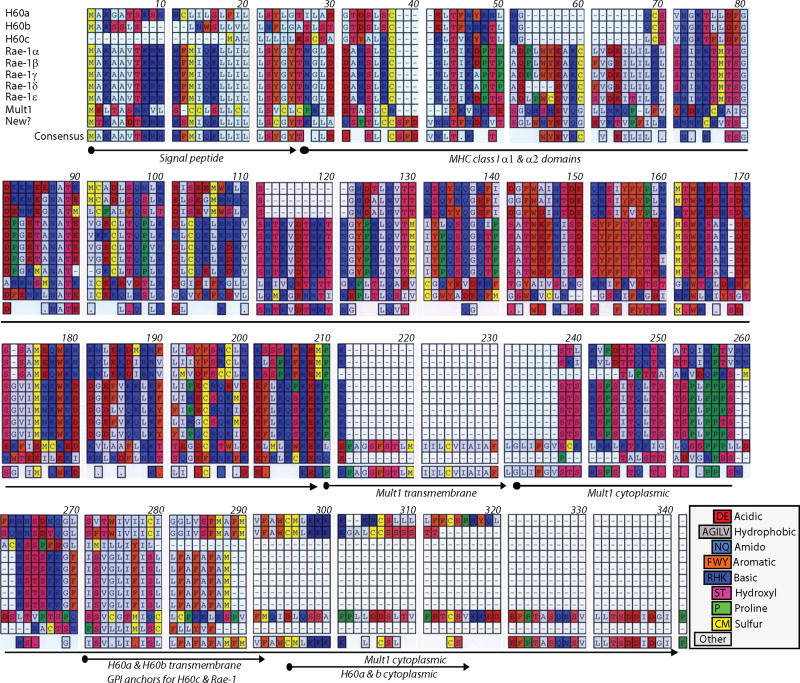
NKG2D ligand family has distant structural homology to the classical MHC class I proteins. Murine NKG2D ligands vary in their length and amino acid sequence (Figure 3). NKG2D ligands H60, Rae-1, Mult1 and the ULBPs have only the α1 and α2 MHC domains. MIC-A and MIC-B contain α1, α2, and α3 MHC domains. Interestingly, none of these ligands are able to bind to the β-2 microglobulin to form the classical MHC class I heterodimeric complexes(Bauer et al., 1998). The α1 and α2 helices of NKG2D ligands lack critical residues that have been shown to interact with the antigenic epitopes that result in the narrowing and partial obstruction of peptide binding grove. Therefore, they are not known to present any antigenic moieties. One of the highly variable features of the murine NKG2D ligands is their cytoplasmic domains. Utilization of data mining programs such as Eukaryotic Linear Motif resource for functional sites in proteins (ELM) reveals the presence of multiple putative sites in the cytoplasmic domains of H60a, H60b and Mult1 proteins (Figure 4). Of particular interest are the sorting/internalization motif in H60a and the di-leucine motif in Mult1 proteins. Presence of these motifs provides another molecular mechanism that may exert a post-translational regulation of their expression. Other motifs that include substrate docking and phosphorylation sites indicate an active involvement of NKG2D ligands in cellular signaling. Two highly conserved seventeen amino acids long repeat motifs are present in the cytoplasmic domain of Mult1, whose cellular functions have yet to be determined.
Figure 3. Amino acid sequence alignment of NKG2D ligands.
All nine murine NKG2D ligands are aligned using MacVector software. Gaps are represented by hyphens. Specific amino acid types are colored in indicate their chemical nature. Consensus amino acid sequence from the aligned sequence is deduced and presented in the bottom. Signal peptide, ectodomains (α1 and α2), GPI anchors, transmembrane and cytoplasmic domains are marked by arrows in the bottom of the sequences.
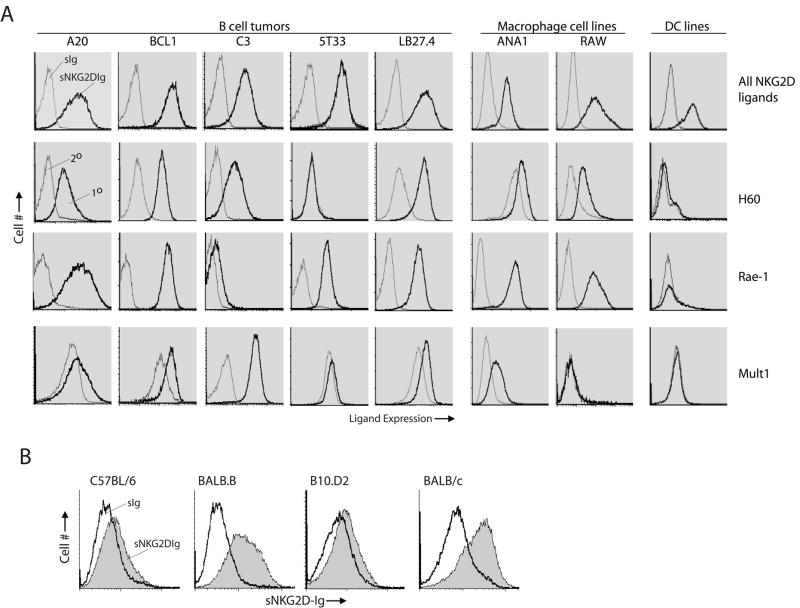
Figure 4. Expression of NKG2D ligands in primary cells and established tumor cell lines.
(A) Expression of NKG2D ligands on tumor cells was analyzed using similar staining procedures. A20 is a widely used tumor cell line and it is a mature B cell lymphoma derived from a spontaneous reticulum cell neoplasm found in an old BALB/cAnN mouse. BCL1 clone 5B1b is immature B lymphoma capable of secreting an IgM of unknown specificity upon LPS induction. C3 is a pro-B lymphoma that is derived from (B10×129/J)F1 mice. 5T33 is a B cell myeloma of unknown developmental stage that is derived from C57BL/6. LB27.4 is a hybridoma generated by fusing A20.2J mature lymphoma with T cell depleted spleen cells from C57BL/6 mice. Ana-1 is a macrophage cell line derived from C57BL/6. RAW264.7 is a macrophage line that is transformed using Abelson murine leukemia virus and DC2.4 is a dendritic cell line. (B) Expression of NKG2D ligands in LPS-stimulated total splenocytes. Single cell suspensions from the spleens of C57BL/6 (H-2b), BALB.B (H-2b), B10.D2 (H-2d) and BALB/c (H-2d) mice were stimulated with 2μg/ml LPS for 48 hours. Stimulated cells were washed, Fc-blocked and stained with soluble NKG2D-Fc followed by an anti-human IgG antibody-conjugated with PE. Cells were also stained with anti-H60a, pan anti-Rae-1 or anti-Mult1 antibodies followed by respective secondary antibodies. Background staining with secondary antibodies alone are shown as controls. sIg, soluble Fc control; sNKG2D-Fc, soluble NKG2D-Fc; 2°, secondary antibody alone and 1°, primary and secondary antibodies.
These murine ligands differ in their binding affinity to NKG2D receptor. The binding affinity (Kd) varies between nM to μM (Mult1 [6 nM] > H60a [20–30 nM] > Rae-1β [345 nM] > Rae-1γ [586 nM] > Rae-1α [690 nM] > Rae-1δ [726 nM] > H60b [310 nM] > H60c [8.7 μM](Takada et al., 2008),(Carayannopoulos et al., 2002a;Smyth et al., 2004),(Carayannopoulos et al., 2002b;O’Callaghan et al., 2001). Affinity of these ligands at the nanomolar range indicates a much tighter binding to NKG2D compared to many other immune receptor-ligand interactions, including peptide-loaded MHC class I to TCR [Kd 1–90 μM] or MHC to CD8 [65–200 μM]. Callaghan et al. demonstrate that H60a-NKG2D interaction is more temperature and electrostatic dependent compared to that of Rae-1γ-NKG2D(O’Callaghan et al., 2001). Therefore, they predict that at higher temperature, such in fever, the relative importance of Rae-1 interaction to NKG2D will increase over other ligands. Currently, the correlation between the ligand affinity and the NKG2D-mediated signaling strength and thereby effector functions is not determined. So far, NKG2D is the only defined receptor for these ligands; however, recent studies by Kriegeskorte et al. showed an NKG2D-independent suppression of T cell proliferation by soluble H60a and MIC-A but not by soluble Mult1, which indicates the existence of other receptors that are ligand-specific(Kriegeskorte et al., 2005).
Expression of NKG2D ligands in tumor cells
Clinically, NKG2D ligands can be used as potential molecular targets in NK cell-mediated immunotherapies to cure malignancies. However, past studies including from our laboratory have exclusively used model tumor cells that have been stably transfected to express the NKG2D ligands(Diefenbach et al., 2000;Cerwenka et al., 2000;Regunathan et al., 2005). Therefore, it becomes important to determine the expression of these ligands on naturally occurring tumor cells. Towards this we selected an array of myeloma, thymoma, macrophage and dendritic cell lines and tested them with soluble NKG2D-immunoglobulin Fc fusion protein (sNKG2D-Fc) or anti-H60a, anti-Rae-1 and anti-Mult-1 antibodies. sNKG2D-Fc stained all the myeloma cell lines (Figure 5A). H60a was expressed on all the myeloma tumors except 5T33, which was derived from C57BL/6 background. Pan-anti-Rae-1 stained all the tested cell lines except C3; however, Mult1 was expressed only on C3 cells. Thus, all the myeloma cells tested were positive for at least one ligand. In contrast, thymoma cell lines EL4 or RMA did not express any of these ligands (not shown). sNKG2D-Fc also stained macrophage cell lines Ana-1 and RAW264.7. Ana-1, derived from C57BL/6, expressed abundant Rae-1 and a low level of Mult1. RAW264.7, which was transformed using Abelson murine leukemia virus, expressed significant amounts of Rae-1 but not other ligands. Interestingly, although sNKG2D-Fc could bind to a dendritic cell line DC2.4, none of these three antibodies could stain this cell line. Thus, the wide expression of NKG2D ligands on a variety of hematological malignant cell lines implies their promise in clinical applications.
Figure 5. Cytoplasmic domains of H60a, H60b and Mult1 possess multiple putative motifs.
Presence of a number of difference motifs indicates a possible post-translational regulation of murine NKG2D ligand expression and their specialized functions. These motifs could facilitate a two way signaling communication or in the sorting, targeting and internalization of these ligands.
NKG2D ligands can be induced in normal cells
Although the expression and function of NKG2D ligands is well documented in tumor cells, their immunological relevance in normal cells is not clear. In BALB mice, H60a mRNA could be detected in multiple tissues including cardiac and skeletal muscles, spleen, thymus and skin, while H60b mRNA is restricted to the cardiac and skeletal muscles. H60c is unique in that its basal transcription was largely limited to the skin(Takada et al., 2008). Although in C57BL/6 strain, H60a gene is not transcribed(Malarkannan et al., 1998), H60b and H60c mRNA are abundantly present(Takada et al., 2008). Rae-1β and Rae-1γ mRNA are predominantly expressed in the embryonic brain of 129J mice(Nomura et al., 1996). Differential transcription of Rae-1 isoforms has also been reported in peritoneal macrophages during MCMV infection, i.e. Rae-1α, Rae-1β, and Rae-1γ in BALB/c and Rae-1δ and Rae-1ε in C57BL/6(Lodoen et al., 2003a). Mult1 mRNA are present in the lung, heart, thymus and kidney from multiple mouse strains(Carayannopoulos et al., 2002b),(Carayannopoulos et al., 2002a). These observations demonstrate that the ligand genes can be transcribed in a variety of tissues; however, their protein expressions have yet to be determined. To determine ligand protein expression, we analyzed resting and LPS-induced splenocytes. The anti-ligand antibodies used for staining tumor cell lines gave high background in mouse splenocytes; therefore, we used only sNKG2D-Fc. Resting splenocytes did not bind to sNKG2D-Fc, indicating that these ligands are not expressed on ‘non-induced’ cells (not shown). However, splenocytes that were induced with LPS for 48 hrs significantly augmented their ability to bind to the sNKG2D-Fc (Figure 5B). Interestingly, only the splenocytes from BALB origin but not C57BL/6 nor B10.D2 had the ability to bind to the sNKG2D-Fc. One explanation for this strain differences is that the H60a gene is not transcribed in the C57BL/6 mice(Malarkannan et al., 1998). Along this line, we could not detect the Rae-1 mRNA in the LPS-stimulated C57BL/6 splenocytes (Samarakoon and Malarkannan, unpublished). These strain variations indicate the presence of pre- and post-translational regulations of NKG2D ligand expression.
The expression of NKG2D ligands in both lymphoid and non-lymphoid tissues indicates that this family plays specialized functions during embryogenesis and immune surveillance. Currently, the cell type specific expression and the immunological relevance of ligands in normal cells are not clear. Most of the studies on NKG2D ligands show that their recognition by NK cells results in the clearance, leading to the assumption that the only functional consequence is the cytotoxicity. However, studies from our laboratory demonstrate that NKG2D-mediated cytotoxicity can be fully inhibited by the Ly49 receptors and therefore the target cells by NKG2D does not always result in cytotoxicity(Regunathan et al., 2005). Is this a novel ‘danger signal’, which does not lead to target cell death? If not cytotoxicity, what other effector functions NK cells exert during NKG2D ligand recognition? We predict that the recognition of the ligands by NKG2D could initiate reciprocal signaling cascades in both NK and target cells. Vital clues are possibly buried in the cytoplasmic domains of NKG2D ligands. Nearly half of the ligand family contains cytoplasmic domains. Among these, Mult1 has the longest cytoplasmic domain with highly sophisticated repeat motifs, whose functions are yet to be deciphered (Figure 4). Other pertinent questions are related to the role of these ligands during the immune ontogeny. In this context it is important to note that many of the molecular machinations used by DNA damage pathway are also shared by the immunoglobulin or T cell receptor gene rearrangement in B and T cells. It is an exciting possibility if B or T cells express NKG2D ligands during their ontogeny in order to get specialized help from NK cells via NKG2D receptor.
Molecular mechanisms that regulate the expression of NKG2D ligands
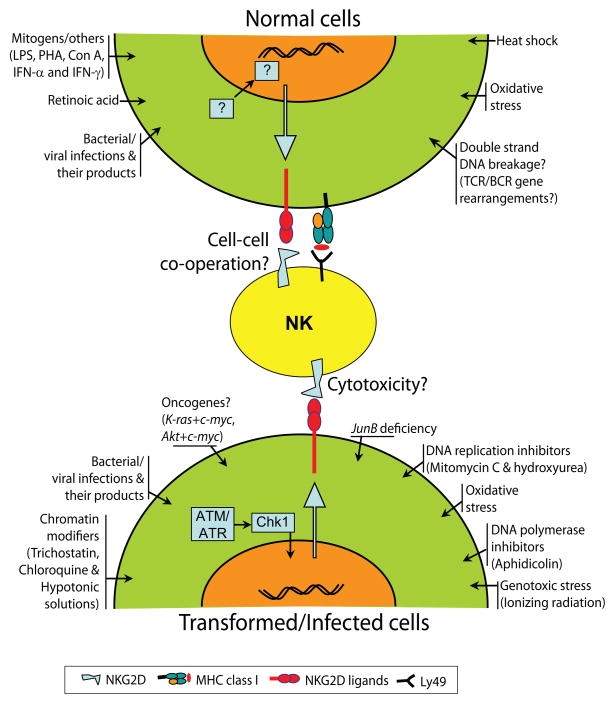
Recent studies have demonstrated that multiple cellular stresses could activate the expression of NKG2D ligands. Therefore, a tightly controlled regulation of NKG2D ligand expression is essential for preventing NK cell recognition of normal cells and their adverse effector functions. In Figure 6, we have summarized multiple stimuli that have the ability to induce the expression of NKG2D ligands in normal and transformed cells. Transcription of human MIC-A and MIC-B can be induced by heat shock; however, the requirements for the expression of ULBPs are not yet clear(Groh et al., 1996). LPS-mediated activation through TLR4 has been shown to induce the transcription of Rae-1, but not H60a or Mult1(Hamerman et al., 2004). More interestinlgly, recent studies from Raulet’s laboratory has made a observations that directly connected the DNA damage caused by stress can lead to the expression of ligands of NKG2D(Gasser et al., 2005). In their studies, the ligands for NKG2D were upregulated by genotoxic stress mediated by ionizing radiation, inhibitors of DNA replication such as mitomycin C, hydroxyurea, and the DNA polymerase inhibitor aphidicolin or chromatin-modifying agents (trichostatin A, chloroquine and hypotonic conditions). DNA damage pathway that is initiated by ataxia telangiectasia mutated (ATM) or ataxia telegiectasia Rad3-related (ATR) kinases has been implicated in the expression of murine NKG2D ligands(Gasser et al., 2005). Moreover, inhibition of ATM, ATR or their downstream mediator, Checkpoint kinase-1 (Chk-1) prevented the expression of NKG2D ligands, confirming the role of this DNA damage pathway. In the same study, authors has excluded the role of antother important member of the DNA damage pathway, tumor suppressor p53; since its absence failed to augment the expression of NKG2D ligands.
Figure 6. Multiple stress stimuli can induce the expression of NKG2D ligands.
Multiple cellular and biochemical events have been shown to induce the expression of murine NKG2D ligands in normal, transformed and infected cells. A comprehensive list of these stimuli is shown. DNA damage pathway that involves ATM/ATR and Chk-1 has been shown to transcriptionally regulate the expression of NKG2D ligands.
Although critical immunological aspects of these ligands are still lacking, the modulatory roles of viral infections on these ligands that leads to viral immune evasions are emerging. In this context, recent studies have demonstrated that the expression of H60, Rae-1 and Mult1 are differentially affected during murine cytomegalovirus (MCMV) infections. For example, MCMV induces the expression of H60b but not H60a or H60c(Takada et al., 2008). Also, it has been shown that gp40 from MCMV can bind to all the five isoforms of Rae-1 and down regulate them(Lodoen et al., 2003b). This down regulation resulted in the inability of the NK cells to recognize the virally infected cells through its NKG2D receptor. On the other hand, another MCMV-derived protein gp48 has been shown to affect the expression of H60a and thereby the effector functions of NK cells(Hasan et al., 2005). Other studies have shown that Mult1 expression and function can be affected by MCMV gene m145-derived glycoprotein. This gene product was sufficient to prevent the transport of Mult1(Krmpotic et al., 2005;Lenac et al., 2006).
Concluding Remarks
NKG2D plays a significant role as an activating receptor for NK, NKT and γδ-T cells and co-stimulating for αβ-T. Tremendous progress has been made in identifying and characterizing multiple ligands for NKG2D receptor. These ligands differ in their genomic organization, amino acid sequence, affinity to NKG2D and more importantly in their expression patterns. A rational explanation for the existence of multiple ligands for a single activating receptor, NKG2D, is still lacking. Future studies should focus on the role of these ligands in regulating the immunological functions of normal cells that have the ability to express them. A better understanding of their cell and tissue type expression patterns is critical for utilizing them as molecular targets for NK cell-mediated immunotherapy to control variety of malignancies.
Acknowledgments
SM is a recipient of ASBMT Young Investigator Award. This work was supported in part by ACS Scholar grant RSG-02-172-LIB; ROTRF grant # 111662730; and NIH grants R01 A1064826-01, U19 AI062627-01, NO1-HHSN26600500032C to S.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- Bauer S, Willie ST, Spies T, Strong RK. Expression, purification, crystallization and crystallographic characterization of the human MHC class I related protein MICA. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 3):451–453. doi: 10.1107/s0907444997015229. [DOI] [PubMed] [Google Scholar]
- Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines 31. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–2194. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J Immunol. 2002a;169:4079–4083. doi: 10.4049/jimmunol.169.8.4079. [DOI] [PubMed] [Google Scholar]
- Carayannopoulos LN, Naidenko OV, Kinder J, Ho EL, Fremont DH, Yokoyama WM. Ligands for murine NKG2D display heterogeneous binding behavior. Eur J Immunol. 2002b;32:597–605. doi: 10.1002/1521-4141(200203)32:3<597::aid-immu597>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- Carayannopoulos LN, Naidenko OV, Kinder J, Ho EL, Fremont DH, Yokoyama WM. Ligands for murine NKG2D display heterogeneous binding behavior. Eur J Immunol. 2002c;32:597–605. doi: 10.1002/1521-4141(200203)32:3<597::aid-immu597>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Hsia JK, Hsiung MY, Raulet DH. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur J Immunol. 2003;33:381–391. doi: 10.1002/immu.200310012. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- Farag SS, VanDeusen JB, Fehniger TA, Caligiuri MA. Biology and clinical impact of human natural killer cells 1. Int J Hematol. 2003;78:7–17. doi: 10.1007/BF02983234. [DOI] [PubMed] [Google Scholar]
- Furano AV, Duvernell DD, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005 doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, Hobby P, Sutton B, Tigelaar RE, Hayday AC. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294:605–609. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci U S A. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- Hamerman JA, Ogasawara K, Lanier LL. Cutting edge: Toll-like receptor signaling in macrophages induces ligands for the NKG2D receptor. J Immunol. 2004;172:2001–2005. doi: 10.4049/jimmunol.172.4.2001. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Hasan M, Krmpotic A, Ruzsics Z, Bubic I, Lenac T, Halenius A, Loewendorf A, Messerle M, Hengel H, Jonjic S, Koszinowski UH. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J Virol. 2005;79:2920–2930. doi: 10.1128/JVI.79.5.2920-2930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. 2002a;55:221–228. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- Karre K. NK cells, MHC class I molecules and the missing self 7. Scand J Immunol. 2002b;55:221–228. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy 115. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte AK, Gebhardt FE, Porcellini S, Schiemann M, Stemberger C, Franz TJ, Huster KM, Carayannopoulos LN, Yokoyama WM, Colonna M, Siccardi AG, Bauer S, Busch DH. NKG2D-independent suppression of T cell proliferation by H60 and MICA. Proc Natl Acad Sci U S A. 2005;102:11805–11810. doi: 10.1073/pnas.0502026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krmpotic A, Hasan M, Loewendorf A, Saulig T, Halenius A, Lenac T, Polic B, Bubic I, Kriegeskorte A, Pernjak-Pugel E, Messerle M, Hengel H, Busch DH, Koszinowski UH, Jonjic S. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J Exp Med. 2005;201:211–220. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenac T, Budt M, Arapovic J, Hasan M, Zimmermann A, Simic H, Krmpotic A, Messerle M, Ruzsics Z, Koszinowski UH, Hengel H, Jonjic S. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J Exp Med. 2006;203:1843–1850. doi: 10.1084/jem.20060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, George T, Devora GA, Sivakumar PV, Davenport C, Lai WC, Schatzle J, Kumar V, Bennett M. Murine natural killer cells and hybrid resistance to hemopoietic cells in vivo 4. Methods Mol Biol. 2000;121:61–71. doi: 10.1385/1-59259-044-6:61. [DOI] [PubMed] [Google Scholar]
- Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003a;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003b;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkannan S, Shih PP, Eden PA, Horng T, Zuberi AR, Christianson G, Roopenian D, Shastri N. The molecular and functional characterization of a dominant minor H antigen, H60. J Immunol. 1998;161:3501–3509. [PubMed] [Google Scholar]
- Moretta L, Biassoni R, Bottino C, Mingari MC, Moretta A. Immunobiology of human NK cells. Transplant Proc. 2001;33:60–61. doi: 10.1016/s0041-1345(00)02790-1. [DOI] [PubMed] [Google Scholar]
- Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123–128. doi: 10.1038/84219. [DOI] [PubMed] [Google Scholar]
- Nomura M, Zou Z, Joh T, Takihara Y, Matsuda Y, Shimada K. Genomic structures and characterization of Rae1 family members encoding GPI-anchored cell surface proteins and expressed predominantly in embryonic mouse brain. J Biochem. 1996;120:987–995. doi: 10.1093/oxfordjournals.jbchem.a021517. [DOI] [PubMed] [Google Scholar]
- O’Callaghan CA, Cerwenka A, Willcox BE, Lanier LL, Bjorkman PJ. Molecular competition for NKG2D: H60 and RAE1 compete unequally for NKG2D with dominance of H60. Immunity. 2001;15:201–211. doi: 10.1016/s1074-7613(01)00187-x. [DOI] [PubMed] [Google Scholar]
- Regunathan J, Chen Y, Wang D, Malarkannan S. NKG2D receptor-mediated NK cell function is regulated by inhibitory Ly49 receptors. Blood. 2005;105:233–240. doi: 10.1182/blood-2004-03-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MJ, Swann J, Kelly JM, Cretney E, Yokoyama WM, Diefenbach A, Sayers TJ, Hayakawa Y. NKG2D recognition and perforin effector function mediate effective cytokine immunotherapy of cancer. J Exp Med. 2004;200:1325–1335. doi: 10.1084/jem.20041522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland CL, Chalupny NJ, Cosman D. The UL16-binding proteins, a novel family of MHC class I-related ligands for NKG2D, activate natural killer cell functions. Immunol Rev. 2001;181:185–192. doi: 10.1034/j.1600-065x.2001.1810115.x. [DOI] [PubMed] [Google Scholar]
- Takada A, Yoshida S, Kajikawa M, Miyatake Y, Tomaru U, Sakai M, Chiba H, Maenaka K, Kohda D, Fugo K, Kasahara M. Two novel NKG2D ligands of the mouse H60 family with differential expression patterns and binding affinities to NKG2D. J Immunol. 2008;180:1678–1685. doi: 10.4049/jimmunol.180.3.1678. [DOI] [PubMed] [Google Scholar]
- Vilarinho S, Ogasawara K, Nishimura S, Lanier LL, Baron JL. Blockade of NKG2D on NKT cells prevents hepatitis and the acute immune response to hepatitis B virus. Proc Natl Acad Sci U S A. 2007;104:18187–18192. doi: 10.1073/pnas.0708968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cherwinski H, Spies T, Phillips JH, Lanier LL. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J Exp Med. 2000;192:1059–1068. doi: 10.1084/jem.192.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, Phillips JH. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zou Z, Nomura M, Takihara Y, Yasunaga T, Shimada K. Isolation and characterization of retinoic acid-inducible cDNA clones in F9 cells: a novel cDNA family encodes cell surface proteins sharing partial homology with MHC class I molecules. J Biochem. 1996;119:319–328. doi: 10.1093/oxfordjournals.jbchem.a021242. [DOI] [PubMed] [Google Scholar]