Abstract
The pace has quickened in circadian biology research. In particular, an abundance of results focused on post-translational modifications (PTMs) is sharpening our view of circadian molecular clockworks. PTMs affect nearly all aspects of clock biology; in some cases they are essential for clock function and in others, they provide layers of regulatory fine-tuning. Our goal is to review recent advances in clock PTMs, help make sense of emerging themes, and spotlight intriguing and perhaps controversial new findings. We focus on PTMs affecting the core functions of eukaryotic clocks, in particular the functionally related oscillators in Neurospora crassa, Drosophila melanogaster, and mammalian cells.
Post-translational modifications emerge on the scene
As our understanding of eukaryotic clocks has shifted focus from organismal behavior to molecular underpinnings, in the broadest description these clocks consist of a positively acting driver that is periodically suppressed by an inhibitory brake (Fig 1). More specifically, heterodimeric positively-acting PAS (named for Per-ARNT-Sim motifs) domain-containing transcription factors (+TFs) activate the transcription of negatively acting factors (−Fs). These, in turn, feed back to inhibit the activity of the +TFs for long periods of time, thus setting the phase of transcription and determining the length of the feedback cycle.1–3
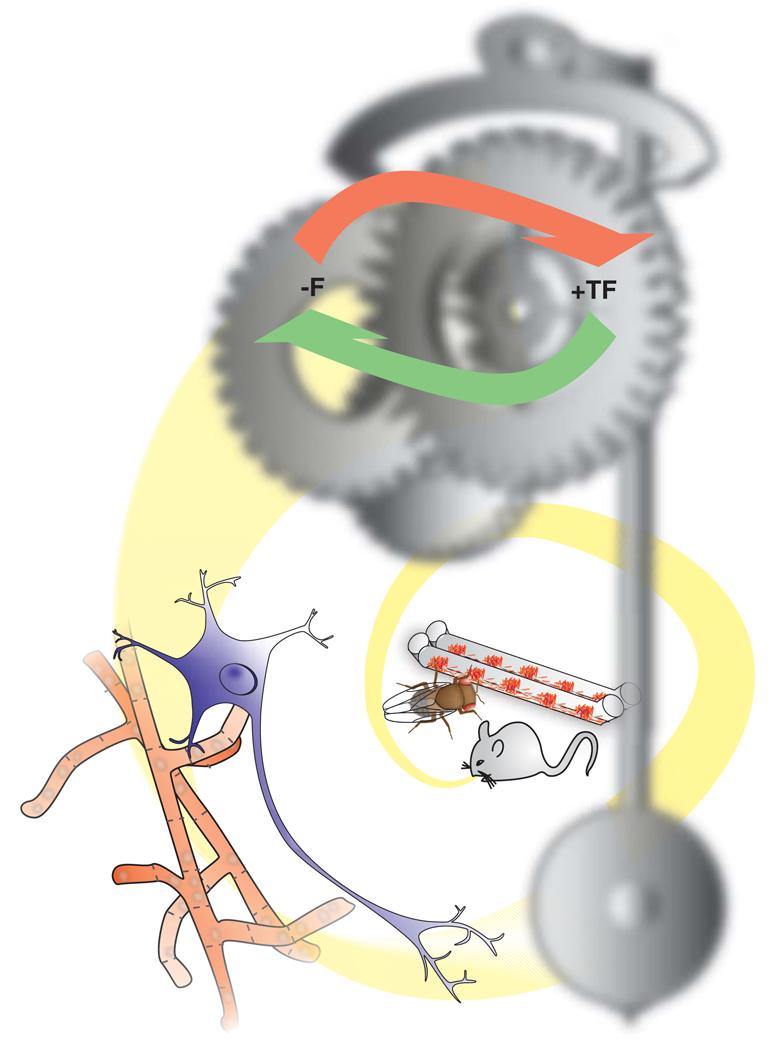
Figure 1. Circadian clocks from organism to mechanism.
Over the last several decades our picture of circadian clocks has spiraled out from a physiological description of clock-controlled activities at the organismal level, through an understanding of clocks as being bound within single cells, to revealing a molecular mechanism that is predominantly constrained within the interplay between positive (+TF) and negative (−F) forces. A clearer picture of the clockworks of these systems is beginning to emerge, showing that many of the processes and components are controlled through post-translational modification.
This core circadian circuit is broadly controlled by post-translational modifications (PTMs). Indeed, recent evidence points to a clock based entirely on PTMs in cyanobacteria (Box 1), although this review does not focus on this system. What did we know about PTMs within these circuits circa 2004? Phosphorylation of −Fs was typical, rhythmic, and phase specific4, 5 and appeared to be the major contribution to determining the very long (circa 24 h) time constant of the feedback loop6. More nuanced studies showed that the activity state of +TFs that shuttled between cytoplasm and nucleus7 was accompanied by changes in histone acetylation8 and chromatin remodeling9, and there were hints that phosphorylation of +TFs10 increased their transcriptional activity11 and targeted them for degradation12. −Fs were known to dimerize and enter the nucleus13–16 and the timing and control of this event was a regulated process17, 18. Importantly, −Fs were known to bind and inhibit +TFs10, 16, 19. Several reports indicated that the joint activity of kinases and phosphatases (some of whose specific roles in the clock are functionally conserved across phyla, e.g., casein kinase 2 (CK2)20–22, casein kinase 1 (CK1)23–25 and protein phosphatase 2A (PP2A)26, 27) caused net progressive phosphorylation of these −Fs4, 5 ultimately leading to their ubiquitylation and proteasome-dependent degradation28–31. These relatively slow processes of −F maturation and degradation were critical for creating a sufficiently long delay required to support a 24 h rhythm. Finally, some clues suggested that not every example of increased phosphorylation of −Fs necessarily resulted in faster degradation and period reduction, e.g. hypophosphorylation of hPER2 (human period homolog 2 (Drosophila)) mutants by CK1 elicited shorter periods32. In another case, kinases were observed to be potentiators of −Fs' inhibitory activity33.
Box 1. An oscillator based only on PTMs in cyanobacteria
An outstanding case study for the importance of PTMs in circadian clocks (albeit one that beyond the scope of this review) comes from recent work by Kondo and co-workers on the in vitro KaiC oscillator (for a topical review see [96]). Briefly, when three cyanobacterial proteins (KaiA, KaiB and KaiC) are incubated in vitro in the presence of ATP, a cycle of phosphorylation is observed on KaiC. This rhythmic cycle of KaiC phosphorylation does not require any new production or destruction of protein and exhibits all the hallmarks of a true circadian rhythm. Although the cyanobacterial clock has evolved independently, this exciting finding has raised interest in the possibility that non-bacterial oscillatory proteins might also be able to support rhythms based solely on PTMs.
An ever-increasing number of PTMs are rapidly becoming implicated in clock control. Given the diversity of experimental systems and variety of PTMs employed by clocks, here we strive to indicate how PTMs are used in common across phyla often acting on related molecules (Table 1), and to highlight occasional discrepancies in the field and real differences in clock regulation. PTMs contribute essential clock functions that are layered atop the canonical transcriptionally-controlled feedback oscillator and thus are an important and topical area of study.
Table 1.
Functional classes of clock proteins across phylaa.
| Class | Ascomycota (Neurospora) |
Arthropoda (Drosophila) |
Chordata (Mus, Homo) |
|---|---|---|---|
| WC-1 | CYC | BMAL1 | |
| +TFs | WC-2 | ||
| CLK | CLOCK | ||
| FRQ | |||
| −Fs | PER | PER 1,2,3 | |
| TIM | |||
| CRY | |||
| CK-1a | DBT | CK1δ/ε | |
| CKA | CK2 | CK2 | |
| PKA | PKA | ||
| Kinases | Shaggy | ||
| PRD-4 | Chk2 | ||
| Chk1 | |||
| ATM | |||
| Phosphatases | PP2A | PP2A | PP2A |
| PP1 | PP1 | ||
| HATs, HDACs | CLOCK SIRT1 |
||
| FWD-1 | SLIMB | β-TRCP | |
| F-boxes | JETLAG | ||
| FBXL3 | |||
Proteins implicated in the operation or resetting of core circadian oscillators from three prominent model organisms are listed. Proteins in line with one another share both sequence/domain structure and serve orthologous functions.
An expanding view of the positive module
Recent data point to a direct link, mediated by +TF PTM activity, between chromatin remodeling and +TF transcriptional activity34. Mouse CLOCK (circadian locomoter output cycles kaput) has histone acetyltransferase (HAT) activity that is required for rhythmic expression of core clock and output genes35. CLOCK HAT activity selectively affects specific targets on histone subunits; however, CLOCK can also acetylate its partner BMAL (brain and muscle aryl hydrocarbon receptor nuclear translocator-(ARNT)-like)36. BMAL acetylation at Lys537 is rhythmic in liver extracts, is essential for clock function and, most interestingly, appears to be required to inactivate BMAL transcriptional activity. Further experimentation is required to determine if +TFs functionally analogous to CLOCK (Drosophila melanogaster CLK and Neurospora crassa WHITE COLLAR-2; WC-2) play similar enzymatic roles; to date there is no evidence that components of the WHITE COLLAR COMPLEX (WCC) are acetylated.
As clock components are acetylated, so too they must be deacetylated. One group has reported that sirtuin1 ((silent mating type information regulation 2, homolog) 1 (S. cerevisiae), SIRT1), an NAD+-dependent deacetylase, has rhythmic deacetylase activity, directly interacts with CLOCK and is required for the rhythmic acetylation pattern of BMAL-Lys537 in cells and in vivo37. But the story is complex: another group shows rhythmic binding, but not activity, of SIRT1 to BMAL–CLOCK and SIRT1-dependent oscillatory PER2 deacetylation38. Interestingly, PER2 acetylation increases its stability. Resolution of these, perhaps not mutually exclusive, observations will require further experimentation. Given the involvement of SIRT1 in metabolic programs, the intriguing new connection between energy metabolism, NAD+ levels and rhythmicity provides ample room for research.
In addition to phosphorylation and acetylation, at least one +TF is modified by the small ubiquitin-like modifier (SUMO). mBMAL1 is rhythmically SUMOylated on a highly conserved lysine residue (K259) in a CLOCK-dependent manner39. In tissue culture cells, BMAL is predominantly polySUMOylated, and this modification leads to ubiquitylation, localization to subnuclear foci and enhanced transcriptional activity40.
An exploding view of the negative module
The negative module is subject to a variety of PTMs that center around the activity of kinases and phosphatases and the array of players is expanding. Together, the net effect of these effectors is to determine the half-life of −F factors through ubiquitin-mediated degradation and to influence period length. Emerging data have consolidated and extended this view. However, not all −F phosphorylation events lead to degradation and we also review non-degradative effects of phosphorylation.
Kinases and phosphatases, old and new
CK1 is the grandfather of circadian-associated kinases23, 41, and as the relationships between CK1 and its −F targets become better understood, additional complexity is being uncovered. In Drosophila, overexpression of dominant negative CK1 (DOUBLETIME; DBT) results in a long period or arrhythmicity, depending on the antimorph dose42. Although this experiment suggests a direct relationship between CK1 dose/activity and period length (i.e., more CK1 activity speeds up the clock), other analyses in flies and mice complicate this concept by the use of mutant alleles that could be hypomorphs, hypermorphs or even mixomorphs. For example, the dbtL and dbtS alleles have opposite period phenotypes in flies but, curiously, both result in decreased CK1 activity in vitro43.
Similar complexities that are currently only partially resolved include two mutations affecting CK1 in mammals, both of which initially appeared to contradict the dogma that increased phosphorylation of −Fs leads to shorter periods. First, although the short period allele CK1ε (tau), was initially shown to be a hypomorph in in vitro assays24, two groups have recently shown that this mutation actually increases in vivo mPER2 phosphorylation, increasing its turnover 44, 45; they argue that the change in kinase activity deriving from this mutation could be substrate-dependent. It is also unclear whether the change in mPER2 phosphorylation is a direct or indirect effect of the altered kinase; thus, it remains an open question whether CK1ε (tau) is indeed a target-specific hypermorph, or alternatively, if the phenotype stems from a change in mPER2 localization46.
A second example involves a hypomorphic version of CK1∂ caused by a T44A amino acid substitution. This mutant causes familial advanced sleep phase syndrome (FASPS)47 and period shortening in mammals. However, transgenic flies which express the mutant kinase did not share the mammalian phenotype; instead the period lengthens. The underlying cause for this dissimilarity is an open question but likely involves differences in regulation between flies and mammals despite conserved clock architectures—one key difference is the role of TIMELESS (TIM) in Drosophila which is not paralleled in mammals. Recent work48 adds to the puzzle: mutations that lower either Drosophila DBT and vertebrate CK1∂ kinase activity led to shortened rhythms. Therefore not all reduced-activity CK1 enzymes elicit opposite effects in mammals and flies, and indeed, functional conservation exists between these homologous enzymes. Overall, reduced kinase activity cannot simply explain the decreased period length, and so further analysis will be required; in the latter study, the authors suggest that changes in site-specific phosphorylation or interaction with a regulator might be responsible for period shortening in the context of reduced kinase activity and this perhaps represents a general solution to the conundrum.
The situation is currently more clear-cut with respect to CK2 and its role in fly clocks. In vitro kinase assays performed using a dPER deletion series implicate key serines which when altered in vivo, lead to period-lengthening49. Although some of these residues might be bona fide CK2 phosphosites, conclusions derived solely from consensus sequence-based approaches or in vitro patterns are fraught with caveats (for example see ref. 50). In addition, overexpression of a dominant negative dCK2 in circadian neurons leads to dramatic period lengthening51; this result helps to distinguish direct from pleiotropic effects of this multifunctional enzyme.
A pressing question about CK1 and CK2 is whether the activity of these kinases is rhythmic. A variety of studies in many organisms have shown that bulk CK1 and CK2 levels do not cycle, as would be expected for such utility kinases. However, in Drosophila, rhythmic aspects of phosphatase activity–regulatory PP2A subunits are expressed in a circadian fashion—provide a nuance to this question26. A finding in Arabidopsis thaliana, on the other hand, could portend possibilities for organisms with multiple CK2-beta subunits: proteasome-dependent degradation of the CKB4 isoform (a newly described fourth beta subunit) is rhythmic52, perhaps indicating that some aspects of kinase regulation are indeed rhythmic.
These veteran kinases have been joined by some clock rookies, the cAMP dependent protein kinase A (PKA) and the DNA damage/checkpoint associated kinases Chk1, Chk2, and ATM (ataxia telangiectasia, mutated). Two independent approaches have implicated PKA in circadian function. Overexpression of the PKA catalytic subunit causes induction of hPER1 expression, and this induction is CREB (cAMP responsive element binding protein) and CCAAT-box binding protein (C/EBP) dependent, suggesting that cAMP-induced CREB and C/EBP are ‘peripheral’ modulators of hPER153. By contrast, Neurospora PKA has also been placed more directly in two parts of the loop: it is a priming kinase for the casein kinase action on the WCC and it stabilizes both WCC and FREQUENCY (FRQ)54. PKA physically associates with WCC and can phosphorylate both WCC and FRQ in vitro. Moreover, strains with reduced PKA activity show increased binding of the WCC to the frq promoter. On first blush, these two studies appear to contradict each other—if PKA inhibits +TF DNA binding, why should overexpression of such a transcriptional inhibitor of CLOCK–BMAL lead to increased hPER1 expression? However, if PKA indeed works at multiple levels, i.e., transcriptional repression and post-translational stabilization, effects of overexpression could be tricky to predict a priori.
Although Chk2 null mutations have not revealed any clock defects under standard conditions, this and perhaps related kinases provide conditional input to clock feedback loops. Indeed, their contribution likely would still be unknown had the co-dominant hypermorphic Chk2 allele, prd-4, not been found to confer a short circadian period in Neurospora55. Molecular analyses revealed direct binding between PRD-4 and the −F FRQ; moreover, DNA damage-dependent FRQ phosphorylation triggers its destabilization—a kinase-based clock input conditional upon DNA damage. It is now clear that this response is widely conserved: Chk1 associates with hPER1 following the irradiation of cells56 and DNA damage results in PER1 phosphorylation57. Moreover, gamma irradiation has been shown to reset the clock of mice and human cell lines in an ATM-dependent manner (see ref. 58 and also shown in MEF cells with both UV or MMS-induced damage59), as predicted from the Neurospora work. Moreover, other clock components, including NPAS2 (neuronal PAS domain protein 2), also appear to be involved in DNA damage responses60.
In addition to the several known kinases, one new player from the universe of phosphatases, protein phosphatase 1 (PP1) has joined the team. Earlier evidence from Neurospora showed that PP1 reduction caused advanced phase and a slightly shorter period27. In mammals, PP1 binds and stabilizes one of the −Fs by dephosphorylation61. Overexpression of PP1 inhibitors and dominant negative forms of PP1 support this conclusion. By contrast, using genetic analysis, PP1 activity appears to decrease period length in flies62. Once again, one possibility is that these apparently contradictory results are reconciled by the presence of the −F binding factor TIM and its binding to PER, which does not appear to play a clock-specific role in mammals.
We expect to continue finding new connections between kinases and phosphatases and the clock. It is becoming clear that the relationships between phosphorylation and degradation of −Fs or activity of +TFs are not as simple as we had initially imagined. Moreover, with a given kinase/phosphatase connected on multiple levels to clock components (as in the case of PKA, which acts upon both +TFs and −Fs), untangling the consequences of simple over- or underexpression will not be trivial. In fact, interconnected relationships might have evolved as an ensemble to encode aspects of post-translational rhythmicity. Along these lines, older evidence indicates that rhythmic cycling of dper and dtim mRNA are dispensable for rhythms63, 64. A recent result consistent with this finding is that PER2-LUC protein from constitutively expressed mPER2-luc, in rat-1 cells, might oscillate only due to PTMs65.
Ubiquitin and the F-box
As mentioned above, a key consequence of progressive phosphorylation by clock-associated kinases is the regulated destruction of their targets, typically −Fs. Seminal findings about degradation in Arabidopsis66 have been recapitulated in Neurospora31. That F-box mediated degradation of −Fs occurs was in itself not surprising; what was perhaps genuinely surprising is that although there are many F-box containing proteins (≅70 in mammals, dozens in flies and a handful in Neurospora), the clock-associated F-box proteins, SUPERNUMERARY LIMBS (SLIMB) in Drosophila29, 30, FWD-1 (F-box and WD-40 repeat-containing protein 1) in Neurospora31, and β-TrCP (beta-transducin repeat containing protein) in mammals67, are all orthologs. Taken together, these results suggest that the choice of a given F-box protein for −F degradation is unlikely to have occurred through convergent evolution. Having said this, it is important to point out that there appears to be a unique aspect to SLIMB regulation in flies. In contrast to other systems, overexpression of SLIMB in the cytoplasm leads to period lengthening29. The authors propose a model in which the SLIMB target, PER, is unable to accumulate to requisite critical levels and is therefore delayed in nuclear translocation.
A novel observation in this context is that phosphorylation of a key dPER phosphosite leads to SLIMB binding in flies. This site, within the first 100 aa of dPER, is rhythmically phosphorylated, leading to F-box binding and rapid degradation68. This result suggests that phosphorylation of specific, critical sites is more important for F-box binding than simple accumulation of electrostatic charge and appears different from another F-box binding design in yeast cell-cycle control69. Moreover, the requirement for phosphorylation on several sites suggests that most phosphorylation on dPER has non-degradative consequences.
Another emerging complexity is that different F-box proteins impact the clock at different levels. For example, in Drosophila, JETLAG resets the clock by causing light-induced TIM degradation70. Mammalian FBXL3 (F-box and leucine-rich repeat protein 3), mediates destruction of an accessory mammalian −F, CRY (cryptochrome)71–73. It will be of interest to determine if these additional F-box protein functions are conserved across species, as observed for the SLIMB homologs.
Non-degradative consequences of −F phosphorylation
In addition to the aforementioned examples of regulated nuclear entry and −F potentiation of negative arm function, there is a growing number of examples of −F phosphorylation leading to effects other than degradation. These include new instances of −F subcellular localization and potency control as well as novel regulation of +TFs and new insight into the regulation of progressive −F phosphorylation.
More evidence regarding −F localization and potency has come from studies in the fly. First, as in mammals, DBT keeps dPER in the cytoplasm74 but interestingly, another kinase, Shaggy, acts to oppose DBT by accelerating PER nuclear accumulation in a TIM-dependent manner. Second, certain dPER phosphosite mutants can lead to short period75—failure to phosphorylate these sites causes dPER to become a potent repressor of dCLK.
Next is the intriguing example of the hPER2S662G variant which leads to FASPS (see above). A decrease in protein stability resulting from changes in the analogous mPER2 residue (S659) was explained by premature nuclear clearance and increased sensitivity to CK1-dependent degradation46. A year later, a different non-degradative explanation emerged when the same mutation, now in hPer2, was introduced into mice. hPER2S662G in mice was associated with decreases in Per2 mRNA levels, leading to the suggestion that phosphorylation of S662, the pertinent residue, decreases the ability of mPER2 to inhibit its own message76.
In Neurospora, phosphorylation of certain key serines in the −F FRQ appears to be required for post-translational up-regulation of +TFs77. When key phosphosites are ablated, FRQ cannot mediate WCC formation and what results resembles a dampened oscillator.
Finally, accumulating evidence suggests that −Fs might harbor ‘clusters’ of sites responsible for distinct functions e.g., stabilization, degradation, nuclear residency and potency of transcriptional inhibition68. Some authors have speculated that clustered phosphorylation within −Fs leads to sequential unfolding and/or changes in the spectrum of interacting partners75. Moreover, a recent study employed heavy isotope labeling and quantitative proteomics to follow the appearance and disappearance of over 75 clustered phosphorylation sites on FRQ78; as seen with dPER, some domains appear to promote turnover and some influence intra- or inter- protein interactions and their loss through mutation can accelerate or decelerate the pace of the clock. Finally, a subset of serines on FRQ appear to facilitate degradation specifically at higher temperatures—thus influencing temperature compensation 79: another case in point that heterogeneous phosphorylation can mediate multiple consequences. Together, these findings are consistent with roles for −Fs as integrators for a variety of cellular signals pertinent to clock function. It will be interesting to see if clustered phosphorylation is a conserved property of −Fs and whether this or some other mechanism underpins progressive phosphorylation. Overall, phosphorylation of −Fs can lead to a variety of non-degradative effects.
Mechanistic details of negative feedback
The original model for circadian negative feedback was that −Fs simply bound +TFs and thereby inactivated them; this view is now being expanded and modified. Specific details of −F action on +TFs as well as information about temporal regulation of +TF promoter occupancy are beginning to take shape.
The consequences of −Fs binding to +TFs
Studies in mammals have recently joined the fold, lending further support to the idea that feedback repression is a key mechanism of clock regulation80. The first suggestion that the model required updating came from a study in Neurospora in which steady-state estimates indicated there was insufficient FRQ to inactivate the WCC in a stoichiometric manner; indeed, WCC phosphorylation81 was associated with decreased DNA binding82. This led to a model in which instead of constitutively binding to WCC, rather FRQ interacts with the WCC transiently and serves as a scaffold for CK1 and CK2 which in turn act enzymatically on the WCC54, 83, 84. More recent data, however, indicate that the feedback loop is closed through dynamic hypophosphorylated-FRQ-mediated export of WCC from the nucleus85. A similar picture has begun to emerge from studies in flies where CK1 (DBT) constitutively binds dPER and phosphorylates dCLK in a dPER-dependent manner86–89. Moreover, DBT is not capable of directly phosphorylating an N-terminal portion of CLK in vitro in the absence of dPER75. As with Neurospora FRQ, a key region on dPER is required for DBT binding; when this region is altered, dPER still binds +TFs but cannot inhibit their activity88, consistent with a scaffold model where dPER connects kinases with +TFs. In a surprising twist, in addition to its kinase activity, DBT might also act as a scaffold for another kinase as DBT catalytic activity appears unnecessary for some dCLK phosphorylation90.
Dynamics of +TF promoter occupancy
Finally, our view of +TFs as being static is changing; instead, we are now beginning to appreciate their dynamic nature. Though not unambiguously so, some evidence indicates that +TFs bind their promoters rhythmically. Such behavior has been shown in mammals34, but other studies10, 36 point to constitutive promoter binding. In flies, DBT-mediated dCLK phosphorylation leads to changes in its binding to E-box-containing promoters86, thereby eliciting changes in chromatin structure91. Neurospora appears to use both strategies; one of the two WCC members, WC-2, binds rhythmically to the frq promoter whereas the other, WC-1, binds constitutively92.
In addition to their DNA binding capacity, the subcellular localization of +TFs is also highly dynamic and likely influenced by PTMs. BMAL1 phosphorylation status is reported to affect its nucleocytoplasmic shuttling, transactivation ability, and turnover93. Likewise, dCLK accumulates in the cytoplasm following DBT-meditated phosphorylation87, and WCC nuclear export is promoted by hypophosphorylated FRQ85, 94. Moreover, active destruction of +TFs appears to provide a previously underappreciated layer of regulation95. Overall, an emerging consensus suggests that binding, localization, and stability of +TFs are dynamic—a situation different from that originally imagined.
Concluding remarks
Our picture of the clockworks and PTMs has been focused (from the simple picture in Fig 1 to the many facets in Fig 2) in a very short time span as appreciation of both the variety and functional significance of PTMs has grown. The clock wiring diagram clearly contains a growing number of post-translational control points and further discovery and refinement should be rapid and exciting.
Figure 2[SC1]. Vignettes of circadian post-translational modifications.
A number of clock PTMs are depicted in a simplified series of cartoons. Representative PTMs are shown and do not reflect the action in all species. For more detail, refer to the text. a) PAS-domain containing positively acting transcription factors (+TFs, green boxes) shuttle (bi-directional arrow) between the nucleus[SC2] on DNA and cytoplasm. b) DNA-bound +TFs activate transcription (bent arrow) of negatively acting factors (−Fs, red ovals). c) Kinases (blue circles) and phosphatases (orange circles) compete to establish a pattern of progressive phosphorylation (P) which can have multiple consequences including cytoplasmic retention of −Fs. d) −Fs transport kinases, which along with independent kinases (blue oval) phosphorylate (curved arrows) +TFs (and/or −Fs) via transient interactions (bi-directional arrow) leading to inactivation of +TFs and export from the nucleus (arrow). e) A mammalian +TF can act as a histone acetyltransferase (curved arrow, Ac; acetyl group) to inactivate its partner. f) In mammals, a deacetylase can remove inhibitory Ac from +TFs and −Fs. g) In mammals, +TFs can be sumoylated (Su) leading to their ubiquitylation (Ub) and degradation. h) Progressive phosphorylation (Ps) on −Fs leads to F-box binding (yellow box), ubiquitylation (Ub) and subsequent degradation. i) In Neurospora mature −F can promote +TF complex formation. j) Upon DNA damage (XXX), DNA-damage dependent kinases (blue hexagon) can phosphorylate −Fs accelerating their degradation. k) Multiple outcomes of −F degradation are depicted. Clusters of phosphosites can mediate different consequences including stabilization, potentiation of −F efficacy and degradation.
One aspect of PTMs that appears destined for more prominence is the role of rhythmic PTMs in nested feedback loops which surround the core that can lead both to stabilization of the core loop and to circadian output. Although the activities of utility kinases such as CK1 and CK2 appear not to be rhythmic, the deacetylase SIRT1 is reported to be clock-controlled and to have a pronounced effect on circadian output 37, 38. This could be the first example of an emerging theme.
Another question that will perhaps dominate our thinking in the near future concerns determining which regulations are ‘core’ controls and which are simply consequences of indirect cascades of secondary effects. For instance, just as synthetic biology has succeeded in assembling components that can oscillate, albeit in a less than fully robust manner, it is clear that evolution can cobble together connections in a seemingly random manner and that those that provide fitness have been maintained—as observed in the separate events which led to the three distinct clock mechanisms in the cyanobacterial (Box 1), plant and fungal/animal lineages. In this context it is perhaps not surprising that the networks are complex. Moreover, research to date, although fruitful, has been not unlike hunting and gathering where each factoid regarding a modifier or modification has been cherished. Looking forward, we can envision the use of genomic and proteomic tools to describe entire clock-interactomes and temporal patterns of PTMs of all clock-relevant molecules.
Acknowledgments
Thanks to R. Lambreghts, M. Shi and H.V. Colot for critical reading of the manuscript. A.M. was supported by a CIHR Postdoc Fellowship. NIH Grants GM34985, GM08336, GM068087 supported work in our laboratory.
Glossary
- antimorph
typically dominant mutations that oppose the wild-type gene action
- ataxia telangiectasia
a rare neurodegenerative disease causing ataxia—coordination
- defects and telangiectasia
small blood vessels
- circadian rhythm
a cycle of length approximately 24 h,from the Latin circa (about) and dies (day)
- negative feedback
a control process that loops back and self-limits
- hypermorph
a mutation that causes an increase in wild-type gene action
- hypomorph
a mutation that causes a partial loss-of-function
- mixomorph
a mutation that has mixed character e.g., it can act as a gain and loss-of-function depending on context
- period
the amount of time required to complete a cycle of activity; the inverse of frequency
- phase
any stereotypic, distinguishable and repeating single part of a cycle
- pleiotropy
the influence of a single gene on many phenotypes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dunlap JC, Loros JJ. How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol. 2006;9:579–587. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 4.Edery I, et al. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garceau NY, et al. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, et al. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci U S A. 2000;97:234–239. doi: 10.1073/pnas.97.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamaru T, et al. Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells. 2003;8:973–983. doi: 10.1046/j.1365-2443.2003.00686.x. [DOI] [PubMed] [Google Scholar]
- 8.Etchegaray JP, et al. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 9.Curtis AM, et al. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- 10.Lee C, et al. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 11.Eide EJ, et al. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase I epsilon. J Biol Chem. 2002;277:17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondratov RV, et al. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vosshall LB, et al. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 14.Luo C, et al. Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 1998;17:1228–1235. doi: 10.1093/emboj/17.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yagita K, et al. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng P, et al. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 2001;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vielhaber E, et al. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao S, et al. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci. 2001;21:7117–7126. doi: 10.1523/JNEUROSCI.21-18-07117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denault DL, et al. WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 2001;20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugano S, et al. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci U S A. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin JM, et al. A role for casein kinase 2 alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, et al. Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev. 2002;16:994–1006. doi: 10.1101/gad.965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kloss B, et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 24.Lowrey PL, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorl M, et al. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 2001;20:7074–7084. doi: 10.1093/emboj/20.24.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sathyanarayanan S, et al. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, et al. Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev. 2004;18:255–260. doi: 10.1101/gad.1152604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keesler GA, et al. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport. 2000;11:951–955. doi: 10.1097/00001756-200004070-00011. [DOI] [PubMed] [Google Scholar]
- 29.Grima B, et al. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- 30.Ko HW, et al. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 31.He Q, et al. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J. 2003;22:4421–4430. doi: 10.1093/emboj/cdg425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 33.Nawathean P, Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- 34.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 35.Doi M, et al. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 36.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 37.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 39.Cardone L, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;19:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price JL, et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 42.Muskus MJ, et al. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol. 2007;27:8049–8064. doi: 10.1128/MCB.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preuss F, et al. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallego M, et al. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A. 2006;103:10618–10623. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng QJ, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanselow K, et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 48.Fan JY, et al. Drosophila and vertebrate casein kinase Idelta exhibits evolutionary conservation of circadian function. Genetics. 2009;181:139–152. doi: 10.1534/genetics.108.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin JM. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J Neurosci. 2005;25:11175–11183. doi: 10.1523/JNEUROSCI.2159-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veluthambi K, Poovaiah BW. In vitro and in vivo protein phosphorylation in Avena sativa L. coleoptiles: effects of Ca2+, calmodulin antagonists, and auxin. Plant Physiol. 1986;81:836–841. doi: 10.1104/pp.81.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith EM, et al. Dominant-negative CK2alpha induces potent effects on circadian rhythmicity. PLoS Genet. 2008;4:e12. doi: 10.1371/journal.pgen.0040012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perales M, et al. The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J. 2006;46:849–860. doi: 10.1111/j.1365-313X.2006.02744.x. [DOI] [PubMed] [Google Scholar]
- 53.Motzkus D, et al. Activation of human period-1 by PKA or CLOCK/BMAL1 is conferred by separate signal transduction pathways. Chronobiol Int. 2007;24:783–792. doi: 10.1080/07420520701672481. [DOI] [PubMed] [Google Scholar]
- 54.Huang G, et al. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 2007;21:3283–3295. doi: 10.1101/gad.1610207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pregueiro AM, et al. The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science. 2006;313:644–649. doi: 10.1126/science.1121716. [DOI] [PubMed] [Google Scholar]
- 56.Gery S, et al. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 57.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 58.Oklejewicz M, et al. Phase resetting of the mammalian circadian clock by DNA damage. Curr Biol. 2008;18:286–291. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 59.Gamsby JJ, et al. A Phylogenetically Conserved DNA Damage Response Resets the Circadian Clock. J Biol Rhythms. 2009 doi: 10.1177/0748730409334748. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman AE, et al. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008;6:1461–1468. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallego M, et al. Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem J. 2006;399:169–175. doi: 10.1042/BJ20060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang Y, et al. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng Y, Hardin PE. Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J Neurosci. 1998;18:741–750. doi: 10.1523/JNEUROSCI.18-02-00741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Z, Sehgal A, et al. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 65.Nishii K, et al. Rhythmic post-transcriptional regulation of the circadian clock protein mPER2 in mammalian cells: a real-time analysis. Neurosci Lett. 2006;401:44–48. doi: 10.1016/j.neulet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 66.Somers DE, et al. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- 67.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiu JC, et al. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borg M, et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koh K, et al. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 73.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 74.Cyran SA, et al. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kivimae S, et al. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Y, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schafmeier T, et al. Phosphorylation-dependent maturation of Neurospora circadian clock protein from a nuclear repressor toward a cytoplasmic activator. Genes Dev. 2006;20:297–306. doi: 10.1101/gad.360906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baker CL, et al. Quantitative Proteomics Reveals a Dynamic Circadian Interactome and Phase-specific Phosphorylation in the Neurospora Circadian Clock. Mol Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mehra A, et al. A role for Casein Kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009;137:749–760. doi: 10.1016/j.cell.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sato TK, et al. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He Q, et al. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J Biol Chem. 2005;280:17526–17532. doi: 10.1074/jbc.M414010200. [DOI] [PubMed] [Google Scholar]
- 82.Schafmeier T, et al. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 83.He Q, et al. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Querfurth C, et al. Posttranslational regulation of Neurospora circadian clock by CK1a-dependent phosphorylation. Cold Spring Harb Symp Quant Biol. 2007;72:177–183. doi: 10.1101/sqb.2007.72.025. [DOI] [PubMed] [Google Scholar]
- 85.Hong CI, et al. Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev. 2008;22:3196–3204. doi: 10.1101/gad.1706908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu W, et al. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim EY, Edery I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim EY, et al. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nawathean P, et al. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu W, et al. DOUBLETIME plays a non-catalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor P, Hardin PE. Rhythmic E-box binding by CLK-CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol. 2008;28:4642–4652. doi: 10.1128/MCB.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belden WJ, et al. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 93.Kwon I, et al. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol. 2006;26:7318–7330. doi: 10.1128/MCB.00337-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cha J, et al. Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. EMBO J. 2008;27:3246–3255. doi: 10.1038/emboj.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schafmeier T, et al. Circadian activity and abundance rhythms of the Neurospora clock transcription factor WCC associated with rapid nucleo-cytoplasmic shuttling. Genes Dev. 2008;22:3397–3402. doi: 10.1101/gad.507408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson CH, Egli M, Stewart PL. Structural insights into a circadian oscillator. Science. 2008;322:697–701. doi: 10.1126/science.1150451. [DOI] [PMC free article] [PubMed] [Google Scholar]



![Figure 2[SC1]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/1c1b/2765057/0c477492baf7/nihms139004f2.jpg)