Many human cardiac diseases have a genetic cause. A large number of these occur in genes coding for the myofibrillar proteins (see [1-8] for reviews). The catalogue of mutations continues to expand. The most extensive observations have been on the beta myosins gene where over 200 different mutations have been found [9]. A wealth of structure-function information has been gleaned from studies of these altered forms.
Studies regarding mutation effects on the giant protein titin have lagged in comparison to the other myofibrillar proteins. It's 12 to 16 fold larger message size (relative to myosin) would suggest it to be a frequent mutation target. There are several reasons why so few titin mutations have been identified. First, the extreme size of titin makes conventional molecular biological approaches impossible. The protein is coded by a single gene in higher organisms [10], and there are 363 exons in the human version [11]. Sequencing a cDNA for full length titin is not possible with current techniques (no one has found ways to deal successfully with an intact 80,000 to 100,000 bp mRNA). Scanning this many exons for sequence variants has only recently been done because of time and expense. Second, there is a large diversity of titin protein forms. The gene is alternatively spliced with different isoform classes in heart and skeletal muscle [10]. The original descriptions identified unique regions near the N line in cardiac (N2B) and skeletal (N2A) that were associated with the tissue specificity. It was subsequently shown that cardiac muscle also contained isoforms with both the N2B unique and N2A unique sequences, and these have been termed N2BA. Further studies have demonstrated a plethora of sub-splicing pathways in the N2BA isoform class [11]. Splicing pathway heterogeneity is particularly diverse in the PEVK region where 10 different clones from a single PCR amplification of human heart cDNA yielded 10 different splicing patterns [12]. Third, much of the protein is believed to function as a structural chain, so unraveling a single link out of potentially 100 immunoglobulin (Ig) domains in the I-band region would change the length very little and have little effect on passive tension [13]. Even drastic changes in the number of exons expressed, as occurs in a recently described rat mutant, produce minimal changes in heart function [12, 14]. Thus changes in titin length alone may not result in serious health consequences. Finally, the understanding of the diverse functions of this protein remains incomplete (see [15-17] for reviews), so its relation to cardiac disease remains even less clear.
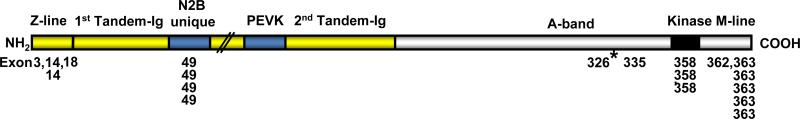
Several early reports linked cardiac disease to the chromosome 2q31 region of the titin gene [18, 19]. The precise locations have now been determined in each case, and a total of 20 different titin mutations have been identified to date (Table1). Of these 10 are located in the 6 most 3’ exons (358 to 363) that code the protein region near the M-line and 4 are found in exon 49 (N2B unique region). Four others occur in the Z-line region (see Figure 1 for a map of these sites to their approximate half sarcomere location). Undoubtedly many other mutations may have occurred with little or no functional consequences. Likewise mutations that do not cause chain termination in the PEVK region would not modify passive tension. Finally the disruption of a single domain from titin's A-band region would also likely not affect the assembly of the thick filament because of the assumed multiple binding sites.
Table 1.
Human titin mutations and their effects. Abbreviations: 1DCM – dilated cardiomyopathy; 2HCM- hypertrophic cardiomyopathy; 3TMD – Tibial Muscular Dystrophy; 4LGMD – Limb Girdle Muscular Dystrophy; 5 – base pair position of mutation in genomic sequence of human titin (GenBank Accession no. AJ277892).
| Disease | Country | Heart | Skeletal muscle | Titin exon | AJ277892 Position5 | Sarcomere position | Mutation type | Cell level effects | Base change | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| DCM1 | Japanese | yes | no | 3 | 17750 | Z-line (Z repeat 1) | 1 base change | decreased T cap binding affinity | G to A | 32 |
| HCM2 | Japanese | yes | no | 14 | 34029 | Z-line (Z repeat 7) | 2 base transversion | increased α-actinin binding affinity | G to T | 33 |
| DCM | Japanese | yes | no | 14 | 34038 | Z-line (Z repeat 7) | 1 base change | decreased α-actinin binding affinity | C to T | 32 |
| DCM | Native American | yes | no | 18 | 37048 | Z-line to I-band | 1 base change | T to C | 19, 31 | |
| DCM | Japanese | yes | no | 49 | 79690 | N2B unique | 1 base change | increased FHL2 binding affinity | C to A | 34 |
| DCM | Japanese | yes | no | 49 | 80313 | N2B unique | 1 base change, stop codon | decreased FHL2 binding affinity | 32 | |
| DCM | Chinese | yes | no | 49 | 82126 | N2B unique | 1 base change | G to A | 35 | |
| DCM | Japanese | yes | no | 49 | 82324 | N2B unique | 1 base change | G to A | 32 | |
| DCM | Australian | yes | no | 326 | 245037 | A-band | 2 base insertion, stop codon | AT insert | 31 | |
| DCM | Australian | yes | no | 335 | 267662 | A-band | 1 base change, stop codon | del G | 36 | |
| Edstrom myopathy | Swedish | no | yes | 358 | 286133 | A-band, kinase | 1 base change | reduced nbr1 binding affinity | C to T | 37 |
| Fatal cardiomyopathy | Sudanese | yes | yes | 358 | 289385 | M-line (Mex 1) | 8 base deletion, stop codon | disrupted M line complex | del 8 bp | 38 |
| DCM | Japanese | yes | no | 358 | 287772 | M-line (Mex 1) | 1 base change | FHL2 binding region | G to A | 34 |
| Fatal cardiomyopathy | Moroccan | yes | yes | 360 | 291297 | M-line (Mex 3) | 1 base deletion, stop codon | disrupted M line complex | del A | 38 |
| TMD3 | French C | no | yes | 362 | 292298 | M-line (Mex 5) | 1 base deletion, stop codon | del T | 39 | |
| TMD/LGMD4 | Finnish maj | no | yes | 363 | 293269−293279 | M-line (Mex 6) | 11 bp deletion/insertion | del/ins | 40 | |
| TMD | Belgian | no | yes | 363 | 293329 | M-line (Mex 6) | 1 base change | T to A | 41 | |
| TMD | French A | no | yes | 363 | 293356 | M-line (Mex 6) | 1 base change | reduced calpain 3 binding | T to C | 39,40 |
| TMD | Spanish | no | yes | 363 | 293378 | M-line (Mex 6) | I base deletion, stop codon | del A | 39 | |
| TMD | French B | no | yes | 363 | 293379 | M-line (Mex 6) | 1 base change | C to T | 39 |
Figure 1.
Diagram of human titin regions and locations of mutations in the half sarcomere. The single titin molecules extend from the Z-line to the M-line in the middle of the sarcomere. The I-band region background is shown in yellow and the A-band region in white. Each number listed refers to a single titin mutation in the corresponding exon annotated in GenBank AJ277892. The mutation for the knockin-mouse report in the current issue [30] is denoted with an asterisk.
A couple animal models with altered titin have also been found. The mdm mouse (muscular dystrophy with myositis) has an 83 amino acid deletion in the N2A region thought to be involved in calpain 3 binding [20, 21]. The lack of cardiac involvement in this model is presumably related to the essential lack of calpain 3 in adult heart [22]. The pickwick mutation in Zebrafish results in a dilated cardiomyopathy like condition with poor sarcomere assembly [23]. The mutation is a T to G transversion in the N2B unique region, but the exact functional effect remains to be determined.
Because obtaining samples from living human hearts is both technically challenging and hard to justify ethically, the development of suitable cellular and animal models has driven the field. Many new mechanistic insights have been gained from transfection of cultured cells and development of transgenic animals, primarily rodents [24-29].
In the current issue Gramlich and coworkers [30] report for the first time the properties of a knock-in mouse containing a titin mutation similar to one reported previously in humans [31]. This autosomal dominant mutation (an AT insertion in the large exon 326 in the myosin binding protein C region of the middle half A band) results in a termination codon and a truncated expressed protein. Heterozygote individuals developed dilated cardiomyopathy with sudden death, but the penetrance was incomplete. Curiously, although this exon is constitutively expressed in skeletal muscle as well, there has been no skeletal muscle mutant phenotype detected to date.
Homozygocity of the current knock-in mouse was found to be lethal by embryonic day 9.5. This was consistent with previous cell culture work showing that near full length titin was essential for myofibrillogenesis [24, 25]. Heterozygotes, however, were essentially normal in regard to health and fertility. Hearts contained normal ratios of titin to myosin, and had similar passive tension properties to wild type. Systolic and diastolic function of heterozygotes was not significant different when compared to normal animals. Echocardiography revealed no dilation or altered cardiac structural dimensions. This lack of the effect of the mutation on the mouse model was probably not too surprising in light of the mild effects in humans. Why would we expect to see changes in a mouse when it takes 20 to 40 years for the phenotype to develop in humans and only in a certain proportion of mutation carriers?
The novelty of the current work was that the investigators found ways to induce the DCM phenotype in the knock-in heterozygotes. Two different approaches were used. In the first angiotensin II was continuously infused into wild type and heterozygote mice to cause arterial hypertension. Both groups responded with increased ejection fraction and fractional shortening after one week, but by two weeks the performance of the heterozygotes had significantly declined for both these parameters. Echocardiography also indicated increased dilation between one and two weeks in the heterozygotes. Microscopic examination showed essentially normal myofibril structure by electron microscopy. However, large increases in interstitial fibrosis (∼12% of area in heterozygotes versus ∼4% in wild type) were observed. The second approach was the continuous infusion of the beta agonist isoproterenol. After one week, the heterozygotes had significantly lower ejection fraction (∼39% vs ∼53%), significantly lower fractional shortening (∼24% vs ∼34%) and significantly enlarged left ventricle chamber size.
Western blotting using antibodies from the expressed titin region and those from the carboxyl end deleted part verified that a truncated titin was expressed in the knock-in heterozygotes. The amount of this shortened titin was much lower than expected, perhaps due to elevated proteolysis. Real time PCR indicated that the message levels for the wild type allele were elevated in heterozygotes, but this compensation was incomplete.
The use of stressors to induce a dilated cardiomyopathy phenotype in rodent models of human disease may prove to be a valuable approach for study of mechanisms related to other mutations. Many unanswered questions remain: what controls the level of up-regulation of wild type alleles in heterozygotes and could we fully express an adequate amount of wild type protein from a single allele? Was the truncated protein fully incorporated into the myofibril structure? Would it be possible to completely shut down expression of the mutant allele, and would this provide a rescue of the wild type cardiac phenotype? Could we further increase the rate and extent of breakdown of mutant protein and thus achieve the normal phenotype? What are the signals that lead a heart to develop dilated cardiomyopathy? These questions must await further studies, but our knowledge appears to have taken a step forward from stressing the giant.
Acknowledgments
Supported by the College of Agricultural and Life Sciences, University of Wisconsin-Madison and NIH grant HL-77196.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- 2.Bos JM, Ommen SR, Ackerman MJ. Genetics of hypertrophic cardiomyopathy: one, two, or more diseases? Curr Opin Cardiol. 2007;22:193–9. doi: 10.1097/HCO.0b013e3280e1cc7f. [DOI] [PubMed] [Google Scholar]
- 3.Marian AJ. Genetic determinants of cardiac hypertrophy. Curr Opin Cardiol. 2008;23:199–205. doi: 10.1097/HCO.0b013e3282fc27d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldfors A, Lamont PJ. Thick filament diseases. Adv Exp Med Biol. 2008;642:78–91. doi: 10.1007/978-0-387-84847-1_7. [DOI] [PubMed] [Google Scholar]
- 5.Tsoutsman T, Bagnall RD, Semsarian C. Impact of multiple gene mutations in determining the severity of cardiomyopathy and heart failure. Clin Exp Pharmacol Physiol. 2008;35:1349–57. doi: 10.1111/j.1440-1681.2008.05037.x. [DOI] [PubMed] [Google Scholar]
- 6.Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77:659–66. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- 7.Chang AN, Parvatiyar MS, Potter JD. Troponin and cardiomyopathy. Biochem Biophys Res Commun. 2008;369:74–81. doi: 10.1016/j.bbrc.2007.12.081. [DOI] [PubMed] [Google Scholar]
- 8.Paul M, Zumhagen S, Stallmeyer B, Koopmann M, Spieker T, Schulze-Bahr E. Genes causing inherited forms of cardiomyopathies. A current compendium. Herz. 2009;34:98–109. doi: 10.1007/s00059-009-3215-8. [DOI] [PubMed] [Google Scholar]
- 9.Buvoli M, Hamady M, Leinwand LA, Knight R. Bioinformatics assessment of beta-myosin mutations reveals myosin's high sensitivity to mutations. Trends Cardiovasc Med. 2008;18:141–9. doi: 10.1016/j.tcm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–6. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 11.Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–72. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 12.Greaser ML, Krzesinski PR, Warren CM, Kirkpatrick B, Campbell KS, Moss RL. Developmental changes in rat cardiac titin/connectin: transitions in normal animals and in mutants with a delayed pattern of isoform transition. J Muscle Res Cell Motil. 2005;26:325–32. doi: 10.1007/s10974-005-9039-0. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol. 2000;7:1117–20. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 14.Greaser ML, Warren CM, Esbona K, Guo W, Duan Y, Parrish AM, Krzesinski PR, Norman HS, Dunning S, Fitzsimons DP, Moss RL. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. J Mol Cell Cardiol. 2008;44:983–91. doi: 10.1016/j.yjmcc.2008.02.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeWinter MM, Wu Y, Labeit S, Granzier H. Cardiac titin: structure, functions and role in disease. Clin Chim Acta. 2007;375:1–9. doi: 10.1016/j.cca.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Linke WA. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc Res. 2008;77:637–48. doi: 10.1016/j.cardiores.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Krüger M, Linke WA. Titin-based mechanical signalling in normal and failing myocardium. J Mol Cell Cardiol. 2009 doi: 10.1016/j.yjmcc.2009.01.004. (in press) (PMID: 19318237) [DOI] [PubMed] [Google Scholar]
- 18.Udd B, Haravuori H, Kalimo H, Partanen J, Pulkkinen L, Paetau A, Peltonen L, Somer H. Tibial muscular dystrophy--from clinical description to linkage on chromosome 2q31. Neuromuscul Disord. 1998;8:327–32. doi: 10.1016/s0960-8966(98)00024-8. [DOI] [PubMed] [Google Scholar]
- 19.Siu BL, Niimura H, Osborne JA, Fatkin D, MacRae C, Solomon S, Benson DW, Seidman JG, Seidman CE. Familial dilated cardiomyopathy locus maps to chromosome. 2q31. Circulation. 1999;99:1022–6. doi: 10.1161/01.cir.99.8.1022. [DOI] [PubMed] [Google Scholar]
- 20.Garvey SM, Rajan C, Lerner AP, Frankel WN, Cox GA. The muscular dystrophy with myositis (mdm) mouse mutation disrupts a skeletal muscle-specific domain of titin. Genomics. 2002;79:146–149. doi: 10.1006/geno.2002.6685. [DOI] [PubMed] [Google Scholar]
- 21.Huebsch KA, Kudryashova E, Wooley CM, Sher RB, Seburn KL, Spencer MJ, Cox GA. Mdm muscular dystrophy: interactions with calpain 3 and a novel functional role for titin's N2A domain. Hum Mol Genet. 2005;14:2801–11. doi: 10.1093/hmg/ddi313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fougerousse F, Anderson LV, Delezoide AL, Suel L, Durand M, Beckmann JS. Calpain3 expression during human cardiogenesis. Neuromuscul Disord. 2000;10:251–6. doi: 10.1016/s0960-8966(99)00107-8. [DOI] [PubMed] [Google Scholar]
- 23.Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, Fishman MC. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–9. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- 24.van der Ven PF, Bartsch JW, Gautel M, Jockusch H, Fürst DO. A functional knockout of titin results in defective myofibril assembly. J Cell Sci. 2000;113:1405–14. doi: 10.1242/jcs.113.8.1405. [DOI] [PubMed] [Google Scholar]
- 25.Musa H, Meek S, Gautel M, Peddie D, Smith AJ, Peckham M. Targeted homozygous deletion of M-band titin in cardiomyocytes prevents sarcomere formation. J Cell Sci. 2006;119:4322–31. doi: 10.1242/jcs.03198. [DOI] [PubMed] [Google Scholar]
- 26.Miller G, Musa H, Gautel M, Peckham M. A targeted deletion of the C-terminal end of titin, including the titin kinase domain, impairs myofibrillogenesis. J Cell Sci. 2003;116:4811–9. doi: 10.1242/jcs.00768. [DOI] [PubMed] [Google Scholar]
- 27.Weinert S, Bergmann N, Luo X, Erdmann B, Gotthardt M. M line-deficient titin causes cardiac lethality through impaired maturation of the sarcomere. J Cell Biol. 2006;173:559–70. doi: 10.1083/jcb.200601014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radke MH, Peng J, Wu Y, McNabb M, Nelson OL, Granzier H, Gotthardt M. Targeted deletion of titin N2B region leads to diastolic dysfunction and cardiac atrophy. Proc Natl Acad Sci USA. 2007;104:3444–9. doi: 10.1073/pnas.0608543104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng J, Raddatz K, Molkentin JD, Wu Y, Labeit S, Granzier H, Gotthardt M. Cardiac hypertrophy and reduced contractility in hearts deficient in the titin kinase region. Circulation. 2007;115:743–51. doi: 10.1161/CIRCULATIONAHA.106.645499. [DOI] [PubMed] [Google Scholar]
- 30.Gramlich M, Michely B, Krohne C, Heuser A, Erdmann B, Klaassen S, Hudson B, Magarin M, Kirchner F, Todiras M, Granzier H, Labeit S, Thierfelder L, Gerull B. Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J Mol Cell Cardiol. 2009 doi: 10.1016/j.yjmcc.2009.04.014. doi:10.1016/j.yjmcc.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerull B, Gramlich M, Atherton J, McNabb M, Trombitás K, Sasse-Klaassen S, Seidman JG, Seidman C, Granzier H, Labeit S, Frenneaux M, Thierfelder L. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30:201–4. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 32.Itoh-Satoh M, Hayashi T, Nishi H, Koga Y, Arimura T, Koyanagi T, Takahashi M, Hohda S, Ueda K, Nouchi T, Hiroe M, Marumo F, Imaizumi T, Yasunami M, Kimura A. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem Biophys Res Commun. 2002;291:385–93. doi: 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- 33.Satoh M, Takahashi M, Sakamoto T, Hiroe M, Marumo F, Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem Biophys Res Commun. 1999;262:411–7. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto Y, Hayashi T, Inagaki N, Takahashi M, Hiroi S, Nakamura T, Arimura T, Nakamura K, Ashizawa N, Yasunami M, Ohe T, Yano K, Kimura A. Functional analysis of titin/connectin N2-B mutations found in cardiomyopathy. J Muscle Res Cell Motil. 2005;26:367–74. doi: 10.1007/s10974-005-9018-5. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Rao L, Zhou B, Zhang BL, Wang YY, Chen B, Wu Y, Huang P. Titin gene mutations in Chinese patients with dilated cardiomyopathy. Zhonghua Xin Xue Guan Bing Za Zhi. 2008;36:1066–9. [PubMed] [Google Scholar]
- 36.Gerull B, Atherton J, Geupel A, Sasse-Klaassen S, Heuser A, Frenneaux M, McNabb M, Granzier H, Labeit S, Thierfelder L. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J Mol Med. 2006;84:478–83. doi: 10.1007/s00109-006-0060-6. [DOI] [PubMed] [Google Scholar]
- 37.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edström L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 38.Carmignac V, Salih MA, Quijano-Roy S, Marchand S, Al Rayess MM, Mukhtar MM, Urtizberea JA, Labeit S, Guicheney P, Leturcq F, Gautel M, Fardeau M, Campbell KP, Richard I, Estournet B, Ferreiro A. C-terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann Neurol. 2007;61:340–51. doi: 10.1002/ana.21089. [DOI] [PubMed] [Google Scholar]
- 39.Hackman P, Marchand S, Sarparanta J, Vihola A, Pénisson-Besnier I, Eymard B, Pardal-Fernández JM, Hammouda el-H, Richard I, Illa I, Udd B. Truncating mutations in C-terminal titin may cause more severe tibial muscular dystrophy (TMD). Neuromuscul Disord. 2008;18:922–8. doi: 10.1016/j.nmd.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Hackman P, Vihola A, Haravuori H, Marchand S, Sarparanta J, De Seze J, Labeit S, Witt C, Peltonen L, Richard I, Udd B. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van den Bergh PY, Bouquiaux O, Verellen C, Marchand S, Richard I, Hackman P, Udd B. Tibial muscular dystrophy in a Belgian family. Ann Neurol. 2003;54:248–51. doi: 10.1002/ana.10647. [DOI] [PubMed] [Google Scholar]