Abstract
To determine specific molecular features of endothelial cells (ECs) relevant to the physiological process of penile erection we compared gene expression of human EC derived from corpus cavernosum of men with and without erectile dysfunction (HCCEC) to coronary artery (HCAEC) and umbilical vein (HUVEC) using Affymetrix GeneChip microarrays and GeneSifter software. Genes differentially expressed across samples were partitioned around medoids to identify genes with highest expression in HCCEC. A total of 190 genes/transcripts were highly expressed only in HCCEC. Gene Ontology classification indicated cavernosal enrichment in genes related to cell adhesion, extracellular matrix, pattern specification and organogenesis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed high expression of genes relating to ECM-receptor interaction, focal adhesions, and cytokine-cytokine receptor interaction. Real-time PCR confirmed expression differences in cadherins 2 and 11, claudin 11 (CLDN11), desmoplakin, and versican. CLDN11, a component of tight junctions not previously described in ECs, was highly expressed only in HCCEC and its knockdown by siRNA significantly reduced transendothelial electrical resistance in HCCEC. Overall, cavernosal ECs exhibited a transcriptional profile encoding matrix and adhesion proteins that regulate structural and functional characteristics of blood vessels. Contribution of the tight junction protein CLDN11 to barrier function in endothelial cells is novel and may reflect hemodynamic requirements of the corpus cavernosum.
Keywords: penis, tight junctions, microarray, gene ontology
the endothelium is a single cell layer lining all vessels of the vascular tree and lymphatics. It has been suggested that the endothelium be considered an organ given the unique and essential functions of this cell layer (1). Regions of the endothelium vary greatly in terms cell surface markers, functional characteristics, and biochemical properties (4, 10, 18, 19, 33). The extensive heterogeneity in the endothelium has been demonstrated through microarray analysis of endothelial cells (ECs) harvested from organs and tissue sites throughout the body (8, 18, 19, 31). Cultured ECs from different vascular beds were found to exhibit very distinct patterns of gene expression, potentially reflecting their unique function in vivo. Chi et al. (2003, Ref. 8) completed the most comprehensive analysis of differing EC lines showing distinct expression profiles for ECs derived from large vessels, microvessels, arteries, and veins. They also demonstrated discrete expression patterns for ECs based on their site of origin within the body's various organs.
Structurally, the erectile apparatus consists of paired corpora cavernosa that run the length of the penis. These specialized vascular compartments consist of lacunar spaces with sinusoidal trabeculae supporting a thick smooth muscle layer lined by ECs. The cavernosal endothelium is uniquely exposed to significant variations in oxygen tension and intracavernosal pressures. The flaccid penis receives minimal arterial inflow and basal oxygen tension ranging from 25 to 40 mmHg (24). During erection there are substantial increases in shear flow, intracavernosal pressure, stretch, and oxygen tension. Cavernosal ECs must be able to withstand this uniquely fluctuating environment, and likely exhibit a distinct gene program (2).
Understanding the transcriptional profile of cavernosal ECs may have relevance to erectile dysfunction (ED) research, because genes related to disease risk, response to therapy, and drug development targets need to be defined. In this study we used microarrays to characterize the global gene expression program of human corpus cavernosum ECs (HCCEC) compared with ECs from human coronary artery (HCAEC) and human umbilical vein (HUVEC).
MATERIALS AND METHODS
Experimental design.
HCCEC, HCAEC, and HUVEC were propagated under identical conditions and grown to confluence. EC cRNA samples were hybridized to Affymetrix Human U133A GeneChips at the University of Washington Center for Array Technology. Statistical filtering and software were used to identify and attribute biological significance to genes with tissue-specific expression highest in the HCCEC. Selected genes were validated with real-time PCR. To determine the physiological significance of CLDN11 expression in ECs, we used a transendothelial electrical resistance (TEER) assay to compare paracellular permeability of control and CLDN11 siRNA-treated cells.
Cell isolation.
Cavernosal tissue was obtained from men undergoing penile surgery or brain-dead male organ donors under an Institutional Review Board-approved protocol. The characteristics of the HCC donors for the array and PCR validation samples are shown in Supplemental Table A.1 The mean age of the donors for the five microarray samples was 58 yr (range 53–65); four of the five had ED with known risk factors (hypertension in four and diabetes in three). The remaining donor did not have ED. For the PCR validation experiment the samples were obtained from two men with ED and from an organ donor without ED. The mean age of this group was 53 yr (range 40–62). HCAEC derived from male donors were purchased from Clonetics; other parameters were unknown in these men. HUVEC were provided by the E. Raines laboratory (University of Washington).
HCCEC were isolated by tissue explant outgrowth as previously described (38). Briefly, cavernosal tissue was minced to ≈1 × 1 mm and spread with a wide-bore pipette into gelatin-coated flasks. Fresh medium was added to cover the explants without allowing them to float. Upon visualization of ECs migrating out of the minced tissue, the explants were removed, colonies monitored for smooth muscle cell contamination, and EC allowed to proliferate. The purity of the cells was determined as previously described (38). Briefly, cell sorting with PE-conjugated anti-CD31 antibody (1:50; Becton Dickinson) provided >95% CD31-positive cells. We confirmed the endothelial origin of the cells by uptake of Alexa-488-acetylated-LDL (Molecular Probes) and immunofluorescence for CD-31 (Pharmingen) and VE-cadherin (9H7, courtesy of Ronald L. Heimark, University of Arizona). Immunostaining for α-smooth muscle cell actin (Sigma) and a human fibroblast protein (Serotec) showed no evidence of contaminating stromal cells.
Cell culture.
ECs were grown on 2% gelatin type B (Sigma)-coated tissue culture dishes in medium 199 (M199, GIBCO) supplemented with 20% FBS, EGM-2MV (Clonetics), l-glutamine, and 10,000 μ/ml penicillin, 10,000 μ/ml streptomycin, and 25 μg/ml Fungizone. All cells were grown in a humidified incubator at 37°C and 5% CO2 with media changed every 2–3 days. For immunohistochemistry, cells were passaged onto glass slides, fixed, and immunolabeled for claudin 11 (Zymed) or VE-cadherin (Alexis) with the appropriate secondary antibody as previously described (38).
RNA isolation.
Total RNA isolation for the arrays was performed using Qiagen RNeasy mini-columns according to manufacturer's instructions (RNeasy Mini Handbook). Briefly, cells grown in T-75 culture flasks were trypsinized with 0.25% trypsin/1 mM EDTA (GIBCO) between the third and seventh passages and centrifuged to pellet the cells. Next, cells were disrupted and homogenized in RNeasy lysis buffer using a 1 ml syringe with 21-gauge needle. Ethanol was added to allow binding of RNA to the silica gel-based membrane of the mini column. Several washes as well as on-column DNase treatment were used to ensure that contaminants and DNA were effectively removed from the column. Finally, RNA was eluted in ∼30 μl of RNase-free water. Concentration and purity were analyzed by spectrophotometry (Biophotometer, Eppendorf), while the integrity of total RNA was confirmed with gel electrophoresis. The RNA samples were divided into 5 μg aliquots and stored at −80°C.
Microarray hybridization and data analysis.
Biotin-labeled target cRNA was prepared according to the Affymetrix eukaryotic target labeling protocol starting with 5 μg total RNA for each sample. Each target was hybridized to an Affymetrix Human U133A GeneChip, scanned at the University of Washington Center for Array Technology and analyzed (37). A variety of performance assessments both pre- and posthybridization were completed including testing cRNA quality using an Agilent Bioanalyzer 2100. In compliance with MIAME standards, Affymetrix data files (CHP and CEL) have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus repository (query accession GSE10804). Raw Affymetrix data files (CEL) were imported into GeneSifter software and robust multiarray average was used to analyze gene expression. Additional assessment of quality and variability of intensity was performed using MAS5. One-way analysis of variance (ANOVA) was used to determine P values for expression across groups, using a 5% false discovery rate (FDR) cutoff derived by a Benjamini and Hochberg correction (35). These parameters were used for statistical filtering of the array data as described below.
Strategies to determine biological significance.
Unsupervised hierarchical clustering of samples was performed using Euclidian distance and complete linkage. To identify genes with highest expression in HCCEC (1.5 fold cutoff and FDR correction of 5%), samples were partitioned using Partitioning Around Medoids/k medoid clustering (12, 20). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway terms were used to attribute biological significance to genes identified as highly expressed in HCCEC. Filtering using Z-scores (11) identified relevant terms from the latest build of GeneSifter using data from Entrez Gene as of May 1, 2007.
Real-time PCR.
For validation of expression levels of selected genes from the arrays, total RNA was extracted from additional different EC lines and prepared as described above. Reverse transcription was carried out with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was carried out as previously described (37) on three samples of cDNA for each group of HCCEC, HCAEC, and HUVEC using an Applied Biosystems 7900 Real-Time PCR system. Target genes were amplified with predesigned Taqman Gene Expression Assay forward and reverse primers (see Table 3 for Assay ID numbers) and probes (Applied Biosystems) using a two-stage cycle of 95°C for 15 s and 60°C for 1 min repeated for 40 cycles. Threshold cycle (CT) values were exported into a spreadsheet and relative changes in gene expression were calculated by the 2−ΔΔCT method (23). Results were given as fold changes in the target gene for HCCEC cDNA relative to HCAEC or HUVEC, with each sample being normalized to β-actin. All samples were prepared and examined in parallel.
Table 3.
Relative Array and PCR expression of selected genes highly expressed in HCCEC compared to HCAEC and HUVEC
| Accession No. | Gene Name | Fold Δ vs. HCAEC |
Fold Δ vs. HUVEC |
Taqman Assay ID | ||
|---|---|---|---|---|---|---|
| Array | PCR | Array | PCR | |||
| NM_001792 | Cadherin 2* | 2.48 | 1.72 | 2.47 | −1.35 | Hs00169953_m1 |
| NM_001797 | Cadherin 11* | 2.12 | 5.11 | 1.97 | −1.16 | Hs00156438_m1 |
| NM_003277 | Claudin 5 | −2.16 | −5.81 | −2.21 | −1.9 | Hs00533949_s1 |
| NM_005602 | Claudin 11 | 5.03 | 25.44 | 4.42 | 2.71 | Hs00194440_m1 |
| NM_004415 | Desmoplakin* | 13.29 | † | 9.76 | † | Hs00189422_m1 |
| NM_004114 | FGF 13 | 5.32 | 8.88 | 4.05 | 16.85 | Hs00182807_m1 |
| NM_013372 | Gremlin 1* | 19.19 | † | 22.19 | † | Hs00171951_m1 |
| AI380298 | Syndecan 2* | 10.9 | 7.5 | 8.04 | 4.58 | Hs00299807_m1 |
| R94644 | Versican* | 8.39 | 25.6 | 4.21 | 4.48 | Hs01007937_m1 |
Array fold change in certain cases is an average because the gene had multiple probe sets called significant.
For Desmoplakin and Gremlin 1, CT values were low for human corpus cavernosum endothelial cells (HCCEC); for human coronary artery endothelial cells (HCAEC) and human umbilical vein endothelial cells (HUVEC) the amplification never reached the linear range, implying very low expression of the gene but making fold change calculations impossible.
siRNA transfection of cavernosal EC.
HCCEC isolated from a single donor were transfected with 30 nM siRNA [either Ambion Silencer Predesigned siRNA constructs for CLDN11 (ID# 16634) or negative control (ID #4635)] according to the manufacturer's protocol using DharmaFECT-1 (Dharmacon). Optimal siRNA concentrations were titrated by pilot experiments (data not shown). Briefly, 3.0 × 105 EC/well in 2 ml antibiotic-free culture media were seeded in six-well plates. siRNA in RNase-free ddH2O and serum-free M199 was incubated in one tube and DharmaFECT-1 and serum-free M199 in another tube at room temperature for 5 min. The contents of the two tubes were mixed by gentle pipetting and incubated at room temperature for 20 min. Antibiotic-free culture medium was added to the mixture to make a final volume of 2.0 ml transfection medium. Culture medium was replaced with 2 ml/well of transfection medium, which was allowed to incubate overnight. The transfection medium was replaced with fresh antibiotic-free culture medium, and cells incubated overnight for recovery.
TEER assay.
The role of CLDN11 in HCCEC permeability was assessed by measurement of TEER. Transfected HCCEC (3 × 104 cells in 100 ml of EC culture medium) were seeded in 6.5-mm Transwell inserts (polyester, 3.0-mm pore, Costar) following pretreatment of the inert with 2% gelatin. EC culture medium (0.6 ml) was added to the lower compartments. Integrity of the EC monolayer was assessed daily by measurement of resistance with an Endohm-6 electrode chamber and an EVOM voltohmmeter (World Precision Instrument, Sarasota, FL). Prior to each measurement, the Endohm-6 chamber was filled with 0.65 ml fresh EC culture medium and incubated in CO2 incubator for 20 min. Transwell inserts with ECs were transferred to the chamber and measured sequentially. TEER was calculated by subtracting background resistant of Transwell insert without ECs, which was measured every third measurement. Each medium in Transwell inserts and lower compartments was replaced for up to 5 days total. Data were expressed as means ± SE from triplicate Transwell units and compared with two-way ANOVA. Aggregate data from three independent experiments were analyzed by calculating mean resistance of control wells for each experiment and designating this value as 100%. Values obtained with CLDN11 transfected cells were then expressed as a percentage of controls and compared with two-way ANOVA.
Western blotting of siRNA transfected HCCEC.
The efficiency of siRNA inhibition of CLDN11 protein expression was assessed by Western blot as previously described (38). Cells were harvested and protein lysates were quantified with the MicroBCA kit (Pierce). Samples (10 μg) were analyzed by SDS-PAGE with rabbit anti-CLDN11 (1:200 Zymed). Secondary antibodies used were donkey anti-rabbit (1:1,000; Amersham) peroxidase-conjugated (1 h, 22°C). Antibodies were stripped with Re-blot Plus reagents (Chemicon) and gels were reblotted with anti-GAPDH (1:1,000, Santa Cruz) as a housekeeping gene.
RESULTS
Microarray hybridization.
Twelve purified EC samples from three distinct vascular origins (5 HCCEC, 4 HCAEC, and 3 HUVEC; all from different donors) were propagated to obtain sufficient RNA for microarray hybridizations. All ECs displayed a cobblestone appearance and were free of contaminating spindle cells as shown in the Data Supplement (Supplemental Fig. A).
The EC cRNA was hybridized to microarrays containing 22,000 elements. Analyzing the aggregate data from all 12 lines revealed that >50% of transcripts were detected as determined by MAS5 present call. A number of well-established markers enriched in ECs were highly expressed in all 12 samples, confirming the endothelial origin of the cultured cells: PECAM (CD31), VE-cadherin (CDH5), von Willebrand factor (VWF), multimerin-1 (MMRN1), EC-specific molecule-1 (ESM1), VEGF receptor-2 (KDR), and endoglin (ENG).
Clustering of cell lines.
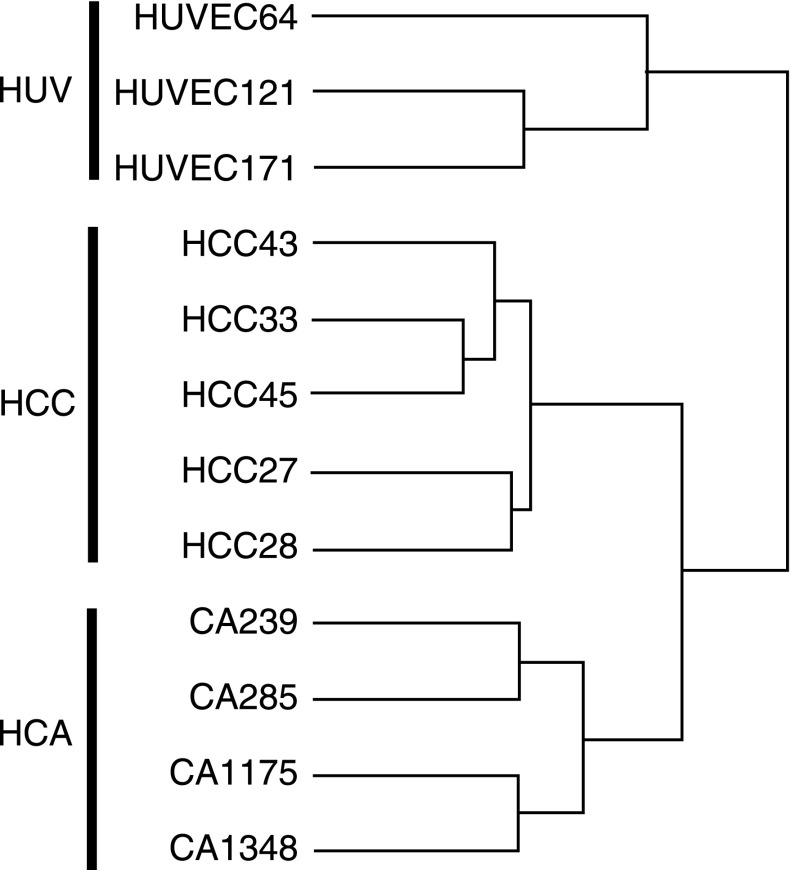
The first significant finding of our analysis was the discrete grouping of the EC lines as revealed by unsupervised hierarchical clustering of the samples (Fig. 1). The individual cell lines grouped into distinct expression patterns based on their in vivo source of origin. This is consistent with previous studies showing distinct clustering of ECs from different vascular regions or organs (8).
Fig. 1.
Unsupervised hierarchical clustering of all genes on the array reveals distinct grouping of endothelial cell (EC) samples by site of origin. The human corpus cavernosal (HCC) ECs aggregated more closely with ECs from human coronary artery (HCA) compared with the human umbilical vein (HUV). GeneSifter software.
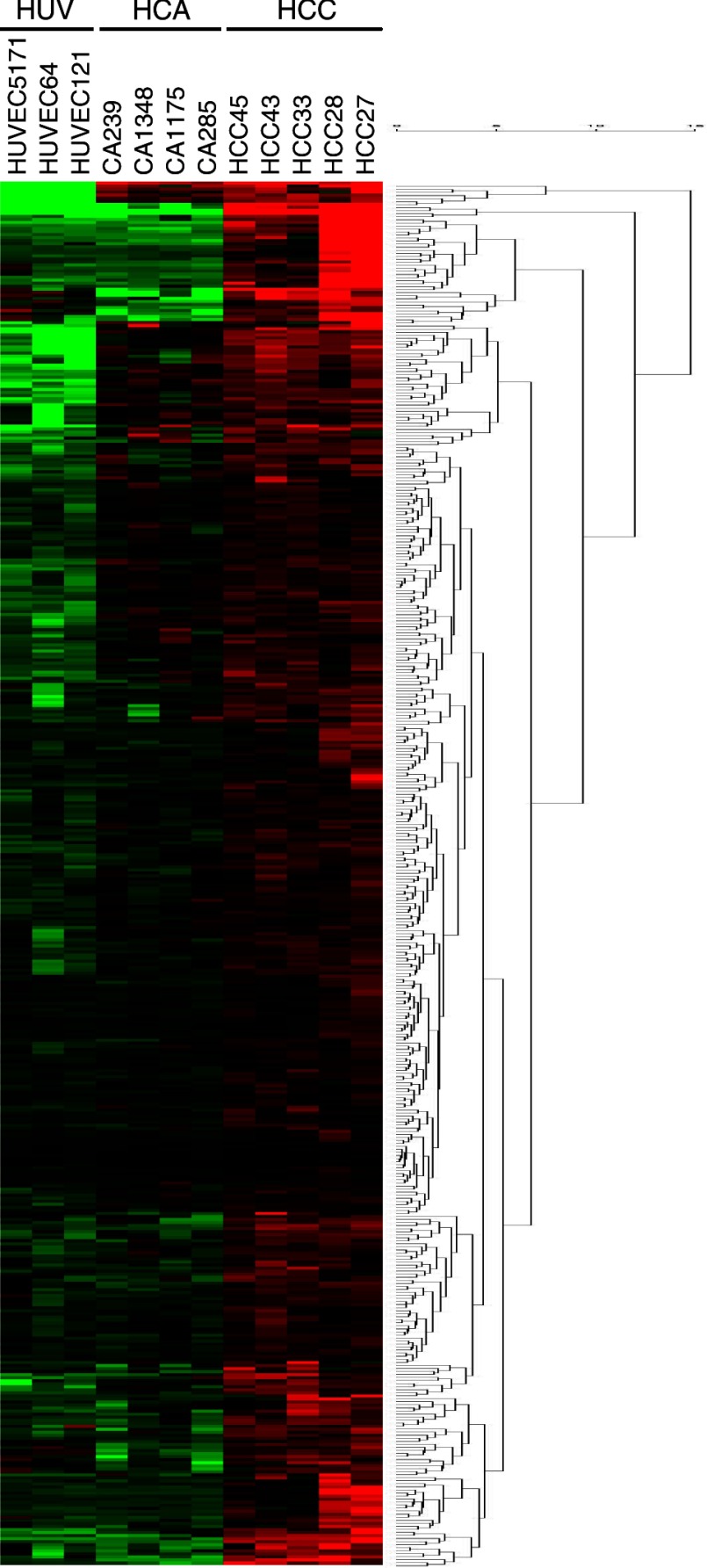
In pairwise comparisons, there were 479 genes highly expressed in HCCEC vs. HCAEC and 696 in HCCEC vs. HUVEC. Partitioning around medoids analysis generated several clusters based on expression levels highest in HCCEC. Figure 2 shows the heat map and dendrogram of 253 probe sets representing 190 unique genes and EST that had the highest expression in the HCCEC. In the online Data Supplement we included an enlarged heat map with gene names (Supplemental Fig. B) as well as the GeneSifter generated values including mean intensity ± SD for all 253 probe sets (Supplemental Table B). Affymetrix data files are accessible in the NCBI Gene Expression Omnibus (GSE10804).
Fig. 2.
Heat map and dendrogram showing partition clustering of genes most highly expressed in HCCEC vs. HCAEC and HUVEC. The heat map shows the intensity of expression as a function of color, with higher expression in red and lower expression in green or black. A complete list of all 253 transcripts identified by GeneSifter as highly expressed in HCCEC is shown in Supplemental Fig. B.
Classification of upregulated genes.
Various strategies were utilized to determine the possible biological relevance of differentially expressed genes to cavernosal function. Using GO terms and filtering described in materials and methods, z-score analysis identified 39 GO categories significantly enriched in the genes that were upregulated in HCCEC compared with the other cell types (see Table 1 for complete list and number of upregulated genes per category). The enriched GO categories with the greatest number of genes associated with those categories included development, cell adhesion, and cell surface receptor signal transduction. Similar z-score analysis using KEGG Pathway terms showed enrichment of only a few pathways in HCCEC including focal adhesion, cell adhesion molecules, and gap junctions (Table 2).
Table 1.
Gene Ontology list of significantly overrepresented gene categories by Z-score
| Ontology | List | Array | Z-Score | |||
|---|---|---|---|---|---|---|
| Biological Process | ||||||
| Cell adhesion | 24 | 564 | 5.26 | |||
| Focal adhesion formation | 2 | 10 | 4.68 | |||
| Cell-cell adhesion | 10 | 181 | 4.3 | |||
| Transmembrane receptor protein tyrosine kinase signaling pathway | 9 | 158 | 4.19 | |||
| Limb morphogenesis | 4 | 42 | 4.14 | |||
| Tissue development | 12 | 251 | 4.13 | |||
| Antiapoptosis | 8 | 144 | 3.86 | |||
| Male sex differentiation | 3 | 29 | 3.79 | |||
| Cell migration | 9 | 192 | 3.49 | |||
| Actin cytoskeleton organization and biogenesis | 8 | 164 | 3.42 | |||
| Calcium-independent cell-cell adhesion | 2 | 17 | 3.37 | |||
| Blood vessel development | 7 | 141 | 3.25 | |||
| Cell-matrix adhesion | 4 | 63 | 3.05 | |||
| Molecular Function | ||||||
| Extracellular matrix structural constituent | 7 | 77 | 5.28 | |||
| Myosin binding | 2 | 11 | 4.4 | |||
| Oxidoreductase activity | 3 | 23 | 4.39 | |||
| Structural molecule activity | 20 | 551 | 3.93 | |||
| Cytokine activity | 9 | 187 | 3.55 | |||
| Rab GTPase binding | 2 | 16 | 3.49 | |||
| Receptor signaling protein activity | 7 | 131 | 3.45 | |||
| Protein binding | 95 | 4,681 | 3.23 | |||
| Calcium ion binding | 20 | 652 | 3.11 | |||
| Transcriptional repressor activity | 8 | 180 | 3.08 | |||
| Cell Component | ||||||
| Collagen type VI | 2 | 2 | 11.24 | |||
| Collagen type I | 2 | 3 | 9.1 | |||
| Extracellular matrix | 16 | 230 | 6.69 | |||
| Anchoring collagen | 2 | 8 | 5.35 | |||
| Extracellular space | 16 | 424 | 3.77 | |||
| Apical junction complex | 4 | 51 | 3.63 | |||
| Cell-cell adherens junction | 2 | 17 | 3.4 | |||
| Intercellular junction | 6 | 109 | 3.34 | |||
| Plasma membrane part | 36 | 1,398 | 3.32 | |||
| Adherens junction | 3 | 37 | 3.22 | |||
Table 2.
KEGG Pathway analysis showing pathways enriched in genes in cavernosal EC and not coronary artery or umbilical vein ranked by Z-score
| Pathway | List | Array | Z-Score |
|---|---|---|---|
| ECM-receptor interaction | 9 | 81 | 6.62 |
| Focal adhesion | 13 | 183 | 5.8 |
| Cell Communication | 7 | 104 | 4.02 |
| Cytokine-cytokine receptor interaction | 10 | 231 | 3.19 |
| Cell adhesion molecules | 6 | 117 | 2.91 |
| Gap junction | 5 | 93 | 2.77 |
| Dorso-ventral axis formation | 2 | 24 | 2.52 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; EC, endothelial cell; ECM, extracellular matrix.
Cell adhesion gene expression.
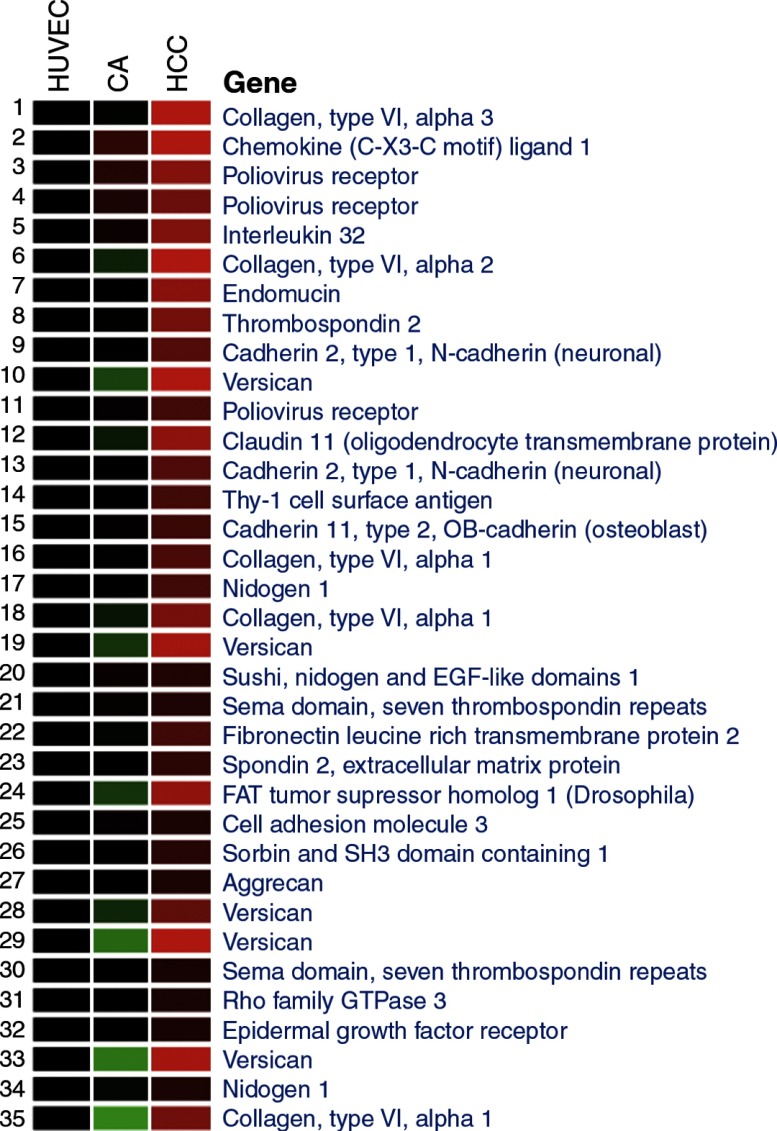
Noting that GO analysis identified cell adhesion genes as significantly enriched among genes upregulated in HCCEC, we clustered all the cell adhesion genes highly expressed in ECs from HCCEC vs. HCAEC and HUVEC (n = 39 of a possible 489 genes, Fig. 3). Numerous cell matrix and cell surface adhesion genes were differentially expressed in the HCCEC, but in contrast few intercellular adhesion molecules exhibited site-specific differential expression. For example, analysis of tight junction genes revealed that CLDN11 was highly expressed in HCCEC, while tight junctional proteins 1 and 2 (zona occludins, TJP), junctional adhesion molecule-3 (JAM3), and occludin showed no differences across cell lines. Of note, significantly lower expression of CLDN5 was seen in HCCEC with a twofold reduction in expression compared with the other two cell lines. A list of all tight junctional proteins and their relative expression on the array is listed in the online supplement (Supplemental Table C).
Fig. 3.
Heat map showing cell adhesion genes highly expressed in HCCEC. We determined by Gene Ontology the numerous cell matrix and cell surface adhesion genes differentially expressed in the HCCEC. Intensity of expression is displayed as mean values as a function of color, with higher expression in red and lower expression in green or black.
Adherens junction genes showed a similar expression pattern in the three different EC groups. Cadherins 2 (CDH2; N-cadherin) and 11 were identified as highly expressed only in HCCEC in this analysis. No difference was seen in expression levels of other genes including VE-cadherin (CDH5), E-cadherin (CDH1), junction plakoglobin (JUP), catenin alpha-1 (CTNNA1), catenin beta-1 (CTNNB1), catenin delta-1 (CTNND1), catenin alpha-like 1 (CTNNAL1), paxillin (PXN), vinculin (VCL), F11 receptor (F11R/JAM1).
Real-time PCR validation of selected genes.
We validated a number of genes differentially expressed on the arrays and relevant to adhesion and vascular function using additional HCCEC, HCAEC, and HUVEC cells from different donors. Genes chosen from the list of highly expressed genes in HCCEC, and their array and real-time PCR expression patterns are shown in Table 3. The only gene expression that was not confirmed by quantitative PCR was the higher expression of cadherin 2 and cadherin 11 in HCCEC vs. HUVEC. These genes were changed only −1.16-fold and −1.35-fold, respectively, and may represent false positive gene changes in the microarray analysis. Another potential explanation for the discrepancy is variability in the characteristics (age, ED status, comorbidities) of the HCCEC donors used in the array and real-time PCR experiments.
Claudin expression in endothelium.
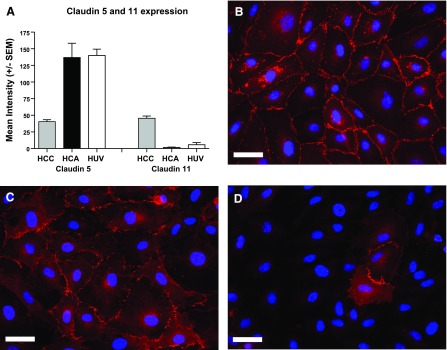
Of 15 claudins on the array, CLDN5 and CLDN11 were differentially expressed amongst the three EC lines. Figure 4A shows the relative intensity of probe sets for these claudins on the microarray. CLDN5 expression was significantly lower in HCCEC vs. HCAEC and HUVEC, whereas CLDN11 was expressed at much higher levels in HCCEC vs. the two other cell types. We confirmed the differential expression of CLDN5 and 11 using real-time PCR (Table 3). Immunohistochemistry also revealed consistent differences in CLDN11 expression between EC based on site of origin (Fig. 4B). All HCCEC isolates expressed high levels of CLDN11 at cell-cell junctions, while HUVEC had lower intensity of immunofluorescence and some areas without expression. HCAEC showed dramatically reduced expression of CLDN11, with only rare cells showing specific junctional immunolabeling of CLDN11.
Fig. 4.
Differential Claudin 5 (CLDN5) and 11 (CLDN11) expression according to cell type. A: mean array intensity values ± SE for CLDN5 and 11 from the different cell types; n = 5 for HCCEC, n = 4 for HCAEC, and n = 3 for HUVEC. Representative immunohistochemistry of CLDN11 in confluent cells showed highest expression levels in HCCEC (B), intermediate in HUVEC (C), and only few sporadic cells expressing CLDN11 in HCAEC (D). These findings were consistent across multiple isolates for all 3 cell types. CLDN11 appears red due to secondary antibody conjugated with Alexa 568 dye and cell nuclei appear blue due to DAPI nuclear staining. Scale bar is 30 microns. Exposure times were identical for all 3 slides, and no post processing of images was used.
CLDN11 regulates endothelial barrier function.
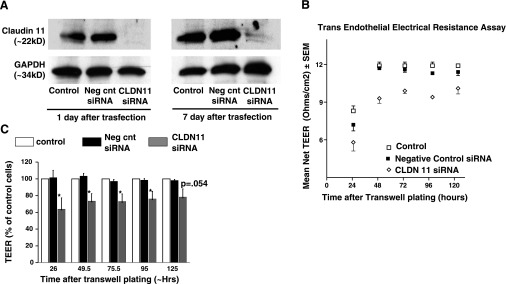
Claudins are important for tight junction structure, and CLDN11 specifically plays a role in the formation of tight junctions in the nervous system and blood testis barrier. In this study, we used siRNA to test the hypothesis that the CLDN11 expression in HCCEC confers enhanced cavernosal EC junctional barrier function. Western blot (Fig. 5A) and immunohistochemistry (data not shown) confirmed the significant reduction in claudin 11 protein expression in HCCEC transfected with siRNA against CLDN11 vs. negative control siRNA-treated cells. The decrease in claudin 11 protein was detected 1 day following siRNA transfection and was verified to remain significantly decreased throughout the length of the barrier function experiment. Results from three independent experiments showed that HCCEC transfected with CLDN11 siRNA had reduced transendothelial electrical resistance compared with negative control siRNA-treated cells (Fig. 5, B and C).
Fig. 5.
siRNA inhibition of CLDN11 reduces transendothelial electrical resistance (TEER) in HCCEC. A: Western blot showing persistent near complete inhibition of CLDN11 protein expression after treatment with CLDN11 siRNA. Note unchanged expression in negative control siRNA. B: graph from a representative experiment showing TEER values for control, negative control siRNA, and CLDN11 siRNA treated HCCEC at various time points. CLDN11 siRNA treatment group had significantly lower net TEER at all time points compared with control and negative control siRNA groups (P < 0.05). C: aggregate data from 3 independent experiments comparing mean TEER as a percentage of controls, with controls expressed as 100%.
DISCUSSION
The corpus cavernosum is a highly specialized vascular bed necessary for normal reproductive and sexual function. In a recent review, Aird (2) postulated that environmental influences in combination with epigenetic programs give rise to the heterogeneity inherent in ECs. HCCECs showed statistically significant differences in several categories of gene expression compared with coronary artery and umbilical vein ECs. This was consistent across a number of methodologies including unsupervised hierarchical clustering, partitioning around medoids, and comparison with previously published vessel-selective gene sets.
Unsupervised hierarchical clustering segregated the samples by site of origin, with cavernosal ECs grouping together. However, the overall Euclidian distance between the samples was small, confirming that few differences in the overall gene program were present between the various vascular beds. In fact, of the 22,000 possible genes on the array, <5% of genes exhibited differential expression between sites of origin, consistent with other reports (8).
Comparison of HCCEC transcripts from the current study with arterial and venous gene set clusters reported by Chi et al. (8) supports a prior suggestion that cavernosal sinusoidal EC express a mixed arterial and venous phenotype (38). Of the genes in the arterial EC selective cluster reported by Chi et al. (8), HCCEC (and HCAEC) in the present study expressed high levels of five of the arterial-specific transcripts, including cytokine like 1 (alias C17), EVA1, integral membrane protein 2A, keratin 7, and notch 4. Endothelial lipase was also expressed more abundantly in the HCCEC and HCAEC vs. HUVEC, while CD44, Hey 2, and ALDH1a1 showed no differences. Interestingly, HCCEC strongly expressed a venous EC-specific gene, myosinB1; expression levels in HCCEC were similar to HUVEC and ninefold greater than HCAEC. There was no differential expression of other members of the vein-specific gene cluster, which included lefty1 and 2, growth differentiating factor1, smoothened, and EphB4.
Overall, we identified 190 genes more highly expressed in HCCECs and validated several of these with real-time PCR. The predominant categories identified using GO and Pathway analysis relate to cell adhesion, extracellular matrix (ECM), pattern specification, and organogenesis (see discussion sections below). The embryological origin of cavernosum, developing from genital tubercle mesenchyme, may influence this EC program (6, 9). Furthermore, the biophysical forces to which cavernosal EC are exposed provide an environmental influence on cell-cell and cell-matrix anchoring as well as the composition of collagen struts. Unlike the limited pressure changes in conduit vasculature, intracavernosal pressure changes are vast. Between the flaccid and erect state, cavernosal pressure changes from ∼10 to 100 mmHg (36). During erection, increases in shear, stretch, and pressure confer a rapid and sustained change in the intracavernosal hemodynamic profile, which then subsides upon detumescence. HCCECs must withstand these vast fluctuations in hemodynamic environment.
Age or ED-related differences in endothelial cell gene expression could also potentially influence the arrays findings. We did not have sufficient numbers of normal donor samples to perform statistical comparisons between ED and normal HCCEC. However, the HCCEC samples in Fig. 1 did not cluster according to donor age or the presence of specific comorbidities such as diabetes. Furthermore, the sample from the donor without ED (HCC43) clustered with the other HCCEC rather than with cells from the other vascular beds. Finally, additional validation with other normal samples confirmed a number of the key findings on the array.
ECM.
The vascular ECM is composed of macromolecules organized by entanglement and cross-linking into a biomechanically active polymer (39). These matrices provide a gel-like form and scaffolding structure with regional tensile strength, elasticity, adhesiveness, and compressibility. The ability of sinusoidal spaces to collapse and distend requires a specific collagen and ECM phenotype. Like epithelia in other compliant organs, such as the lung or urinary bladder, cavernosal ECs elaborate some of these matrix proteins. Compared with HCAEC and HUVEC, HCCEC expressed high levels of mRNA transcripts encoding types I, III, IV, and VI collagen (Fig. 3), reflecting the composition of the human cavernosal matrix (34). The high expression of collagens in HCCECs is of potential relevance to erectile function. Not only is collagen a critical component of the structural backbone of the cavernosal sinusoids, penile collagen is also reported to be important for integrity of the venoocclusive mechanism, a requirement for rigid erection (27). Of note, the laminins, key noncollagenous constituents of the basement membrane, were not differentially expressed across the different EC lines.
Other genes encoding ECM and proteins of the extracellular space that were highly expressed in cavernosal ECs included a glycoprotein (nidogen), membrane-associated proteoglycans (syndecan 2 and glipican), and two chondroitin sulfate proteoglycan core proteins (aggrecan and versican). While the role of these genes and their gene products in normal cavernosal function is unknown, recent studies suggest alterations in these intercellular signaling and cellular regulation genes may play a role in disease-related abnormalities in matrix production or degradation.
Cell adhesion, cadherins.
Several homotypic cell-cell adhesion molecules were identified on the array as differentially expressed in HCCEC, including cadherins that contribute to adherens junctions. The blood-trapping function of sinusoids, which is required to pressurize the cavernosum during penile erection, necessitates robust intercellular adhesion properties. Polarity and integrity of cell-cell adhesion is provided by adherens junctions that may be needed in epithelia that experience strong contractile or mechanical forces at their apices (26). CDH5, the major homotypic adherens junction protein in ECs, was expressed equally in all three cell lines. However, two additional cadherins were expressed two- to fivefold higher in HCCEC compared with HCAEC. CDH2 (N-cadherin), a type 1 cadherin, contributes to homotypic endothelial cell-to-cell adhesion but also has been shown to mediate cancer cell migration and heterotypic adhesion to EC (32). Increased TEER in microvascular ECs (as compared with pulmonary ECs) was demonstrated to be associated with higher CDH2 expression (29). Cadherin 11 (CDH11, OB-cadherin), a type II cadherin similar in structure to CDH5, has been identified in ECs in 60 GEO entries, but in general expression intensity is low (GEO entry: GDS1402/NM_001797.1_PROBE1/CDH11/Homo sapiens). Interestingly, CDH11 was shown to be upregulated by shear stress in ECs (3). It is possible that the heightened expression of CDH11 in HCCECs is associated with the hemodynamic stress conferred to ECs of the penis.
Desmoplakin, a molecular element of cytoplasmic plaques associated with desmosomes, was also highly expressed in HCCEC. Of note, desmoplakin shares sequence similarity with intermediate filament proteins. Desmosome-intermediate filament interactions are known to be significant in other tissues that must withstand high levels of mechanical stress, such as the epidermis (13).
Tight junctions, claudins.
Claudins, a molecular component of tight junctions or zonula occludens, is an adhesive element most important for the formation of permeability barriers in epithelia and endothelia (17). Tight junctions regulate movement of macromolecules, small solutes, and ions and are composed of the integral membrane proteins such as claudins, occludins, and junctional adhesion molecules (30). The blood-brain barrier is the prototypic selective permeability barrier, and its tight junctions comprise claudin 5 [the predominant claudin (26)], claudin 12, occludin, ZO1, and JAM1 (30, 40). Claudin 5-deficient mice exhibit size-selective (<800 Da) opening of the blood-brain barrier (28), and in vitro studies also suggest that claudin 5 regulates paracellular permeability in brain microvascular ECs (30), although not in umbilical vein EC (14). We found that claudin 5 was present in HCCEC, although at a significantly lower expression level than that HCAEC or HUVEC.
The tight junction protein most uniquely and highly expressed in HCCECs in our study was CLDN11 (alias OSP, oligodendrocyte-specific protein). CLDN11 is associated with the myelin sheath, stria vascularis, choroids plexus, and blood-testis barrier (21, 25). In searching GEO Profiles for CLDN11 and human ECs, we found 19 records (See Supplemental Table D; accession date July 30, 2008) reporting CLDN11 gene expression in ECs, including dermal lymphatic endothelial cells, umbilical artery and vein endothelial cells, pulmonary artery endothelial cells, uterine microvascular endothelial cells, and aortic endothelial cells. However, these data had not been validated by other methodologies. In contrast, we confirmed our findings by real-time PCR in additional cell replicates (Table 3) and by conventional PCR in normal human cavernosal tissue (data not shown).
CLDN11 and HCCEC barrier function.
Claudins create the seal-forming elements in tight junctions, through either homotypic or heterotypic adhesions (7). Thus the physical absence of these molecules may directly impair tight junction seal formation and lead to increased paracellular permeability. We found that knock-down of CLDN11 by siRNA resulted in a decrease in transendothelial resistance in HCCECs, suggesting an important role for CLDN11 in barrier function. The elevated expression of CLDN11 in HCCECs may reflect the requirement for junctional integrity and barrier integrity due to the high intracavernosal pressures during erection.
CLDN11 may impart barrier function through its atypical formation. Unlike other claudins, CLDN11 arranges in unbranched parallel fibrils, which may confer additional adhesiveness. In fact, the blood testis barrier, of which CLDN11 is the principal claudin, represents one of the most adhesive tight junction barriers (7). Interestingly, one study in CLDN11-deficient mice reported that tight junctions in the strial ECs within the cochlea of the inner ear appeared intact despite the absence of CLDN11 (16). In our study, immunohistochemistry for VE-cadherin demonstrated no changes in the appearance of cell-to-cell contacts in control and CLDN11 siRNA-treated cells (data not shown), indicating the loss of CLDN11 did not appear to disrupt total junctional stability. We did not assess possible compromise in junctions with more in-depth analysis such as electron microscopy. However, altogether these data suggest that the loss of CLDN11 does not alter barrier function through the complete loss of junctional integrity.
It is still possible that downregulation of CLDN11 may prevent or disrupt interaction of other junctional or ECM molecules (5) in recruitment of proteins such as occludins to the tight junction complex. Furthermore, claudins in one cell can associate with claudins in adjacent cells via heterotypic (binding other claudin isoforms) or homotypic (binding claudins of the same isoform) interactions (15). Claudins can also bind to other claudin proteins located within the same cell (cis-interactions) (22). It is unknown whether CLDN11 interacts with other claudins such as CLDN5 in HCCECs and if this interaction is important for barrier function.
Limitations.
Certain limitations exist in the present study that merit discussion. Because most of the HCCEC donors had ED, changes in gene expression due to comorbid disease or ED itself may have confounded our analyses and require further investigation in a larger number of normal samples. The expression pattern of cultured cells may include artifacts of their in vitro conditions. While one would expect such bias to be the same across vascular beds, unrecognized differences in growth rates, confluence, or other issues are possible. Use of laser capture microdissection could potentially allow analysis of cavernosal ECs obtained from their in vivo environment. Comparison of the corresponding cells in vitro would provide insight into the artifactual effects of tissue culture. We culture cavernosal ECs from the spongy erectile tissue of the cavernosum, and it is possible that some ECs from penile resistance arteries grow out of our minced specimens and have a different expression pattern than sinusoidal EC. However, observations from our laboratory indicate a very poor rate of cell outgrowth from isolated cavernosal arteries vs. sinusoidal tissue, suggesting that the majority of cells in our present study are of sinusoidal origin (unpublished observations, K. L. Engel and I. Luttrell). Nonspecific effects of transfection or off-target effects potentially confound the TEER findings using siRNA, although the use of scrambled controls and consistent findings with three separate replicates limit this possibility. Additional strategies to confirm the findings of the TEER with a CLDN11 rescue experiment would require novel retroviral constructs that are beyond the scope of the present studies.
Conclusions.
HCCEC express a small subset of genes that differ significantly from HCAEC and HUVEC. The most prominent categories of transcripts highly expressed in HCCEC included genes encoding collagen, ECM proteins, and cell junction proteins. These data imply that the validity of investigations of EC signaling underlying erectile function/dysfunction would benefit greatly from the specific study of cells isolated from the cavernosal vascular bed. In this study, we detected expression of CLDN11 in endothelial cells. Furthermore, HCCEC expressed significantly more CLDN11 than ECs from the other vascular beds studied. We also demonstrated the importance of this tight junction protein for barrier functions of the sinusoidal endothelium, as downregulation of CLDN11 by siRNA increased HCCEC paracellular permeability in vitro.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants 1R21DK-69686 (H. Wessells), 1RO1DK-55017 (H. Wessells), 5T32DK-007779 (C. J. Sullivan), 5U24DK-058813, and 5P30 DK-17047.
Supplementary Material
Footnotes
1The online version of this article contains supplemental material.
REFERENCES
- 1. Aird WC. Endothelium as an organ system. Crit Care Med 32: S271–S279, 2004. [DOI] [PubMed] [Google Scholar]
- 2. Aird WC. Mechanisms of endothelial cell heterogeneity in health and disease. Circ Res 98: 159–162, 2006. [DOI] [PubMed] [Google Scholar]
- 3. Andersson M, Karlsson L, Svensson PA, Ulfhammer E, Ekman M, Jernas M, Carlsson LM, Jern S. Differential global gene expression response patterns of human endothelium exposed to shear stress and intraluminal pressure. J Vasc Res 42: 441–452, 2005. [DOI] [PubMed] [Google Scholar]
- 4. Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Steps toward mapping the human vasculature by phage display. Nat Med 8: 121–127, 2002. [DOI] [PubMed] [Google Scholar]
- 5. Bronstein JM, Tiwari-Woodruff S, Buznikov AG, Stevens DB. Involvement of OSP/claudin-11 in oligodendrocyte membrane interactions: role in biology and disease. J Neurosci Res 59: 706–711, 2000. [DOI] [PubMed] [Google Scholar]
- 6. Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, Brown PO. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci USA 99: 12877–12882, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng CY, Mruk DD. Cell junction dynamics in the testis: Sertoli-germ cell interactions and male contraceptive development. Physiol Rev 82: 825–874, 2002. [DOI] [PubMed] [Google Scholar]
- 8. Chi JT, Chang HY, Haraldsen G, Jahnsen FL, Troyanskaya OG, Chang DS, Wang Z, Rockson SG, van de Rijn M, Botstein D, Brown PO. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci USA 100: 10623–10628, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chi JT, Rodriguez EH, Wang Z, Nuyten DS, Mukherjee S, van de Rijn M, van de Vijver MJ, Hastie T, Brown PO. Gene expression programs of human smooth muscle cells: tissue-specific differentiation and prognostic significance in breast cancers. PLoS Genet 3: 1770–1784, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Craig LE, Spelman JP, Strandberg JD, Zink MC. Endothelial cells from diverse tissues exhibit differences in growth and morphology. Microvasc Res 55: 65–76, 1998. [DOI] [PubMed] [Google Scholar]
- 11. Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol 4: R7, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dudoit S, Fridlyand J. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol 3: RESEARCH0036, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fagotto F, Gumbiner BM. Cell contact-dependent signaling. Dev Biol 180: 445–454, 1996. [DOI] [PubMed] [Google Scholar]
- 14. Fontijn RD, Rohlena J, van MJ, Pannekoek H, Horrevoets AJ. Limited contribution of claudin-5-dependent tight junction strands to endothelial barrier function. Eur J Cell Biol 85: 1131–1144, 2006. [DOI] [PubMed] [Google Scholar]
- 15. Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol 147: 891–903, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gow A, Davies C, Southwood CM, Frolenkov G, Chrustowski M, Ng L, Yamauchi D, Marcus DC, Kachar B. Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J Neurosci 24: 7051–7062, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84: 345–357, 1996. [DOI] [PubMed] [Google Scholar]
- 18. Ho M, Yang E, Matcuk G, Deng D, Sampas N, Tsalenko A, Tabibiazar R, Zhang Y, Chen M, Talbi S, Ho YD, Wang J, Tsao PS, Ben Dor A, Yakhini Z, Bruhn L, Quertermous T. Identification of endothelial cell genes by combined database mining and microarray analysis. Physiol Genomics 13: 249–262, 2003. [DOI] [PubMed] [Google Scholar]
- 19. Kallmann BA, Wagner S, Hummel V, Buttmann M, Bayas A, Tonn JC, Rieckmann P. Characteristic gene expression profile of primary human cerebral endothelial cells. FASEB J 16: 589–591, 2002. [DOI] [PubMed] [Google Scholar]
- 20. Kaufam L, Rousseeuew PJ. Finding Groups in Data: an Introduction to Cluster Analysis. New York: Wiley, 1990. [Google Scholar]
- 21. Kitajiri S, Miyamoto T, Mineharu A, Sonoda N, Furuse K, Hata M, Sasaki H, Mori Y, Kubota T, Ito J, Furuse M, Tsukita S. Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J Cell Sci 117: 5087–5096, 2004. [DOI] [PubMed] [Google Scholar]
- 22. Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta 1778: 631–645, 2007. [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 24. Moreland RB. Is there a role of hypoxemia in penile fibrosis: a viewpoint presented to the Society for the Study of Impotence. Int J Impot Res 10: 113–120, 1998. [DOI] [PubMed] [Google Scholar]
- 25. Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S. Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J Cell Biol 145: 579–588, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 147: 185–194, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nehra A, Goldstein I, Pabby A, Nugent M, Huang YH, de las Morenas A, Krane RJ, Udelson D, Saenz de Tejada I, Moreland RB. Mechanisms of venous leakage: a prospective clinicopathological correlation of corporeal function and structure. J Urol 156: 1320–1329, 1996. [DOI] [PubMed] [Google Scholar]
- 28. Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol 161: 653–660, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ofori-Acquah SF, King J, Voelkel N, Schaphorst KL, Stevens T. Heterogeneity of barrier function in the lung reflects diversity in endothelial cell junctions. Microvasc Res 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohtsuki S, Sato S, Yamaguchi H, Kamoi M, Asashima T, Terasaki T. Exogenous expression of claudin-5 induces barrier properties in cultured rat brain capillary endothelial cells. J Cell Physiol 210: 81–86, 2007. [DOI] [PubMed] [Google Scholar]
- 31. Podgrabinska S, Braun P, Velasco P, Kloos B, Pepper MS, Skobe M. Molecular characterization of lymphatic endothelial cells. Proc Natl Acad Sci USA 99: 16069–16074, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qi J, Chen N, Wang J, Siu CH. Transendothelial migration of melanoma cells involves N-cadherin-mediated adhesion and activation of the beta-catenin signaling pathway. Mol Biol Cell 16: 4386–4397, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest 102: 430–437, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raviv G, Kiss R, Vanegas JP, Petein M, Danguy A, Schulman C, Wespes E. Objective measurement of the different collagen types in the corpus cavernosum of potent and impotent men: an immunohistochemical staining with computerized-image analysis. World J Urol 15: 50–55, 1997. [DOI] [PubMed] [Google Scholar]
- 35. Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375, 2003. [DOI] [PubMed] [Google Scholar]
- 36. Shafik A, Shafik I, El SO, Shafik AA. On the pathogenesis of penile venous leakage: role of the tunica albuginea. BMC Urol 7: 14, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sullivan CJ, Teal TH, Luttrell IP, Tran KB, Peters MA, Wessells H. Microarray analysis reveals novel gene expression changes associated with erectile dysfunction in diabetic rats. Physiol Genomics 23: 192–205, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wessells H, Teal TH, Engel K, Sullivan CJ, Gallis B, Tran KB, Chitaley K. Fluid shear stress-induced nitric oxide production in human cavernosal endothelial cells: inhibition by hyperglycaemia. BJU Int 97: 1047–1052, 2006. [DOI] [PubMed] [Google Scholar]
- 39. Wight TN. The extracellular matrix and atherosclerosis. Curr Opin Lipidol 6: 326–334, 1995. [DOI] [PubMed] [Google Scholar]
- 40. Wolburg H, Lippoldt A. Tight junctions of the blood-brain barrier: development, composition and regulation. Vascul Pharmacol 38: 323–337, 2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.