Abstract
The first telomerase cofactor identified was the budding yeast protein Est1, which is conserved through humans. While it is evident that Est1 is required for telomere DNA maintenance, understanding its mechanistic contributions to telomerase regulation has been limited. In vitro, the primary effect of Est1 is to activate telomerase-mediated DNA extension. Although Est1 displayed specific DNA and RNA binding, neither activity contributed significantly to telomerase stimulation. Rather Est1 mediated telomerase upregulation through direct contacts with the reverse transcriptase subunit. In addition to intrinsic Est1 functions, we found that Est1 cooperatively activated telomerase in conjunction with Cdc13 and that the combinatorial effect was dependent upon a known salt-bridge interaction between Est1 (K444) and Cdc13 (E252). Our studies provide insights into the molecular events used to control the enzymatic activity of the telomerase holoenzyme.
Keywords: telomere biology, telomerase regulation
Telomeres are nucleoprotein complexes found at chromosome ends that maintain DNA termini at an appropriate length to preserve genome stability and cell viability (1). Most eukaryotes use the telomerase enzyme to sustain telomeric DNA. The telomerase core enzyme is comprised of a reverse transcriptase (Est2 in yeast) and an RNA template (TLC1) and the yeast holoenzyme includes the Est1 and Est3 cofactors. EST1 was the first protein-encoding gene identified that was speculated to be a telomerase component, as est1Δ cells display the ever shorter telomere phenotype (2). While Est1 is an established telomerase cofactor, its direct mechanistic contribution to telomerase activity has not been understood.
Genetic experiments suggest two potential Est1 roles with telomerase, as a bridging factor coupling telomerase to the telomeric DNA binding protein Cdc13 during telomere recruitment and as an activator for telomerase enzymatic activity once the holoenzyme is telomere-bound. A seminal study, which exploited protein fusions between Est1, Cdc13 or telomerase, suggested an Est1 activation function since telomeric DNA is hyperelongated in an Est1-dependent manner upon expression of a Cdc13-Est2 fusion protein (3). In addition, it was shown that covalent linkage of normally deleterious Est1 or Cdc13 mutants abrogates telomere defects implying that the mutations affect recruitment. Supporting a requisite Est1-Cdc13 interaction for proper telomerase regulation are reciprocal-charge mutants [e.g., est1–60 (K444E) suppresses cdc13–2 (E252K)] that separately lead to telomere DNA shortening (4). Since Est1–60 can bind telomerase in vivo, it was proposed that the K444E mutation disrupts Cdc13 association (4, 5). However, other studies suggest that the Cdc13–2 mutation does not disrupt Est1 interactions since wild-type Est1 interacts with Cdc13 and Cdc13–2 in vivo (6–8). Although in the absence of a direct biochemical assessment it is difficult to delineate the mechanistic effects of the various Est1 and Cdc13 mutations.
To modulate telomerase it has been speculated that Est1 relies on its two known activities, DNA and RNA binding. Prior biochemical studies demonstrate that yeast and human Est1 bind single-stranded G-rich DNA and apparently recognize RNA non-specifically in vitro (9–11). In contrast to the in vitro work, in vivo studies indicate that yeast Est1 targets a bulged-stem loop in the TLC1 telomerase RNA (12, 13). The Est1-TLC1 interaction appears to be direct since it occurs in the absence of the Est2 protein in vivo (14). Despite these reports, it was not apparent how Est1 modulates telomerase. We have attempted to understand the Est1 regulatory mechanism by investigating: 1) the Est1 impact on telomerase DNA extension activity; 2) the necessity of the TLC1 bulged-stem loop for Est1 RNA binding; 3) the Est1 DNA binding determinants; 4) the influence of Est1 nucleic acid binding on telomerase regulation; 5) the activities of various Est1 point mutants; and 6) combined telomerase regulatory functions of Est1 and Cdc13.
Results
Est1 Activates Telomerase DNA Extension Activity.
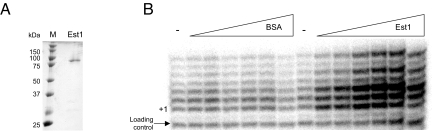
To study the mechanism of Est1 action the protein was purified to near homogeneity (Fig. 1A). As an initial functional test, we titrated the Est1 protein into telomerase DNA extension reactions. We found that Est1 stimulated activity approximately 14-fold using telomerase extracts prepared from either est1Δ or parental cells (Fig. 1B and Fig. S1). As the DNA banding pattern is not reproducibly altered (e.g., enhancement of longer products) upon Est1 addition but rather the relative product intensities are elevated, we suggest that Est1 increases the percent of active enzyme. Based upon the activation potential, Est1 displayed a high affinity for telomerase (Kd 3.5 ± 1 nM in est1Δ and 27 ± 4 nM in wild-type telomerase extract) (Fig. S1). The variance in Est1-telomerase affinities might result from the presence of Est1 protein, albeit limiting, within the telomerase extracts or from a telomerase structural difference. We suspect telomerase produced in the absence of Est1 is in a distinct conformation since the Est1 stimulatory effect is comparable using either wild-type and est1Δ telomerase extracts and even limiting Est1 amounts would diminish the fold increase over the unsupplemented DNA extension activity. To understand the Est1-mechanism used to modulate telomerase we examined the impact of Est1 properties on the stimulatory effect.
Fig. 1.
Purified Est1 enhances telomerase DNA extension activity in vitro. (A) Recombinant Est1 (1 μg) was resolved on a 12% SDS/PAGE and stained with Coomassie Blue. (B) The Est1 effect on telomerase-mediated DNA extension was tested using extracts prepared from est1Δ yeast with an immobilized 7-base 3′-overhang DNA substrate. The DNA extension reactions were supplemented with BSA or Est1 (1, 2.5, 5, 10, 25, and 50 nM), as marked. To serve as a loading control an end-labeled 27-base primer was added before precipitation of the extension products. The positions of the loading control primer and + 1 extension product are marked.
Est1 Selectively Binds To the TLC1 RNA Bulged-Stem Loop.
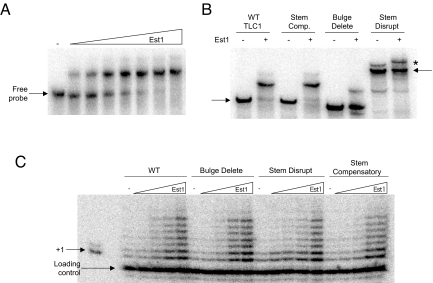
It has been suggested that a central bulged-stem loop within the TLC1 RNA is critical for telomerase regulation by Est1 (12, 13). Therefore, we generated the Est1-TLC1 RNA binding site (bulged-stem loop fragment) and examined Est1 binding in vitro (Fig. 2A). By electro-mobility shift analysis (EMSA) Est1 displayed a relatively high affinity for the bulged-stem loop RNA (Kd ≈40 nM), albeit lower than its affinity for telomerase (Fig. 1). The variation in RNA affinity relative to a prior report likely reflects the use of different RNA probes (9). The Est1 RNA binding was dependent upon the bulged-stem loop structure, as a disrupted-stem RNA displayed no apparent interaction and a bulge-delete RNA had a decreased affinity (Kd ≈300 nM). TLC1 RNA with compensatory mutations to reestablish the stem structure partially recovered the binding affinity (Kd ≈150 nM) (Fig. 2B). Thus, Est1 is dependent upon the bulged-stem loop structure to bind to the central TLC1 region and the in vitro binding determinants correlate well with prior in vivo studies (13).
Fig. 2.
Est1 RNA binding is dependent upon the central TLC1 bulged-stem loop. (A) To assess the Est1 RNA binding activity EMSA was used in conjunction with a radiolabeled TLC1 RNA probe. To measure the interaction affinity between Est1 and TLC1 the wild-type bulged-stem loop RNA (250 pM) was incubated alone or with varying Est1 concentrations (30, 60, 120, 240, 480, 960, and 1,440 nM). All binding reactions were resolved by native polyacrylamide electrophoresis. The arrows mark free probe locations. (B) The Est1 RNA binding specificity was determined using established TLC1 RNAs that included stem disruption, bulge deletion and stem compensatory mutants (13). The slower migration for the stem-disruption RNA indicates that it is in a distinct conformation. The ability of Est1 (480 nM) to bind to each TLC1 derivative (250 pM) was determined by RNA EMSA. The asterisk marks the position of a minor slower migrating, distinctly folded RNA that occurs with the stem disrupt RNA. Although the migration of minor species reproducibly slows further following incubation with Est1, the change in migration is not dependent upon a stable Est1 interaction as it is apparent even after degradation of the Est1 following protease addition. Hence, the slight migration shift in the minor RNA species present in the stem disrupt experiment does not result from a stable association with Est1. (C) The Est1 effect on telomerase enzymes with the full-length TLC1 bulged-stem loop derivatives was tested using in vitro DNA extension assays and a 7-base 3′-overhang DNA substrate. Reactions were supplemented with increasing Est1 amounts (1, 10, 100, or 200 nM), as designated.
The role of the TLC1 bulged-stem loop in conducting Est1 telomerase activation was determined by titrating Est1 protein into extension reactions with telomerase extracts prepared from yeast expressing full-length wild-type, stem-disrupt, bulge-delete or stem-compensatory TLC1 RNAs (13). Unexpectedly, Est1 comparably activated the telomerase TLC1 derivatives (Fig. 2C). Although the stem-disrupt telomerase displayed an approximate 2-fold Est1 affinity decrease, the effect is mild relative to the decline in binding to the stem-disrupt TLC1 RNA binding (Fig. 2B). Based on these differences, we suggest that the bulged-stem loop is one determinant in telomerase association but that other, likely protein-protein interactions between Est1 and Est2, are important for controlling telomerase activity.
Est1 Binds Single-Stranded Telomeric DNA.
The initial Est1 biochemical characterization demonstrated that the protein preferentially binds DNAs with 3′ single-stranded G-rich sequence (9). We wished to extend these observations by delineating the DNA length requirements for Est1 binding. As an initial substrate we examined Est1 interactions with a 30-base, single-stranded G-rich oligonucleotide by EMSA and found that Est1 bound with an apparent Kd of 75 nM (Fig. S2A); no binding was observed using a C-rich 30-base primer. Using a series of single-stranded G-rich primers that varied from 15 to 30 bases we determined a minimal length requirement between 18 and 22 nucleotides for binding to a fully single-stranded DNA (Fig. S2B).
We also tested the single-stranded DNA length requisite using a hybrid DNA substrate comprised of a single-stranded 3′ end and a double-stranded 5′ section. We found that Est1 bound well to 22- and 15-base 3′-overhang substrates but was unable to bind to shorter 3′-overhangs (Fig. S2C). The ability of Est1 to bind hybrid DNAs with a 15-nucleotide overhang suggests that double-stranded DNA can be tolerated at the 5′ end. Hence, depending upon the nature of the substrate (i.e., fully single-stranded or hybrid) Est1 varied slightly in its DNA length requirements but still required single-stranded DNA for binding.
Next, we determined whether Est1 might bind RNA and DNA simultaneously, as a prior report had indicated (9). We added Est1 to reactions containing a radiolabeled DNA probe alone or with increasing amounts of unlabeled TLC1 RNA (Fig. S2D). If Est1 can concurrently bind DNA and RNA then a higher order, slower migrating complex would be expected; however, if binding were competitive then a decline in the Est1-DNA complex signal should occur. Since the Est1-DNA signal decreased with increasing TLC1 RNA levels (Fig. S2D), Est1 DNA and RNA binding is mutually exclusive. Contrary to the efficient competitive ability of TLC1 RNA for Est1 DNA interactions, we found that DNA competed ineffectively for RNA binding. While the differential competition outcomes were unanticipated given the comparable Est1 binding affinities for RNA and DNA, the observed results likely show that TLC1 RNA is the preferred Est1 substrate. Minimally, our data indicate that telomerase regulation by Est1 involves only one bound nucleic acid.
Est1 Telomerase Activation Is Independent of Est1 DNA Binding.
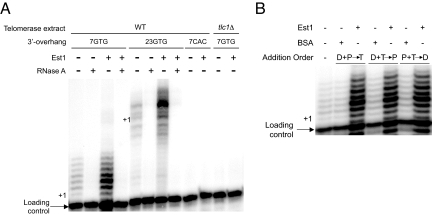
To address whether Est1 exploits single-stranded DNA to effect telomerase action we performed extension assays using DNAs with relatively short or long single-stranded lengths. We found that Est1 equivalently upregulated telomerase extension activity using substrates with 3′-overhangs of 7- or 23-nucleotides (Fig. 3A). Importantly, the basal and Est1-stimulated DNA extension products from either substrate were telomerase-dependent since RNaseA treatment abolished activity (Fig. 3A) (15). The products below the + 1 position for each DNA substrate are likely produced by a previously described telomerase-associated endonuclease activity (16). Given the minimum 15-base requirement for Est1 DNA binding, it is unlikely that Est1 associates with the DNA during telomerase-mediated DNA lengthening.
Fig. 3.
Est1 upregulates the telomerase DNA extension activity independent of DNA interactions. (A) The capacity of Est1 (50 nM) to stimulate telomerase-mediated DNA extension using substrates with either 7- or 23-base single-stranded 3′-overhang lengths was determined. To demonstrate that the DNA extension products were telomerase-dependent the telomerase extracts were pretreated with RNaseA, a DNA substrate with a C-rich 3′-overhang was incorporated or extract prepared from tlc1Δ yeast was used in the extension reactions, as indicated. (B) The Est1 ability to affect free or DNA-bound telomerase was determined by altering the order of addition for Est1/BSA protein (P) (50 nM), 7-base DNA substrate (D) and telomerase extract (T). The two components that were preincubated (2 min r.t.) are group followed by the factor that was added with free nucleotide to initiate the reaction. The positions of the loading control primer and the various + 1 DNA extension products are marked.
To directly test whether Est1-DNA interactions influence telomerase regulation before assembly with telomeric DNA, we performed order-of-addition experiments (Fig. 3B). We preincubated Est1/BSA protein (P) with the DNA (D) before adding telomerase (T), telomerase with the DNA before adding the Est1/BSA or Est1/BSA with telomerase before adding the DNA substrate. The reactions were initiated upon addition of the third component and free nucleotide. For the reactions where telomerase was preincubated with the DNA substrate, the immobilized DNA-bound complexes were washed to remove any unbound telomerase before initiating the reactions. Of note, similar reactions were performed with the 23-base DNA substrate where the DNA-bound complexes were washed to remove any unbound Est1/BSA/telomerase before initiating the reactions and comparable stimulatory effects were observed. Since Est1 stimulated the DNA extension activity equivalently before or after telomerase DNA assembly, we suggest that Est1 tethers to DNA-bound telomerase and modulates the enzymatic activity independent of direct DNA binding by Est1.
Est1 Point Mutants Display Differential Activities.
Genetic analysis has generated a large Est1 point mutant collection that have been speculated to affect telomere maintenance by a number of means including altered Cdc13 association, defective TLC1 RNA interactions or declined telomeric DNA binding (4, 17). In an attempt to better understand Est1 function, we have purified and characterized a select set including Est1–38, Est1–41, Est1–42, Est1–49, Est1–50, and Est1–60. Prior in vivo analysis showed that Est1–38, Est1–41, and Est1–42 have lowered telomerase interactions but only altered telomeric DNA length slightly, Est1–49 and Est1–50 on the other hand displayed near wild-type telomerase binding but shortened telomeric DNA and Est1–60 does not support telomere DNA maintenance as it has been speculated to no longer interact with Cdc13 (4, 17).
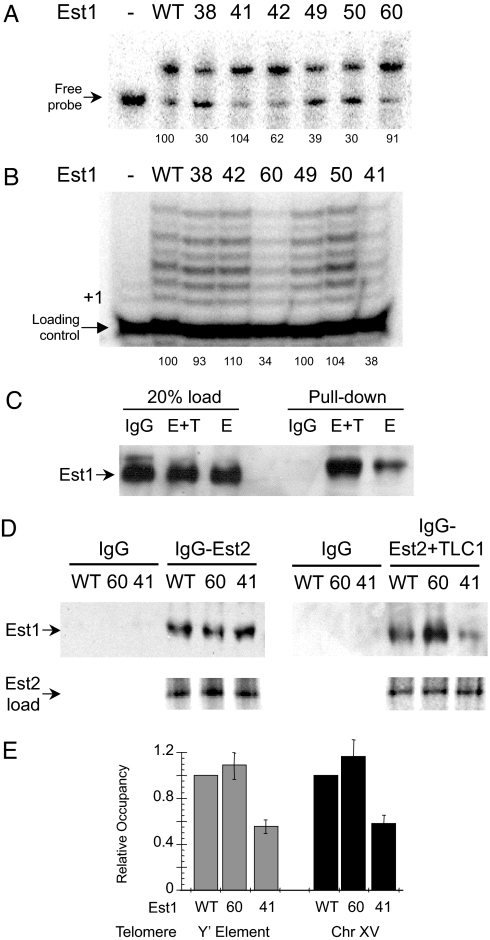
We found that Est1–38, Est1–42, Est1–49, and Est1–50 had decreased RNA binding (≈30, 62, 39, and 30% of wild type) and that Est1–38 and Est1–50 had declined DNA binding (≈25 and 43% of wild type) and whereas the remaining mutants were functionally comparable to wild-type Est1 for nucleic acid interactions (Fig. 4A and Table S1). Notably, Est1–38, Est1–49, and Est1–50 comparably stimulated telomerase activity relative to wild-type Est1 (Fig. 4B). In contrast, Est1–60 and Est1–41 had decreased (≈34% and 38% of wild type, respectively) abilities to activate telomerase (Fig. 4B); yet, neither showed an apparent nucleic acid binding defect. Thus, in agreement with our wild-type protein data, Est1 nucleic acid binding and telomerase activation potentials are non-correlative.
Fig. 4.
Defects in nucleic acid binding and telomerase stimulation are non-correlative for Est1 derivatives. The Est1 point mutants Est1–38, Est1–41, Est1–42, Est1–49, Est1–50, and Est1–60 were purified to near homogeneity and characterized. (A) The RNA binding activity of each derivative (100 nM) was determined by EMSA using a radiolabeled TLC1 RNA (250 pM). (B) The ability of the various Est1 proteins (50 nM) to enhance telomerase DNA extension activity was examined. The positions of + 1 and the loading control primer are specified. (C) The capacity of Est1 to interact with Est2 independent of Tlc1 RNA was examined in vitro. Est1 (500 ng) was added to either IgG Sepharose alone or IgG Sepharose preincubated with either ProA-Est2 + Tlc1 (E+T) or ProA-Est2 (E). Est1 loading (20% input) control data are shown. (D) The abilities of wild-type Est1, Est1–60 and Est1–41 to interact with Est2 in the absence or presence of TLC1 RNA were determined using Est2 protein immobilized on IgG Sepharose. Representative data from three independent experiments is shown. For the pull-down experiments the Est1 proteins were visualized by Western blot analysis using an antibody directed against the His6-tag. Est2 loading control data are shown where the [35S]Met-labeled Est2 translation products were resolved by SDS/PAGE and visualized using a PhosphoImager. (E) Telomere association by the Est1 derivatives was monitored using the ChIP assay. Est1 residencies at a telomere population were determined using PCR and primers select for a subtelomeric Y' element found at 11 chromosomal termini (gray bars) or at a single telomere using primers specific for a subtelomeric region of chromosome XV (black bars); values were normalized to the signal from an internal non-telomeric DNA. All data represent average values (mean ± SD) from three independent assays.
Since RNA and DNA binding did not have a significant influence on telomerase regulation by Est1 we suspected a direct protein-protein interaction between Est1 and Est2 was required. To determine if Est1 can interact with Est2 independent of TLC1 RNA, we synthesized Est2 using rabbit reticulolysate in the absence of TLC1 RNA. The Est2 protein was produced as an amino-terminal protein A fusion protein to allow isolation with IgG Sepharose (18). We found that Est1 can interact with Est2 independent of Tlc1 RNA (Fig. 4C). Our data agree with prior in vivo studies showing that Est1 and Est2 interact in a tlc1-null background and that human Est1A associates directly with the reverse transcriptase subunit in vitro (5, 11). However, the interaction is less efficient in the absence of Tlc1, which suggests that the Tlc1 RNA fosters assembly of the complex, as has been previously suggested (12, 13).
Next, we examined the interaction patterns for wild-type Est1, Est1–60, or Est1–41 (Fig. 4D). We did not observe a significant change in association between the Est1 derivatives and Est2 alone. If, however, we synthesized the Est2 in the presence of TLC1 and then performed the pull-down we observed an enhanced Est1–60 association (≈2.5-fold increase) and a declined Est1–41 interaction (≈40%) relative to wild-type Est1 (Fig. 4D). The reduced telomerase association by Est1–41 is comparable to the decreased in vivo interactions previously observed for Est1–41 and telomerase (17). Since neither Est1 mutant displayed an inherent RNA binding defect (Fig. 4A), we suggest that TLC1 binding has an allosteric effect on either or both Est1 and Est2, which alters Est1 interactions with the core enzyme.
To determine the relative telomere occupancy levels of Est1, Est1–41, and Est1–60 in vivo we used the chromatin immunoprecipitation assay. We found a similar telomere interaction pattern for the Est1 derivatives as we had observed for RNA binding—Est1–41 levels were reduced but Est1–60 and wild-type Est1 occupancies were comparable (Fig. 4E). While our in vitro and in vivo data are consistent, the Est1–60 telomere binding was unexpected since prior genetic work predicted that Est1–60 (K444E) would not associate due to a disrupted salt-bridge interaction with Cdc13 (4). Based upon the presented data, we suggest that the reported telomeric DNA shortening in the est1–60 yeast involves a defect in telomerase stimulation, in addition to a perturbed interaction between Cdc13 and Est1 that is essential for proper telomerase function at a telomere. To understand if the Est1–60 mutation affects functional interactions involving Cdc13 and telomerase we investigated the combinatorial effects of Est1 and Cdc13 on telomerase DNA extension activity.
The Est1-Cdc13 Interaction Is Critical for Modulating Telomerase DNA Extension Activity.
Prior genetic work showed that the telomere DNA length defect in the est1–60 background is alleviated by the second-site suppressor mutation cdc13–2 (4). Therefore, if the inability of Est1–60 to fully stimulate telomerase in vitro is related to the in vivo telomere phenotype then inclusion of the Cdc13–2 mutant should reestablish proper telomerase upregulation with Est1–60 in vitro. To determine whether interactions between Est1 and Cdc13 impact telomerase we tested the individual and combined effects of purified Est1 and Cdc13 proteins on telomerase DNA extension activity in vitro. Of note, we found that Cdc13 was capable of stimulating telomerase DNA extension activity independent of Est1 and we have followed this observation in a separate study (19).
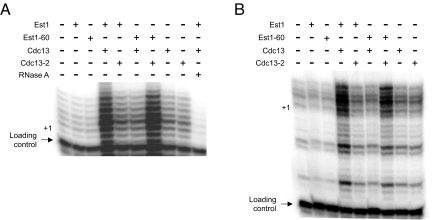
Although DNA extension was enhanced using relatively high Est1 or Cdc13 protein concentrations (0.1–1.0 μM), we did not observe a significant combinatorial effect of Est1 and Cdc13 on telomerase activity at the relatively high protein concentrations. However, at protein amounts (2.5 nM) suboptimal for activation by either individual protein the combination of Est1 and Cdc13 led to upregulation of telomerase isolated from wild-type yeast (Fig. 5). While the cellular level of Est1 (a telomerase holoenzyme subunit) has not been determined, if we assume an amount comparable to the TLC1 RNA core subunit than the Est1 cellular concentration would be in the lower nanomolar range (20). Significantly, the combinatorial Est1-Cdc13 effect was not apparent when one of the wild-type proteins was substituted with a mutant derivative. Yet, in the presence of both Est1–60 and Cdc13–2 telomerase activation was apparent (Fig. 5). Since the combinatorial Est1-Cdc13 effect was apparent using either a short (7-base) or long (23-base) 3′-overhang substrate, we suggest that DNA binding by Est1 and Cdc13 is not imperative to stimulate the DNA-bound telomerase enzyme. Hence, in a manner paralleling the in vivo phenotypes the Est1–60 and Cdc13–2 mutants compensate and reestablish activity comparable to the wild-type condition. While the interaction between Est1 and Cdc13 might influence a number of telomere events, our data demonstrate that the association is minimally required to properly control the enzymatic function of telomerase.
Fig. 5.
The Est1 and Cdc13 association impacts telomerase activation. The influence of purified Est1, Est1–60, Cdc13, or Cdc13–2 proteins (2.5 nM) alone or in the indicated combinations on telomerase DNA extension activity was determined using DNA substrates with either a 7-base (A) or 23-base (B) single-stranded 3′-overhang and telomerase extract prepared from wild-type yeast. The positions of the loading control primer and the + 1 DNA extension product are marked.
Discussion
Est1 is a fundamental telomere DNA maintenance factor that is conserved from yeast to human (10, 21). Genetic experiments in yeast have suggested an Est1-role in both recruiting and activating telomerase (3). Yet evidence demonstrating a direct Est1 regulatory function with telomerase had been lacking. Here we show that purified Est1 protein stimulates telomerase DNA extension activity in vitro, which parallels recent work with the human factors (11). However, in contrast to prior suggestions, the direct Est1 regulatory mechanism does not significantly rely on either telomeric DNA or TLC1 RNA binding.
While Est1 TLC1 interactions are dependent upon the bulged-stem loop structure both in vitro and in vivo (Fig. 2) (12, 13), alteration of the bulged-stem loop only had a minor effect on telomerase stimulation in vitro suggesting Est1 recognizes TLC1 but the association is not a major activation-determinant. Given the requisite for the TLC1 bulged-stem loop for telomerase binding in vivo, why isn't there a more significant effect in vitro? Perhaps the RNA interaction has an important role during telomerase holoenzyme assembly that is essential for efficient telomerase action in vivo but is dispensable in vitro. Presumably holoenzyme assembly is required for efficient telomere-recruitment in vivo; the presented in vitro assays likely are not impacted by recruitment events. The Est1 association with TLC1 RNA in the absence of the Est2 protein indicates that the Est1-TLC1 interaction might represent the first step in holoenzyme assembly in vivo (14).
In addition to RNA interactions, it has been suggested that single-stranded DNA binding by Est1 might contribute to telomerase modulation (9). However, Est1 does not appear to rely on DNA interactions to affect telomerase in vitro (Fig. 3 and Fig. S2). Perhaps the Est1 DNA binding ability provides a means to telomere-associate independent of telomerase, but once telomerase arrives Est1 switches ligands and binds to TLC1—comparable DNA/RNA swapping has been previously postulated for other factors (22). Although the necessity of the replication protein A (RPA) for Est1 telomere loading suggests Est1 DNA binding activity is insufficient for telomere interactions (23). Alternatively Est1 DNA binding might be important for other non-telomeric cellular functions.
In our telomerase regulatory model (Fig. S3), Est1 joins the core enzyme by selectively binding the TLC1 bulged-stem RNA loop—assembly of the holoenzyme before telomere association is requisite for downstream control events including telomerase upregulation (8, 12, 13). Once the holoenzyme engages telomeric DNA the contacts between Est1 and Est2 are critical for efficacious telomerase DNA extension activity. In addition to Est1, telomerase regulation in vivo requires Cdc13—minimally interactions between Est1 and Cdc13 are used to modulate the holoenzyme. Although it had been suggested that a salt-bridge linkage between Est1 and Cdc13 is required for Est1 nucleation at a telomere, it appears that this is not the case. Prior studies had interpreted the telomeric DNA shortening in the est1–60 background as an inability to recruit Est1 to the telomere and the phenotype suppression in est1–60 cdc13–2 yeast further supported this notion (4). However, the Est1–60 protein displayed no apparent telomere recruitment defect (Fig. 4D). In conjunction with previous reports showing that Est1 interacts at telomeres in both wild-type and cdc13–2 yeast (7, 8), the necessity for the Est1(K444)-Cdc13(E252) salt-bridge for Est1 telomere recruitment is unlikely. While interactions between Est1 and Cdc13 might be essential for stabilizing telomerase at a telomere in vivo, our studies have not addressed this potential function.
Minimally, the Est1-Cdc13 salt-bridge is important for regulating telomerase enzymatic activity (Fig. 5). While the absolute Est1 requisite for telomere DNA maintenance in vivo can be bypassed using a Cdc13-Est2 fusion protein, the significant reduction in telomeric DNA lengthening upon est1 deletion in the Cdc13-Est2 fusion background supports an Est1 role independent of telomere recruitment (3). The covalent linkage between the telomerase Est2 protein subunit and Cdc13 likely bypasses any potential telomere stabilization role by Est1. The inability of Est1–60 to stimulate telomerase in vitro despite its capacity to interact with Est2 suggests that the mutant does not properly shift telomerase to a stimulated state (Fig. 4). Perhaps in the presence of the Cdc13–2 protein, which suppresses the Est1–60 defect (Fig. 5), the conformational switch in telomerase occurs. We suggest the cdc13–2 and est1–60 mutations separately interfere with a critical contact point between the two proteins that is necessary for telomerase stimulation and for potentially stabilizing telomerase to a telomere. Presumably, in the presence of both protein mutants the charge-swap reestablishes the Est1-Cdc13 interaction and permits proper telomerase control. Further studies will be necessary to delineate the potential structural changes that occur within the various telomere-binding proteins to mediate these regulatory steps. Nevertheless, the presented data provide an understanding of Est1-telomerase control and a foundation for future mechanistic studies on the telomerase holoenzyme.
Materials and Methods
Est1 and Cdc13 Purification.
The Est1 proteins were expressed as amino-terminal (His)6 fusions using a T7 expression vector (pET28) in codon optimized Rosetta Escherichia coli cells (Novagen Inc.). At 18 °C, 12 L LB media were seeded with transformants from an overnight culture to an O.D.595 = 0.1 and induced for 30 min with IPTG after the O.D.595 = 0.3. The cultures were clarified by centrifugation, resuspended in ice-cold 1× Talon binding buffer, lysozyme (0.5 μg/mL) was added and the samples were nutated for 30 min at 4 °C before snap freezing. Est1 was purified as a soluble protein using metal affinity chromatography (Talon resin) as per the manufacturer's instructions (Clontech Inc.). The eluted protein was diluted 2:1 with TEN0G Buffer (20 mM Tris pH 6.9, 0.1 mM EDTA, and 10% glycerol), applied to ResourceQ resin (GE Healthcare Inc.), the column was washed with TEN0.45G buffer (450 mM NaCl) and the protein was eluted with a 450–1,000 mM NaCl gradient. The eluted protein was concentrated to approximately 100 μL using a microconcentrator, 2 mL TMG30 (sodium acetate pH 7.0) was added to adjust the pH and salt concentration, the volume was reduced to approximately 50 μL, cold 50% glycerol (50 μL) was added, and the sample was stored at 4 °C. The soluble purified protein identity was confirmed by mass spectrometry (Fig. S4).
The Cdc13 proteins were expressed as amino-terminal (His)6 fusions using T7 expression vectors (pET28) in streptomycin pseudoresistant E. coli cells CH184 (19). Expression in CH184 was essential for obtaining sufficient quantities of soluble, purified full-length Cdc13 and Cdc13–2. CH184 transformants were grown overnight at 24 °C in LB media supplemented with streptomycin (0.2 mg/mL) and kanamycin (35 μg/mL), LB media supplemented with kanamycin (35 μg/mL) was seeded to an O.D.595 = 0.1 with cells from the overnight culture, the cells were grown to an O.D.595 = 0.4, protein expression was induced with IPTG and the cells were incubated for an additional 4 h. After IPTG induction the cultures were clarified by centrifugation, resuspended in ice-cold 1× Talon binding buffer (Clontech Inc.) supplemented with 1% Nonidet P-40, the cells were lysed and the Cdc13 proteins were purified using metal affinity chromatography (Talon resin) as per the manufacturer's instructions (Clontech Inc.) followed by further resolution over MonoQ anion exchange resin and a Superdex-200 size exclusion column (GE Healthcare Inc.).
Telomerase Extract.
Telomerase extracts were prepared using wild-type (YPH499) or est1Δ Saccharomyces cerevisiae as described (24). In brief, yeast extract was sequentially resolved over DEAE and MonoQ resins (GE Healthcare Inc.) using TMG buffer (20 mM Tris pH 8.0, 1.1 mM MgCl2, 0.1 mM EDTA, 1.5 mM DTT, 10% glycerol, 0.1% Triton X-100, and NaOAc pH 5.0). Following elution from the MonoQ resin the telomerase fraction was concentrated to approximately 100 μL using a microconcentrator, 2 mL of TMG30 (sodium acetate pH 7.0) was added to adjust the pH and salt concentration, the volume was reduced to approximately 50 μL, 50 μL of 80% cold glycerol was added, and the mixed sample was stored at −20 °C before quantification of protein and TLC1 RNA levels.
TLC1 Derivatives.
To prepare telomerase extracts with TLC1 RNA derivatives we used an established system (13). In short, yeast (TCy43) expressing wild-type TLC1 from the pTLC1-LYS2-CEN plasmid were transformed with vectors for bulge-delete, stem-disrupt or stem-compensatory TLC1 RNAs. Transformants were replica plated onto α-aminoadipate media to counter select pTLC1-LYS2-CEN. Colonies were used to seed simple media lacking tryptophan (WT TLC1 cultures lacked lysine), yeast were grown at 30 °C, the cells were harvested at O.D.595 = 1.0 and telomerase extracts were prepared. Genomic DNA was prepared from each culture to monitor telomere lengths and, as expected, telomere lengths decreased in the cells expressing the stem-disruption and bulge-deletion TLC1 RNA but not in the other yeast.
Telomerase DNA Extension Assays.
Telomerase DNA extension reactions were performed as described (22). In brief, the DNA extension reactions used MonoQ telomerase extract containing equivalent TLC1 RNA levels in extension buffer (50 mM Tris pH 8.0, 1.0 mM MgCl2, 1.0 mM spermidine, 1.0 mM DTT, 0.5% glycerol, 50 μM dTTP, and 10 mCi [α-32P] dGTP) supplemented with 2.0 pmol telomeric DNA substrate (7-base 3′overhang GTGTGTG; 23-base GTGTGTGGGTGTGGTGGTGTGTG) immobilized on paramagnetic beads (24). For the Est1 order of addition experiments (Fig. 3B) the DNA-bound complexes were washed 3× with nucleotide free extension buffer before adding the indicated third components and free nucleotide. Extension reactions were incubated for 30 min at 30 °C, the beads were collected by magnetic separation, washed 2× with 1× EcoRI buffer, resuspended in 50 μL of 1× EcoRI buffer supplemented with 100 μg/mL BSA and 10 units EcoRI, and incubated 1 h at 37 °C. The cleaved DNA fragments were magnetically separated from the beads, an end-labeled precipitation/loading control primer was added, the DNAs were subjected to ethanol precipitation, the products were reconstituted in formamide/NaOH loading buffer, subjected to denaturing gel electrophoresis and visualized using a Molecular Dynamics PhosphoImager.
RNA EMSA.
To analyze RNA binding activity we exploited the defined TLC1 RNA Est1 binding site (nucleotides 535—707) (12). We prepared T7 in vitro expression constructs in pBLUESCRIPT for the wild-type, bulge-deletion, stem-disruption and stem-compensation TLC1 RNAs by amplifying the central TLC1 region by PCR. The RNA transcripts were synthesized using the T7 Maxishortscript in vitro transcription kit (Ambion Inc.) in the presence of [α-32P] UTP or with unlabeled ribonucleotide for use in the DNA/RNA competition experiments and purified using RNeasy mini kit per manufacturer's instructions (Qiagen Inc.). The transcripts were diluted to 32 nM, heat denatured (5 min at 95 °C) and snap cooled before use. The RNA EMSAs were performed in 10 mM HEPES (pH 7.8), 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 3% Ficoll, heparin (1 mg/mL), and RNasin inhibitor (9). Incubations were for 20 min on ice and the samples were resolved on 6% native polyacrylamide gels using 1× GTG buffer. After resolution, the polyacrylamide gels were dried and the products were visualized with a PhosphoImager.
Est2-Est1 Coprecipitation Assay.
The Est2 protein was generated using the TnT Quick Coupled system per manufacturer's instructions (Promega Inc.) using [35S]-Methionine, pET-Est2 plasmid (T7-ProA Est2) and TLC1 plasmid (T7-Minit 500) where indicated (18). For the coprecipitation, either the Est2 TnT reaction or an equivalent aliquot of TnT Quick Master mix was added to the IgG beads and nutated overnight at 4 °C, the beads were washed 4× with cold TMG-30 buffer, resuspended to a final volume of 30 μL with TMG-30 and split into three 10-μL aliquots. The indicated Est1 proteins (50 ng) were added, the reactions were nutated for 5 min at 4 °C, the beads were clarified (3,000 × g, 1 min, 4 °C), the supernatants were removed, the beads were washed 3× with cold TMG-30 buffer, sample were resuspended in SDS buffer, boiled for 3 min, and resolved on a 12% SDS/PAGE. The Est1 proteins were visualized by immunological detection using anti-His monoclonal mouse antibody (Novagen Inc.).
Chromatin Immunoprecipitation Assay.
The ChIP experiments were performed as previously described (24). To determine the relative telomere occupancy by Est1 derivatives, est1Δ transformants expressing wild-type Est1, Est1–41, or Est1–60 were grown to an O.D.595 = 0.5 before formaldehyde crosslinking and immunoprecipitation. Following the ChIP procedure, target DNA levels were determined by quantitative PCR as per the manufacturer's instructions (Bio-Rad Inc.) using primers specific for a Y' element found within 11 subtelomeric regions, oligonucleotides that amplify a single subtelomeric region on chromosome XV or a primer set specific for an internal control, non-telomeric DNA region (YJL052W). The fold-enrichments were determined by normalizing the ratio of the specific/nonspecific telomeric values at the telomeric DNA targets to the ratio of the specific/nonspecific YJL052W signals.
Supplementary Material
Acknowledgments.
We thank Dr. Victoria Lundblad (Salk Institute For Biological Studies) for the multiple Est1 plasmid constructs; Dr. Jose Barral (University of Texas, Galveston) for the streptomycin resistant bacteria; Dr. Thomas Cech (University of Colorado, Boulder) for the Est2 and TLC1 vectors; Dr. David Zapulla for his technical advice; and Drs. Joachim Lingner, Yun Wu, and Virginia Zakian for helpful comments on the manuscript. This work was supported by a Computational Molecular Biology-National Institutes of Health training grant (to D.C.D.) and Public Service Grant DK074270 (to B.C.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905703106/DCSupplemental.
References
- 1.Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 2.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1993;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 3.Evans SK, Lundblad V . Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 4.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 7.Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 8.Chan A, Boulé JB, Zakian VA. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4:e1000236. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Virta-Pearlman V, Morris DK, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 10.Snow BE, et al. Conservation of the telomerase protein Est1p in humans. Curr Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 11.Redon S, Reichenbach P, Lingner J. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 2007;35:7011–7022. doi: 10.1093/nar/gkm724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol Cell Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulge stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steiner BR, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu H, Xia J, Lue NF. Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol Cell Biol. 2000;20:6806–6815. doi: 10.1128/mcb.20.18.6806-6815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans SK, Lundblad V. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics. 2002;162:1101–1115. doi: 10.1093/genetics/162.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 19.DeZwaan DC, Toogun OA, Echtenkamp FJ, Freeman BC. The Hsp82 molecular chaperone can promote the switch between unextendable and extendable telomere states. Nat Struct Mol Biol. 2009;16:711–716. doi: 10.1038/nsmb.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mozdy AD, Cech TR. Low abundance of telomerase in yeast: Implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner JA. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 22.Suswam EA, Li YY, Mahtani H, King PH. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005;33:4507–4518. doi: 10.1093/nar/gki763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schramke V, et al. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- 24.Toogun OA, Zieger W, Freeman BC. The p23 molecular chaperone promotes functional telomerase complexes through DNA dissociation. Proc Natl Acad Sci USA. 2007;104:5765–5770. doi: 10.1073/pnas.0701442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.