Abstract
“Soft” and “hard” are the two main market classes of wheat (Triticum aestivum L.) and are distinguished by expression of the Hardness gene. Friabilin, a marker protein for grain softness (Ha), consists of two proteins, puroindoline a and b (pinA and pinB, respectively). We previously demonstrated that a glycine to serine mutation in pinB is linked inseparably to grain hardness. Here, we report that the pinB serine mutation is present in 9 of 13 additional randomly selected hard wheats and in none of 10 soft wheats. The four exceptional hard wheats not containing the serine mutation in pinB express no pinA, the remaining component of the marker protein friabilin. The absence of pinA protein was linked inseparably to grain hardness among 44 near-isogenic lines created between the soft variety Heron and the hard variety Falcon. Both pinA and pinB apparently are required for the expression of grain softness. The absence of pinA protein and transcript and a glycine-to-serine mutation in pinB are two highly conserved mutations associated with grain hardness, and these friabilin genes are the suggested tightly linked components of the Hardness gene. A previously described grain hardness related gene termed “GSP-1” (grain softness protein) is not controlled by chromosome 5D and is apparently not involved in grain hardness. The association of grain hardness with mutations in both pinA or pinB indicates that these two proteins alone may function together to effect grain softness. Elucidation of the molecular basis for grain hardness opens the way to understanding and eventually manipulating this wheat endosperm property.
Keywords: Triticum aestivum L., grain softness protein
Wheat (Triticum aestivum L. em Thell.) is the single most important food crop in the world. Although best known for its gluten-forming properties, the primary basis for discriminating different end uses in wheat is not protein content but grain hardness. Grain hardness refers to the texture of the kernel (caryopsis), that is, whether the endosperm is physically hard or soft. Nearly all of the world production and trade in wheat (≈550 and 100 mmt annually, respectively) is identified as being either soft or hard. Generally speaking, hard wheat is used for bread whereas soft wheat is used for cookies, cakes, and pastries (1). This difference in grain texture results from the expression of one major gene, designated Hardness (Ha) (2, 3), located on the short arm of chromosome 5D (4, 5). The presence of a single major gene is contrary to the allohexaploid nature of wheat (T. aestivum) (2n = 6x = 42 chromosomes; genomes AABBDD) because most genes exist in triplicated homoeologous sets, one from each genome. Alleles of the hardness gene are present on the 5A and 5B chromosomes of hexaploid wheat but are not expressed. For this reason, durum wheats (T. turgidum L. var. durum) (2n = 4x = 28 chromosomes; genomes AABB), which lack the D genome, are generally harder textured than hard hexaploid wheat.
A 15-kDa marker protein for grain softness, termed “friabilin,” is present on the surface of water-washed starch from soft wheats in high amounts and on hard wheat starch in small amounts and is absent on durum wheat starch (6). N-terminal sequence analysis of friabilin indicated a mixture of two or more discrete polypeptides (7, 8). The two major component polypeptides were found to be identical to the two lipid binding proteins termed “puroindolines” (9), pinA and pinB, respectively. The transcripts of pinA and pinB are controlled by chromosome 5D (10). To date, no evidence exists that friabilin consists of any additional proteins.
PinA and pinB are unique among plant proteins because of their tryptophan-rich, hydrophobic domains, which have affinity for binding lipids (11, 12). The association of friabilin (pinA and pinB) with the surface of the starch granule apparently is mediated by polar lipids. In fact, the occurrence of membrane structural lipids, glyco- and phospho-lipids, with the surface of water-washed starch follows that of friabilin (13): High amounts are present on soft wheat starch, low amounts are present on hard wheat starch, and there is none on durum.
A glycine-to-serine change (Gly-46 to Ser-46) in the tryptophan-rich domain of pinB recently was reported in two hard wheat varieties (10). This sequence change was linked inseparably to grain hardness and could lessen the strength of lipid binding because of the inherent decrease in hydrophobicity of a glycine to serine change (14). The complete linkage between this mutation in pinB and hard grain texture among 83 chromosome 5D recombinant substitution lines suggested that this protein is involved in the control of grain softness (10). The change present in pinB was identical in two different hard textured varieties and absent in two soft textured reference varieties. This change simply could have represented a tight linkage between the friabilin component pinB and the Hardness gene. In this paper, we characterize additional hard textured varieties and show that they either contain the serine mutation in pinB or exhibit an absence of pinA protein and transcript. The evidence suggests that mutations in either component of friabilin are responsible for the manifestation of grain hardness and that the Hardness phenotype is due to the tightly linked genes pinA and pinB. In addition, we present evidence that a previously reported grain softness protein (15, 16) (GSP-1) reported to be linked to grain texture is not associated with grain hardness.
MATERIALS AND METHODS
Plant Culture and Measurement of Grain Hardness.
Soft and hard near isogenic lines (NILs) of Falcon and Heron (17) were grown near Lind, WA in the summer of 1995. Grain hardness was measured by using a near-infrared (NIR) spectrometer (model 450, Technicon, Tarrytown, NY), on UDY ground grain (Method 39–70A; American Association of Cereal Chemists 1995). Single kernel hardness readings (Single Kernel Characterization System) were obtained by analyzing 300 kernel samples of grain for hardness by using the Perten Model Single Kernel Characterization System 4100 following the manufacturer’s suggested operating procedure (Perten Instruments, Reno, NV).
DNA Isolation and PCR Amplification of PinA, PinB, and GSP-1.
DNA was isolated by the procedure of Dellaporta et al. (18). Amplification of pinB sequences specific for the Gly-46 or Ser-46 is described elsewhere (10). Amplification of pinA sequences were performed as described (9). Annealing temperature for both sets of primers was maintained at 58°C. Individual PCRs were replicated two or more times. A GSP-1 probe was amplified by reverse transcription PCR from RNA extracted from kernels of Chinese Spring 14 days after flowering. The 5′ primer consisted of the DNA sequence 5′-GTAGTGAGCACTACTATTGC-3′, and the 3′ primer was the reverse complement of 5′-GAGCCTTCCCTCCAAGTGC-3′. The PCR annealing temperature was 58°C. These primers amplified an internal 400-bp fragment described as GSP-1b (15). This 400-bp fragment was used to probe at high stringency total RNA from Falcon/Heron NILs and total RNA from Chinese Spring chromosome 5D recombinant substitution lines (10).
Northern Blot Analysis.
RNA isolation, preparation of Northern gel blots, and probing were done by standard methods as described (10). PinA, pinB, and GSP-1 probes were prepared by the random primer method (GIBCO/BRL) to a specific activity of >1 × 109 cpm/μg DNA. Two high stringency washes at 67°C were done for each Northern blot before autoradiogram exposure.
Isolation of Triton X-114 Soluble Proteins and Protein Electrophoresis.
Triton-soluble proteins were isolated by phase partitioning of Triton X-114 (19). Crushed whole kernels were added to 1% (vol/vol) Triton X-114 in Tris-buffered saline (10 mM Tris/150 mM NaCl, pH 7.5) at 4°C and were mixed for 30 min. After a brief centrifugation (10,000 × g, 5 min), the supernatant was transferred to 37°C for 30 min and re-centrifuged. The lower detergent phase was transferred to a new tube, and the phase partitioning was repeated. After phase partitioning, the proteins in the detergent rich phase were precipitated with 80% (vol/vol) acetone. Pellets were washed with acetone and then ether and dried. SDS sample buffer (no added reducing agents) was added to adjust the protein load to 1 mg whole-kernel equivalents/lane. SDS/PAGE was performed by standard methods by using 13.5% T and 2.6% C and 0.75-mm thick SE600 gels (Bio-Rad), and gels were silver stained by a trichloroacetic acid fixation method (7).
RESULTS
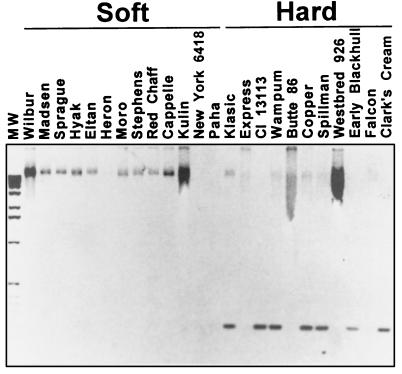
A primer designed to recognize the serine sequence change present in pinB of the hard wheat cultivars Wanser and Cheyenne was used on 13 additional soft and 11 additional hard-textured wheats (Fig. 1). The serine mutation in pinB is a change of amino acid 46 glycine (9) (GGC) to serine (AGC). This sequence change was exploited to make glycine- or serine-specific PCR primers (10). “Hard” or serine-specific 3′ pinB primers end with T, and the “soft” or glycine-specific primer 3′ ends with C. A size-specific product (250 bp) detected with the hard serine specific primer is taken as indicating the presence of serine in position 46. Although the glycine-to-serine change in pinB is quite common among hard wheats (Fig. 1), some exceptions were found. The majority (7 of 11) of the hard wheats exhibited a serine-specific 250-bp band, whereas four hard wheat varieties, Express, Butte 86, Westbred 926, and Falcon, did not. Each of these four exceptions exhibited a glycine-specific “soft” wheat pinB band (data not shown). All 13 soft wheats produced the glycine-specific band and no serine-specific band (data not shown). Amplification of the entire pinB coding sequence was possible for each of the genomic DNAs used in Fig. 1.
Figure 1.
A Gly-46 to Ser-46 sequence change in pinB is common among hard wheat varieties and is absent in soft wheat varieties. Gel photo (negative image) of PCR products obtained by using a serine-specific PCR primer on genomic DNA of soft and hard hexaploid wheat varieties. Soft (S) indicates that the variety has soft-textured grain, and hard (H) indicates hard-textured grain. Lane marked MW is a 1 kb molecular mass marker ladder.
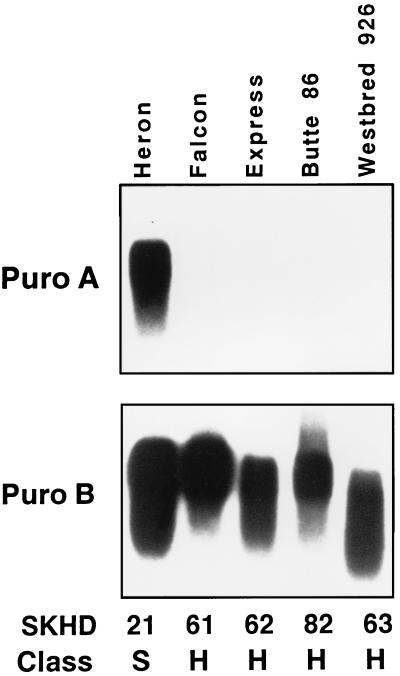
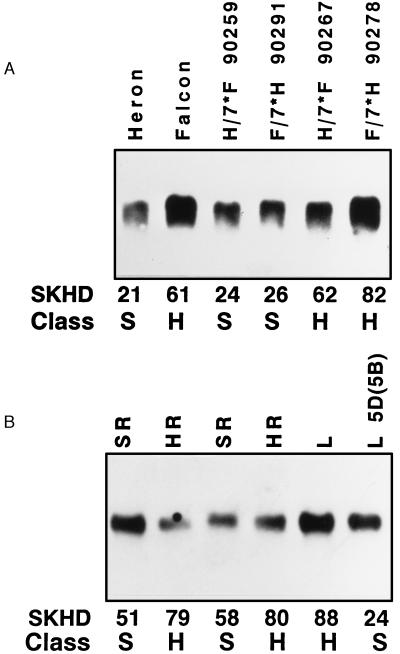
Friabilin component transcript and protein analysis was performed on the four hard textured wheat varieties that did not contain the glycine-to-serine sequence change in pinB. Fig. 2 shows the results of probing Northern blots prepared with total RNA from developing seeds of the soft textured variety Heron and the four glycine-type hard-textured varieties. None of these four hard varieties contains any detectable transcripts for pinA. Transcript levels of pinB in each of these hard genotypes were similar to the soft genotypes.
Figure 2.
PinA transcripts (Puro A) are absent in hard wheat varieties that lack the pinB Ser-46 mutation. PinB transcripts (Puro B) are present in all wheats irrespective of hardness. Each lane was loaded with 10 μg of total RNA isolated from immature seeds at 14 days after flowering. Replicate blots were probed with either pinA or B. SKHD and Class represent the Single Kernel Characterization System phenotypic grain hardness reading and hardness class, respectively (S, soft class; H, hard class).
Friabilin components can be separated on SDS/PAGE gels into two distinct components (7), tentatively identified as pinA and pinB. Separation of the friabilin components from Chinese Spring (soft), Chinese Spring disomic chromosome 5D substitution lines, CS-CNN DS5D (hard) (Cheyenne hard wheat variety as donor of the pair of 5D chromosomes), Heron, and Falcon was performed. Close examination of the bands shown in Fig. 3 reveals that the upper band is missing in Falcon and is most likely pinA because the absence of this friabilin band corresponds to the absence of the pinA transcript. This pinA band is present in all soft wheats such as Chinese Spring and Heron and all pinB serine-type hard wheat mutants, such as Cheyenne and the hard Chinese Spring/Cheyenne disomic substitution derivative, CS-CNN DS5D. The protein band corresponding to pinA was also absent from Express, Butte 86, and Westbred 926 (data not shown). Langdon durum lacks any trace of friabilin protein, whereas substituting the pair of 5B chromosomes of Langdon with the 5D from Chinese Spring LGD-CSDS 5D(5B) restores friabilin and grain softness (Fig. 3).
Figure 3.
Friabilin component pinA is absent in Falcon hard wheat variety but present in the soft wheat varieties Chinese Spring (CS), Heron, and the disomic substitution line of Langdon durum containing the 5D chromosome of Chinese Spring [LGD-CSDS5D(5B), abbreviated L5D(5B)] and the serine-type hard wheat disomic substitution line of Chinese Spring containing the 5D of Cheyenne, a hard wheat [CS-CNN DS5D, abbreviated CS (CNN5D)]. Langdon (LGD, abbreviated L) durum has neither pinA nor pinB. Photo of silver-stained SDS/PAGE gel of 14- to 16-kDa region showing Triton X-114-isolated starch surface proteins. Arrows indicate position of friabilin component proteins pinA and pinB (A and B, respectively).
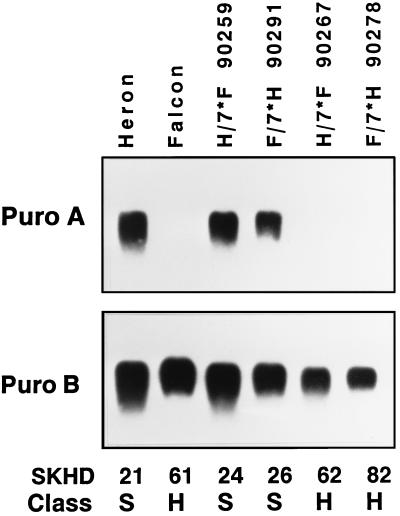
One of the four hard wheat varieties lacking the pinB serine mutation, Falcon, was chosen for further study. Complementary soft and hard NILs have been derived by using Falcon as the hard allele donor and Heron as the soft allele donor (2). In this set of NILs, soft and hard lines exist where either Falcon or Heron served as the recurrent parent. NILs of each set were created by seven generations of backcrossing and selecting for either soft or hard textured grain. Transcript levels of pinA and pinB were studied by using total RNA blots prepared from developing grain of Heron, Falcon, and four NILs. The NILs shown in Fig. 4 represent one hard and one soft NIL each created by using either Falcon or Heron as the recurrent parent. PinA transcripts were undetectable from Falcon and the two hard-textured NILs (Fig. 4). Transcript levels of pinA were similar in the two soft NILs as compared with the original soft parent Heron. Transcript abundance of pinB was similar in each of these varieties.
Figure 4.
PinA transcripts (Puro A) are absent in the hard wheat variety Falcon (F) and two hard NILs derived from Falcon and Heron (H) soft wheat, one each, after 7 backcrosses (90267 and 90278). PinA transcripts are present in Heron and the two soft NILs (90259 and 90291). PinB transcripts (Puro B) are present in all wheats irrespective of hardness. Each lane was loaded with 10 μg of total RNA isolated from immature seeds at 14 days after flowering (DAF). Replicate blots were probed with either pinA or pinB. SKHD and Class are as described in Fig. 2.
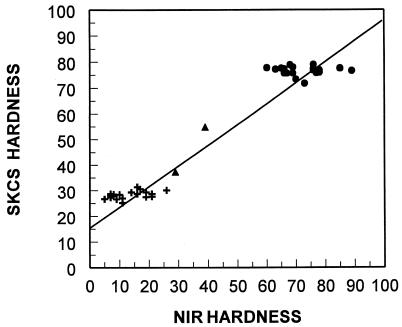
We tested the linkage between the presence or absence of pinA protein and grain softness/hardness. Each of the 44 Falcon/Heron hard/soft NILs was characterized by separating friabilin component proteins on SDS/PAGE and scoring presence of pinA protein. The NILs also were characterized as to pinA gene content based on the capability of amplifying pinA from genomic DNA. Individual lines were classified as being Falcon type with pinA absent (null) or Heron type with pinA present. A graph of phenotypic grain hardness of these NILs by two different methods is shown in Fig. 5 with the pinA type indicated. Two NILs were found to be mixtures of seeds containing pinA and those lacking the protein. Ten individual kernels each of these two NILs (AUS 90077 and AUS 90254) were assayed for the presence of pinA. NIL AUS 90077, NIR hardness of 39, had six of 10 kernels containing pinA. Assay of NIL AUS 90254, NIR hardness of 29, showed that seven of 10 kernels contained pinA. Based on the intermediate hardness values of these two NILs, the percent mixture closely fits the expected frequency. {Soft NIL average NIR equals 15. Hard NIL average NIR equals 71. NIL 90077 expected equals [(6 × 15) + (4 × 71)]/10 = 37.4. NIL 90254 expected equals [(7 × 15) + (4 × 71)]/10 = 31.8.} The observed percent mixture for each of these NILs then likely explains their somewhat intermediate hardness values. Consequently, there was no recombination between the presence of pinA and grain softness in this set of genetic stocks.
Figure 5.
No evidence for recombination between pinA and grain softness. Phenotypic grain hardness of 44 Falcon/Heron hard/soft NILs as measured by Single Kernel Characterization System (SKCS) vs. NIR hardness. Presence or absence of pinA is as shown (+, pinA present; •, pinA absent; ▴, NILs consisting of physical mixtures of seeds containing and lacking pinA).
Additional genes, termed “GSP-1,” have been suggested to be involved in grain softness (15, 16). A GSP-1-related clone, SR3.1, detects a restriction fragment length polymorphism that apparently is linked with grain hardness (16); however, no transcript or gene expression analysis has been reported. We performed Northern analysis on Heron, Falcon, and the four hard/soft NILs used in our experiments and found no consistent transcript level differences for GSP-1 related to grain hardness (Fig. 6A). Additionally, four individual hard and soft recombinant lines derived from Chinese Spring and CS-CNN DS5D were examined (Fig. 6B). As with the Falcon/Heron NILs, GSP-1 transcripts were present irrespective of grain hardness. Langdon durum and the soft disomic substitution lines LGD-CS DS5D(5B) also both had GSP-1 transcripts present (Fig. 6B). Although GSP-1 has been mapped physically to the short arm of each of the three group 5 homologous chromosomes (20), transcripts are not controlled solely by 5D. Based on these data, we consider a direct role of GSP-1 in effecting grain softness unlikely.
Figure 6.
Presence of GSP-1 transcript is not associated with grain hardness and is not controlled by chromosome 5D. (A) Transcript levels of GSP-1 in the soft wheat variety Heron, the hard wheat variety Falcon, and two soft and two hard NILs derived, one of each hardness class, from Falcon and Heron after seven backcrosses. (B) Transcript levels of GSP-1 in soft and hard chromosome 5D recombinant lines (SR and HR, respectively) derived from Chinese Spring and the hard disomic substitution line (CS-CNN DS5D), the durum variety Langdon (L), and the soft disomic substitution line LGD-CSDS 5D(5B) [abbreviated, Langdon (L5D(5B)]. Each lane was loaded with 10 μg of total RNA isolated from immature seeds at 14 days after flowering, and the blots were probed with a GSP-1-derived probe. SKHD and Class are as described in Fig. 2.
DISCUSSION
In each of the 11 hard varieties presented in Fig. 1, a mutation in either pinA or pinB was found. The results presented here and elsewhere (10) characterize 13 hard textured wheat varieties. A glycine-to-serine mutation is present in pinB in nine of the 13 hard wheats. The four remaining varieties have mutations in the other component of friabilin. As such, the two components of friabilin together appear to comprise the Hardness gene. The specific molecular role(s) of friabilin proteins in controlling grain softness is not known. However, among all soft grain members of the Triticeae tribe surveyed, friabilin is known to be present (8, 21). It is possible that the friabilin component genes pinA and pinB evolved to effect soft texture together. A mutation in either gene, as demonstrated here, is sufficient to produce a hard genotype and phenotype. The more common mutation, occurring in 9 of 13 hard wheat varieties, is the serine mutation in pinB. Each of the remaining four hard wheats contain apparent null mutations for pinA. We suggest the new molecular marker and allelic designations of Pina-D1a for the “wild-type” pinA glycine-type allele present in Chinese Spring and Pina-D1b for the null allele present in the above identified hard wheats such as Falcon. These designations for wild type follow the revised Guidelines for Nomenclature of Biochemical/Molecular Loci in Wheat and Related Species (22) and are consistent with the nomenclature established for the pinB alleles (10).
The difference in presence/absence of pinA in the Falcon/Heron NILs was independent of genetic background because results were consistent for the two soft NILs (one each, Falcon and Heron backgrounds) and the two hard NILs (one each, Falcon and Heron backgrounds). These NILs were created by seven generations of backcrossing to a recurrent parent and would be expected to be >99.5% genetically identical to the recurrent parent. Sequence analysis of pinB from both the soft and hard NILs failed to reveal any significant sequence changes in comparison to earlier reported sequences, and no amino acid sequence changes were found within the tryptophan-rich domain (data not shown). Among these four exceptional hard textured varieties lacking the pinB serine mutation, each is missing pinA transcripts and proteins. The lack of recombination between the presence of pinA and grain softness among the 44 NILs indicates a linkage from 0 to 6.3 cM (P = 0.05). This result confirms the results of others (23) who found that a pinA-derived probe detected a restriction fragment length polymorphism that was linked with grain hardness.
The characterization of this pinA “null” mutation within one of the two components of the starch surface protein friabilin is more significant than the level of linkage between this protein and the Hardness gene. Indeed, we reported earlier (10) that the serine-type mutation also exhibits no recombination with grain hardness such that the maximum map distance between pinB and Hardness is 4.3 cM. Rather than simply exhibiting tight linkage, we postulate that puroindolines play a direct role in effecting grain softness. It is surprising that all hard hexaploid wheats surveyed to date contain one of two “hardness” mutations. This observation may relate to the relatively recent addition of the D-genome to create hexaploid wheat (24) or to the relatively narrow genetic base of modern wheat varieties (25–27). We currently are conducting a broad survey of hard hexaploid wheats. Preliminary results indicate that one of the two mutations presented here, namely, either the serine mutation in pinB or the null mutation of pinA, is found in >95% of greater than 140 U.S. hard wheat varieties surveyed (unpublished data). Additional as yet unknown mutations in either pinA or pinB are expected in the exceptional varieties not having one of the two characterized mutations.
An additional gene, termed “GSP-1,” also is linked to the Hardness gene (16). However, we find no evidence that this gene is functionally involved in the development of grain texture. In fact, GSP-1 is not controlled solely by chromosome 5D, as are pinA and pinB, and its transcripts are present in the durum variety Langdon. Although transcripts of a gene involved in grain hardness may not differ between soft and hard wheats, Hardness gene components likely are controlled solely by chromosome 5D. Transcripts of the friabilin component genes pinA and pinB are absent in durum varieties consistent with the lack of friabilin protein (Fig. 3 and ref. 10). Because friabilin is not found in durum wheats (21), it and GSP-1 must be distinct starch-associated proteins.
The results suggest that the Hardness gene is comprised of the friabilin component genes pinA and pinB. The products of these genes likely act together, possibly as a heterodimer, because a mutation in either gene is equally severe. However, experiments designed to detect a native protein of 30 kDa have been unsuccessful (unpublished data). Further experiments designed at understanding how pinA and pinB work together in the molecular basis of grain hardness could provide better tools for discrimination among the market classes of wheat.
Acknowledgments
We thank Saul Bosley and Skip King for expert technical assistance and Dr. Leonard Joppa for the durum variety Langdon and the substitution line LGD-CS DS5D(5B).
ABBREVIATIONS
- GSP
grain softness protein
- NIR
near infrared reflectance
- NILs
near isogenic lines
References
- 1.Morris C F, Rose S P. Cereal Grain Quality. New York: Chapman and Hall; 1996. pp. 3–54. [Google Scholar]
- 2.Symes K J. Aust J Agric Res. 1965;16:113–123. [Google Scholar]
- 3.Baker R J. Crop Sci. 1977;17:960–962. [Google Scholar]
- 4.Mattern P J, Morris R, Schmidt J W, Johnson V A. Proceedings of the 4th International Wheat Genetics Symposium. Columbia, MO: Univ. Missouri; 1973. pp. 703–707. [Google Scholar]
- 5.Law C N, Young C F, Brown J W S, Snape J W, Worland J W. Seed Protein Improvement by Nuclear Techniques. Vienna, Austria: International Atomic Energy Agency; 1978. pp. 483–502. [Google Scholar]
- 6.Greenwell P, Schofield J D. Cereal Chem. 1986;63:379–380. [Google Scholar]
- 7.Morris C F, Greenblatt G A, Bettge A D, Malkawi H I. J Cereal Sci. 1994;21:167–174. [Google Scholar]
- 8.Jolly C J, Rahman S, Kortt A A, Higgins T J V. Theor Appl Genet. 1993;86:589–597. doi: 10.1007/BF00838714. [DOI] [PubMed] [Google Scholar]
- 9.Gautier M F, Aleman M E, Guirao A, Marion D, Joudier P. Plant Mol Biol. 1994;25:43–57. doi: 10.1007/BF00024197. [DOI] [PubMed] [Google Scholar]
- 10.Giroux M J, Morris C F. Theor Appl Genet. 1997;95:857–864. [Google Scholar]
- 11.Blochet J E, Kaboulou A, Compoint J P, Marion D. In: Gluten Proteins 1990. Bushuk W, Tkachuk R, editors. St. Paul, MN: American Association of Cereal Chemists; 1991. pp. 314–325. [Google Scholar]
- 12.Wilde P J, Clarke D C, Marion D J. J Sci Food Agric. 1993;41:1570–1576. [Google Scholar]
- 13.Greenblatt G A, Bettge A D, Morris C F. Cereal Chem. 1995;72:172–176. [Google Scholar]
- 14.Thorgeirsson T E, Russel C J, King D S, Shin Y K. Biochemistry. 1996;35:1803–1809. doi: 10.1021/bi952300c. [DOI] [PubMed] [Google Scholar]
- 15.Rahman S, Jolly C J, Skerrit J H, Wallosheck A. Eur J Biochem. 1994;223:917–925. doi: 10.1111/j.1432-1033.1994.tb19069.x. [DOI] [PubMed] [Google Scholar]
- 16.Jolly C J, Glenn G M, Rahman S. Proc Natl Acad Sci USA. 1996;93:2408–2413. doi: 10.1073/pnas.93.6.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Symes K J. Aust J Agric Res. 1969;20:971–979. [Google Scholar]
- 18.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 19.Bordier C. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 20.Gill K S, Gill B S, Endo T R, Boyko E V. Genetics. 1996;143:1001–1012. doi: 10.1093/genetics/143.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison M R, Greenwell P, Law C N, Sulaiman B D. J Cereal Sci. 1992;15:143–149. [Google Scholar]
- 22.McIntosh R A, Hart G E, Gale M D. Proc. 8th International Wheat Genetics Symposium. Beijeng, People’s Republic of China: China Agric. Scientech. Press; 1995. pp. 1333–1500. [Google Scholar]
- 23.Sourdille P, Perretant M R, Charmet G, Leroy P, Gautier M F, Joudrier P, Nelson J C, Sorrells M E, Bernard M. Theor Appl Genet. 1996;93:580–586. doi: 10.1007/BF00417951. [DOI] [PubMed] [Google Scholar]
- 24.Gill K S, Lubbers E L, Gill B S, Raupp W J, Cox T S. Genome. 1991;34:362–374. [Google Scholar]
- 25.Cox T S. Use of Plant Introductions in Cultivar Development, Part 1. Spec. Publ. 17. Madison, WI: Crop Science Society of America; 1991. pp. 25–47. [Google Scholar]
- 26.van Beuningen L T, Busch R H. Crop Sci. 1997;37:580–585. [Google Scholar]
- 27.Mercado L A, Souza E, Kephart K D. Theor Appl Genet. 1996;93:593–599. doi: 10.1007/BF00417953. [DOI] [PubMed] [Google Scholar]