Abstract
Recent structures of Escherichia coli catabolite activator protein (CAP) in complex with DNA, and in complex with RNA polymerase α subunit C-terminal domain (αCTD) and DNA, have yielded insights into how CAP binds DNA and activates transcription. Comparison of multiple structures of CAP-DNA complexes has revealed contributions of direct readout and indirect readout to DNA binding by CAP. The structure of the CAP-αCTD-DNA complex has provided the first structural description of interactions between a transcription activator and its functional target within the general transcription machinery. Using the structure of the CAP-αCTD-DNA complex, the structure of an RNAP-DNA complex, and restraints from biophysical, biochemical, and genetic experiments, it has been possible to construct detailed three-dimensional models of intact Class I and Class II transcription activation complexes.
Keywords: catabolite activator protein (CAP), cAMP receptor protein (CRP), RNA polymerase, σ70, promoter, DNA binding, DNA bending, transcription activation
Introduction
The Escherichia coli catabolite activator protein (CAP; also known as the cAMP receptor protein, CRP) activates transcription at more than one hundred promoters. CAP functions by binding, in the presence of the allosteric effector cAMP, to specific DNA sites in or near target promoters and enhancing the ability of RNA polymerase holoenzyme (RNAP) to bind and initiate transcription (reviewed in [1]). Transcription activation by CAP is a classic model system for structural and mechanistic studies of transcription activation. Thus, CAP was the first transcription activator to have been purified, was the first transcription activator to have its three-dimensional structure determined, and has been the subject of extensive biophysical, biochemical, and genetic investigations. Transcription activation by CAP at the simplest CAP-dependent promoters requires only three macromolecular components--CAP, RNAP, and promoter DNA--and requires only one DNA site for CAP [1]. Transcription activation by CAP at such promoters is simpler than most examples of transcription activation in bacteria (which require more numerous macromolecular components and/or DNA sites [2]), and very substantially simpler than examples of transcription activation in eukaryotes (which require tens of macromolecular components and DNA sites [3]). Accordingly, it has been possible to develop structural and mechanistic descriptions of transcription activation by CAP at such promoters that are more nearly complete than descriptions of any other examples of transcription activation.
The immediate scope of this article is to review results of recent structural and functional studies addressing the physical basis of DNA binding by CAP and the mechanism of transcription activation by CAP. Readers interested in cAMP binding by CAP and the cAMP-mediated allosteric transition in CAP are referred to recent articles reporting structures of the cAMP-liganded state of CAP [4,5] and the unliganded state of the CAP homolog CooA [6], recent articles describing effects of cAMP binding on the structure and dynamics of CAP in solution [7,8], and a recent review article [9].
DNA binding by CAP
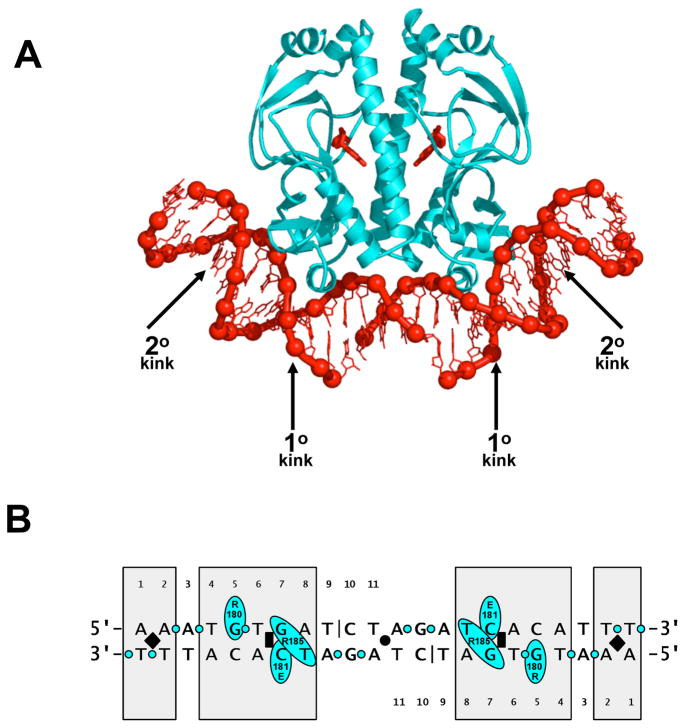
CAP is a dimer of two identical subunits, each of which is 209 residues in length and contains a helix-turn-helix DNA-binding motif [10]. CAP interacts with a 22 bp two-fold-symmetric DNA site, 5′-AATGTGATCTAGATCACATTT-3′ [11]. The CAP-DNA complex is two-fold symmetric: one subunit of CAP interacts with one half of the DNA site; the other subunit of CAP interacts with the other half of the DNA site (Fig 1 [12,13,14]). The initial structures of CAP-DNA complexes revealed two distinctive features [12,13,14]: First, CAP recognizes its DNA site through a combination of “direct readout” (DNA-sequence recognition mediated by direct hydrogen-bonded or van der Waals interactions with DNA base pairs) and “indirect readout” (DNA-sequence recognition mediated by sensing of DNA-sequence-dependent effects on DNA-phosphate position, DNA-phosphate solvation, or susceptibility to DNA deformation). Second, CAP bends DNA by ~80°, wrapping DNA toward and around the sides of the CAP dimer. DNA bending is localized to two phased kinks in each DNA half-site: a “primary kink” of ~40°compressing the DNA major groove between positions 6 and 7, and a “secondary kink” of ~9° compressing the DNA minor groove between positions -1 and 2. Recent crystallographic and biophysical studies have shed further light on these issues.
Figure 1.
DNA binding by CAP: structure of the CAP-DNA complex.
(A) Structure of CAP in complex with its consensus DNA site (PDB 1RUN) [14], showing primary- and secondary-kink sites. CAP is in cyan; DNA and cAMP bound to CAP are in red. The crystallization DNA fragment contained a single-phosphate gap between positions 9 and 10 of each DNA half-site (Fig 1b).
(B) Summary of CAP-DNA interactions. Shaded boxes indicate positions at which CAP exhibits strong sequence preferences [11,15,16,17]. The black circle, black rectangles, and black diamonds indicate, respectively, the two fold-symmetry axis, the primary-kink sites; and the secondary-kink sites. The black vertical lines indicate the positions of single-phosphate gaps present in the crystallization DNA fragment. The cyan ovals and cyan circles indicate, respectively, amino acid-base contacts and amino acid-phosphate contacts.
Direct vs. indirect recognition
CAP exhibits strong sequence preferences at seven positions within each DNA half-site: positions 1, 2, and 4–8 (boxed positions in Fig 1b [11,15,16,17]). Sequence preferences at three positions within each DNA half-site--positions 5, 7, and 8--are accounted for by direct amino acid-base contacts (Fig 1b [12,13,14,18,19,20]). The guanidinium side chain of Arg180 forms hydrogen bonds with the guanine O6 and N7 atoms of the consensus base pair G:C at position 5; the carboxylate side chain of Glu181 forms a hydrogen bond with the cytosine N4 atom of the consensus base pair G:C at position 7; and the guanidinium side chain of Arg185 forms a hydrogen bond with thymine O4 atom of the consensus base pair A:T at position 8 (and, in some structures, also with the guanine O6 and/or N7 atoms of G:C at position 7). In contrast, sequence preferences at the remaining positions occur in the absence of amino acid-base contacts and thus must involve indirect readout (Fig 1b [12,13]).
DNA kinking and indirect recognition
The primary kink is located between positions 6 and 7 (Fig 1b). Positions 6 and 7 are part of a T:A/G:C base-pair step, a base-pair step that is associated with high roll angles and exceptional susceptibility to roll deformation [21]. It has been proposed that specificity for T:A at position 6 is a consequence of formation of the DNA kink between positions 6 and 7, and of effects of the T:A/G:C step on the geometry of DNA kinking, the energetics of DNA kinking, or both [13].
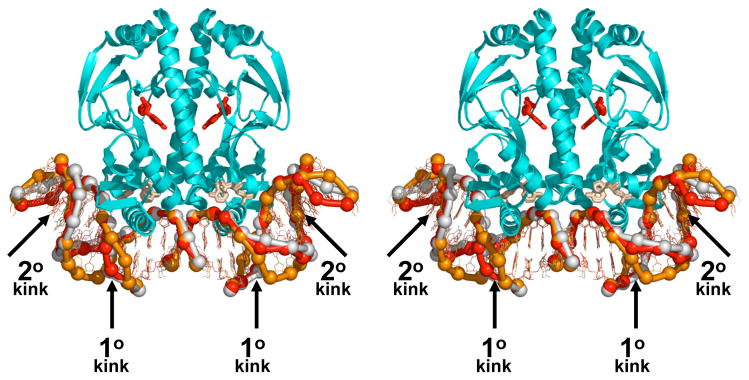
A recent study, involving a combination of biochemical and crystallographic approaches, showed that an amino-acid substitution in CAP that eliminates specificity for T:A at position 6 also eliminates formation of the primary kink (DNA structure in orange in Fig 2 [22*]). The results provide strong support for the proposed connection between specificity for T:A at position 6 and formation of the primary kink [22*].
Figure 2.
DNA binding by CAP: structures of CAP-DNA complexes with substitutions in the primary-kink site. Superimposed structures of CAP in complex with the consensus DNA site (DNA in red; PDB 1O3Q [23*]), CAP in complex with DNA having C:G in place of T:A at position 6 of each DNA half-site (DNA in yellow; PDB 1O3R [23*]), and [Asp181]CAP in complex with DNA having C:G in place of T:A at position 6 of each DNA half-site (DNA in orange; PDB 1O3S [22*]). The structures were obtained from isomorphous crystals with space-group symmetry P3121. Structures of CAP-DNA complexes with this space-group symmetry exhibit two molecules of cAMP per CAP subunit: one in the high-affinity site for cAMP (red), and one in the low-affinity site for cAMP (beige) [22*,23*,24].
A companion study showed that complexes of CAP with DNA sites having the consensus base pair T:A or the nonconsensus base pair C:G at position 6 exhibit similar overall DNA bend angles and local geometries of DNA kinking (DNA structures in yellow and red in Fig 2 [23*]). The results suggest that indirect readout in this system does not involve differences in the geometry of DNA kinking but, rather, solely differences in the energetics of DNA kinking [23*]. (However, it will be necessary to obtain structural data for complexes of CAP with DNA sites having G:C or A:T at position 6 in order to make a definitive statement.)
The results of both studies imply that the main determinant local DNA geometry in this system is protein-DNA interaction, and not DNA sequence [22*,23*].
DNA kinking and DNA smooth bending
In the first study described in the preceding section, protein-DNA interactions were shown to be similar in the structure of a CAP-DNA complex and in the structure of an [Asp181]CAP-DNA complex (wherein [Asp181]CAP is a CAP derivative having a Glu181→Asp181 substitution, a substitution that shortens the residue-181 side chain by one methylene group)--including even the hydrogen bond between the side-chain carboxylate of residue 181 and the cytidine N4 atom of G:C at position 7 [22*]. The overall DNA bend angles also were shown to be similar (DNA structures in orange and red in Fig 2 [22*]). However, the local DNA geometries at the primary-kink sites were shown to be radically different, with the CAP-DNA complex exhibiting a kink, and the [Asp181]CAP-DNA complex exhibiting a smooth bend (DNA structures in white and orange in Fig. 2 [22*]). The results indicate that a given overall DNA bend angle can be achieved through very different local DNA-helical parameters at the primary-kink site. The results further indicate that, in this case, the main determinant of local DNA-helical parameters at the primary-kink site is protein-DNA interaction, and not DNA sequence--with the protein in essence “bending DNA to its will.”
We note, however, a complexity in the kinking vs. bending story. Two structures of wild-type CAP-DNA complexes solved in space group P3121 also exhibit radically different local DNA-helical parameters at the primary kink site: a kink in one structure [23*], and a smooth bend in the other structure [24]. The DNA fragments in the two structures differ in multiple respects, including lengths (38 vs. 46 base pairs), sequences (consensus vs. nonconsensus at positions 1, 9, and 10 of each half-site) and positions of single-phosphate gaps (top strand vs. bottom strand at position 10 of each half-site). Therefore, in this case, it is not possible to deduce the basis for the difference in local DNA-helical parameters at the primary kink site.
DNA bending in solution
Nanosecond time-resolved fluorescence resonance energy transfer measurements between a probe incorporated in CAP and a complementary probe incorporated at each of a series of sites in the DNA indicate that the mean DNA bend angle in the CAP-DNA complex in solution is 77(±3)° degrees--a value consistent with the mean DNA bend angle observed in crystal structures, 80(±12)° [25*]. Lifetime-distribution analysis indicates that the distribution of DNA bend angles is relatively narrow, with <10% of DNA bend angles exceeding 100° [25*]. Millisecond time-resolved luminescence resonance energy transfer experiments provide independent evidence that the upper limit of the distribution of DNA bend angles is ~100° [25*].
What factors are responsible for DNA bending by CAP?
One factor is the DNA sequence of the consensus DNA site. Three studies find that the consensus DNA site for CAP contains an intrinsic bend in the absence of CAP [26,27,28]. Estimates of the DNA bend angle in the absence of CAP vary. An estimate of 15o was obtained in cyclization assays employing tandem arrays of DNA sites [26]. A much higher estimate of 52° was obtained in electrophoretic mobility shift phasing assays [28]. Millisecond time-resolved luminescence resonance energy transfer experiments indicate that the upper limit of the DNA bend angle in the absence of CAP is 40–50° [27].
Another, likely more important, factor is formation of electrostatic interactions between positively charged residues on the sides of the CAP dimer (Lys22, Lys26, Lys44, Lys166, His199, and Lys201) and negatively charged DNA phosphates (positions -5 to -2) [12,15,25*,27,28]. These electrostatic interactions are proposed to contribute to DNA bending in two ways: (i) by stabilizing the bent state through amino acid-phosphate interactions [12,15,25*,27], and (ii) by destabilizing the unbent state through asymmetric phosphate neutralization [28]. Substitution of positively charged residues on the sides of the CAP dimer (Lys22, Lys26, Lys44, and Lys166) reduces the mean DNA bend angle in the CAP-DNA complex in solution by ~5° per residue per half-complex, as assessed in fluorescence resonance energy transfer and fluorescence anisotropy assays [27].
Transcription activation by CAP
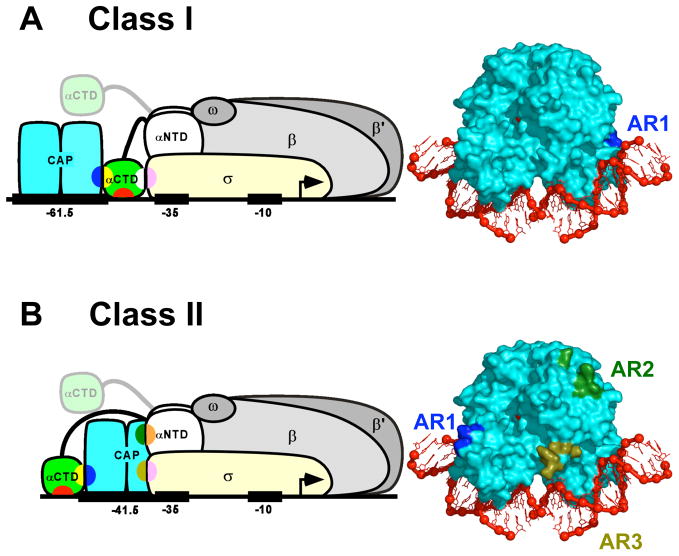
Simple CAP-dependent promoters--i.e. promoters that require only CAP for transcription activation--can be grouped into two classes based on the position of the DNA site for CAP and the corresponding mechanism for transcription activation [1].
At Class I CAP-dependent promoters, the DNA site for CAP is located upstream of the core promoter. The best-characterized Class I CAP-dependent promoters are the lac promoter and the artificial promoter CC(−61.5), each of which has a DNA site for CAP centered at position −61.5. Transcription activation at Class I CAP-dependent promoters involves a single protein-protein interaction between CAP and RNA polymerase holoenzyme (RNAP; subunit composition αIαIIββ’ωσ70 [29]), and proceeds through a simple “recruitment” mechanism, whereby CAP facilitates binding of RNAP to the promoter to yield the RNAP-promoter closed complex (Fig 3a [1]).
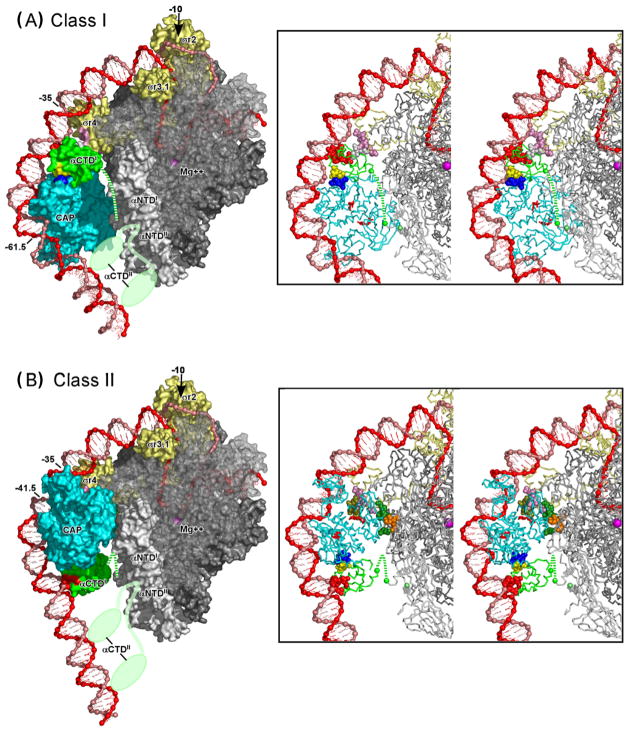
Figure 3.
Transcription activation by CAP: schematic models and activating regions.
(A) Transcription activation at a Class I CAP-dependent promoter [1,40**]. Left: Ternary complex of CAP, RNAP, and a Class I CAP-dependent promoter having the DNA site for CAP centered at position -61.5 [e.g., lac or CC(-61.5)]. Transcription activation involves interaction between AR1 of the downstream subunit of CAP (blue) and the “287 determinant” of one αCTD protomer (yellow). The AR1-αCTD interaction facilitates binding of αCTD, through its “265 determinant” (red), to the DNA segment immediately downstream of CAP and, through its “261 determinant” (white), to residues 573–604 within σR4 (pink). The second αCTD protomer (positioned arbitrarily in figure) interacts non-specifically with upstream DNA [1,39,41]. Right: Structure of the CAP-DNA complex showing AR1 of the downstream subunit (blue).
(B) Transcription activation at a Class II CAP-dependent promoter [1,58,59]. Left: Ternary complex of CAP, RNAP, and a Class II CAP-dependent promoter having the DNA site for CAP centered at position −41.5 [e.g., gal or CC(−41.5)]. Transcription activation involves three sets of CAP-RNAP interactions: (i) interaction between AR1 of the upstream subunit of CAP (blue) and the “287 determinant” of one αCTD (yellow), an interaction that facilitates binding of αCTD, through its “265 determinant” (red), to the DNA segment immediately upstream of CAP; (ii) interaction between AR2 of the downstream subunit of CAP (dark green) and residues 162–165 of αNTDI (orange); and (iii) interaction between AR3 of the downstream subunit of CAP (olive green) and residues 593–603 of σR4 (pink). The second αCTD protomer (positioned arbitrarily in figure) interacts non-specifically with upstream DNA [1,62,63]. Right: Structure of the CAP-DNA complex showing AR1 of the upstream subunit (blue), AR2 of downstream subunit (dark green), and AR3 of downstream subunit (olive green).
At Class II CAP-dependent promoters, the DNA site for CAP overlaps the core promoter, overlapping the core-promoter −35 element. The best-characterized Class II CAP-dependent promoters are the galP1 promoter and the artificial promoter CC(−41.5), each of which has a DNA site for CAP centered at position −41.5. Transcription activation at Class II CAP-dependent promoters involves three sets of protein-protein interactions between CAP and RNAP, and proceeds through both “recruitment” and “post-recruitment” mechanisms, whereby CAP both facilitates binding of RNAP to the promoter to yield the RNAP-promoter closed complex and facilitates isomerization of the RNAP-promoter closed complex to yield the RNAP-promoter open complex (Fig 3b [1]).
Transcription activation at Class I CAP-dependent promoters
At Class I CAP-dependent promoters, CAP activates transcription by binding to a DNA site located upstream of the core promoter and interacting with the RNAP α subunit C-terminal domain (αCTD),an 85 amino-acid residue independently folded domain that is flexibly tethered to the remainder of RNAP (Fig 3a [1,30]). Interaction of CAP with αCTD facilitates binding of αCTD--and, through it, the remainder of RNAP--to promoter DNA, and thereby stimulates transcription initiation.
Biochemical and genetic results indicate that the interaction between CAP and αCTD is mediated by “activating region 1” of the downstream subunit of CAP (AR1; blue in Fig 3a [31,32,33,34,35]) and the “287 determinant” of αCTD (yellow in Fig 3a [36]). Biochemical and genetic results further indicate that the interaction between αCTD and DNA is mediated by the “265 determinant” of αCTD (red in Fig 3a [36,37,38]) and the DNA minor groove [39]. At Class I CAP-dependent promoters where the DNA site for CAP is centered at position −61.5, such as the lac and CC(−61.5) promoters, interaction between CAP and αCTD places αCTD adjacent to σ70, and permits functional protein-protein interaction between αCTD and σ70 (Fig 3a [40**]). The interaction between αCTD and σ70 is mediated by the “261 determinant” of αCTD (white in Fig 3a [36,40**]) and residues 573–604 within the module of σ70 responsible for recognition of the promoter −35 element, σ70 region 4 (σR4; pink in Fig 3a [40**]). RNAP contains two copies of αCTD: αCTDI and αCTDII (Fig 3a [29]). At Class I CAP-dependent promoters, one αCTD protomer--interchangeably αCTDI or αCTDII--interacts with CAP [1,39,41,42]. The other αCTD protomer interacts non-specifically with upstream DNA [1,39,43].
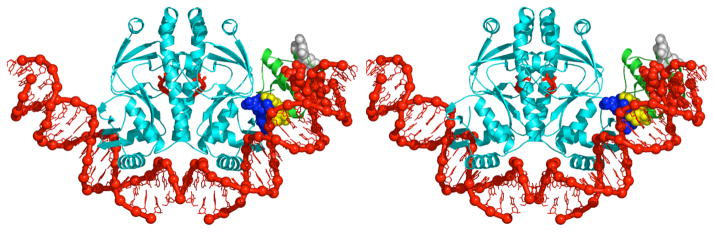
The recently determined crystal structure of a complex containing CAP, αCTD, and DNA has provided the first high-resolution structural description of the interaction between a transcription activator and its functional target within the general transcriptional machinery (Fig 4) [44**]. The interactions in the structure confirm, point-by-point, interactions predicted by biochemical and genetic results [44**]. Thus, CAP makes protein-protein interactions with αCTD, and αCTD makes protein-DNA interactions with the DNA minor groove adjacent to the DNA site for CAP. The interaction between CAP and αCTD is mediated by AR1 of CAP and the 287 determinant of αCTD (blue and yellow in Figs 3a, 4). The interaction between αCTD and DNA is mediated by the 265 determinant of αCTD (red in Figs 3a, 4) and the backbone and spine of hydration of the DNA minor groove adjacent to the DNA site for CAP. The 261 determinant of αCTD (white in Figs 3a, 4) is located on the face of αCTD opposite from CAP and is prominently exposed, consistent with availability to participate in interactions with σR4.
Figure 4.
Transcription activation by CAP: structure of the CAP-αCTD-DNA complex. CAP-αCTD-DNA interactions representative of those at Class I and Class II CAP-dependent promoters (PDB 1LB2 [44**]). CAP is in cyan; αCTD is in green; DNA and cAMP bound to CAP are in red. AR1 of CAP (blue), the “287 determinant” of αCTD (yellow), the “265 determinant” of αCTD (red), and the “261 determinant” of αCTD (white) are in van der Waals representations.
Significantly, in the structure of the CAP-αCTD-DNA complex, there are no conformational changes in CAP and αCTD, and the interface between CAP and αCTD is small (six residues each of CAP and αCTD; 630 Å2 of buried surface area [44**]). The small size of the interface, and the absence of conformational change in activator and target, are consistent with the proposal that transcription activation at Class I CAP-dependent promoters involves a simple “recruitment” mechanism--i.e. simple “adhesive” interactions between activator and target that facilitate and/or stabilize interaction of the general transcription machinery with promoter DNA [1,30,44**,45,46]. (Activation by recruitment does not requires conformational signalling within or through the target, does not require extensive, high-information-content interactions between activator and target, and entails modest net interaction energies between activator and target--interaction energies comparable to the magnitude of activation [45,46].) By joining the crystal structures of the CAP-αCTD-DNA complex [44**] and the σR4-(−35 element) complex [47**]--simply superimposing DNA segments of the two structures onto a single, continuous DNA segment having a site for CAP, a site for αCTD, and a −35 element, spaced as at lac or CC(−61.5)--it has been possible to construct a provisional structural model for the CAP-αCTD-σR4-DNA complex at a Class I CAP-dependent promoter such as at lac or CC(−61.5) [40**,48**]. The resulting model places the 261 determinant of αCTD adjacent to the 573–604 determinant of σR4, permitting favorable electrostatic interaction between the determinants (which have, respectively, high net negative charge and high net positive charge) and permitting, with modest adjustment of side-chain torsion angles, direct contact between experimentally defined interacting residues. The fit between modelled and experimentally defined interactions is striking and can be further improved by moderate compression of the DNA major groove immediately upstream of the −35 element (i.e., at positions −38 and −39 [40**,48**]). Figure 5a presents a structural model of the intact, full Class I CAP-RNAP-promoter complex at lac. The model was constructed in three steps: (i) joining the crystal structures of the CAP-αCTD-DNA complex [44**], the σR4-(−35 element) complex [47**], and the RNAP-DNA complex [49**]--superimposing DNA segments of the three structures to generate a single, continuous DNA segment having a site for CAP, a site for αCTD, a −35 element, and a −10 element, spaced as at lac; (ii) refining local DNA-helix parameters immediately upstream of the −35 element (positions −36 to −41), using experimentally defined αCTD-σR4 interactions [40**] and non-interpenetration as constraints, and using sequence-dependent DNA-deformation energies [50] as restraints; and (iii) modelling downstream DNA segments as in published models of the RNAP-promoter open complex [39,51**]. The model proposes moderate compression of the DNA major groove immediately upstream of the −35 element (~5° roll at the −38/−39 base-pair step). The model is consistent with all available experimental information and provides a structural framework for understanding Class I CAP-dependent transcription. An important feature of the model is that--due to consecutive phased DNA bends in the −35 element, the DNA immediately upstream of the −35 element, and the DNA site for CAP--essentially the entire upstream-promoter region between positions −40 and −100 is proposed to be in proximity to RNAP, and, in particular, the DNA minor grooves at positions −43, −53, −63, −73, −83, and −93 are proposed to be in proximity to αCTDI and αCTDII (Fig 5a). The proposed positions of upstream-promoter DNA minor grooves relative to αCTDI and αCTDII account for results indicating that the αCTD protomer not in contact with CAP can be crosslinked to the DNA minor groove at positions −73, −83, and −93 [39] and is available in principle to interact with a second activator in the −93 or −103 region [42,52].
Figure 5.
Transcription activation by CAP: structural models of intact Class I and Class II CAP-RNAP-promoter complexes.
(A) Structural model of the intact Class I CAP-RNAP-promoter complex at lac.
(B) Structural model of the intact Class I CAP-RNAP-promoter complex at CC(−41.5).
In each panel, a molecular surface representation is shown at left; and a stereodiagram with a ribbon representation is shown at right. Colors of CAP and RNAP are as in Fig 3: CAP is in cyan; αCTDI is in green; αCTDII is in light green (shown in two alternative positions in surface representations; omitted for clarity in ribbon representations); σ70 is in light yellow; αNTDI and αNTDII are in light gray; β is in medium gray (semi-transparent in surface representations, to permit view of DNA strands in RNAP active-center cleft); and β’ and ω are in dark gray. Colors of determinants of CAP and RNAP also are as in Fig 3: AR1, AR2, and AR3 of CAP are in dark blue, dark green, and olive green; the 287, 265, and 261 determinants of αCTDI are in yellow, red, and white; the 162–165 determinant of αNTDI is in orange; and the 593–604 determinant of σ70 is in pink. The DNA template and nontemplate strands are in red and pink. The C-terminus of αNTDI (green) the C-terminus of αNTDII (light green), and the active-center Mg++ (magenta) are indicated by spheres. The linker connecting αCTDI and αNTDI is indicated by a dashed green line. The linker connecting αCTDII and αNTDII is indicated in each of two alternative positions as a light green line.
Methods: Models were constructed by: (i) joining crystal structures of the CAP-αCTD-DNA complex (PDB 1LB2 [44**]), the σR4-(−35 element) complex (PDB 1KU7 [47**]), and an RNAP-DNA complex (PFB 1L9Z [49**]; residues 150–160 and 164–170 of αNTDI modelled as in PDB 1BDF [65]; residues 161–163 of αNTDII modelled along shortest sterically allowed path; side chains modelled using MaxSprout [http://www.ebi.ac.uk/maxsprout/] )--superimposing DNA segments of the three structures onto a single, continuous DNA segment having sites spaced as at lac (panel A) or CC(−41.5) (panel B); (ii) deforming conformations of DNA positions −13 to −31 and −41 to −36 (panel A) or −13 to −30 and −38 to 33 (panel B) to minimize the elastic energy of DNA at the base-pair level [50] while satisfying DNA anchoring conditions, non-interpenetration constraints (Cα-Cα distance ≥3.5 Å for all residue pairs), and proximity constraints (Cα-Cα distance ≤12 Å for residue pairs specified below); and (iii) modelling DNA template-strand positions −11 to +20 and nontemplate-strand positions −7 to +20 as in published models of the RNAP-promoter open complex [39,51**]. For panel A, the following proximity constraints were used: proximity of residues 257, 258, 259, and 261of αCTD to at least one of residues 593, 596, 597, 600, 601, and 604 of σR4, and vice versa (mutational analysis [36,37,40**]); and proximity of residue 261of αCTD to residues 596 and 600 of sR4 (suppression analysis [40**]) (residues numbered as in E. coli RNAP). For panel B, the following proximity constraints were used: proximity of residues 19, 21, 96, and 101 of the downstream CAP subunit to at least one of residues 162, 163, 164, and 165 of αNTDI, and vice versa (mutational analysis [56]); proximity of residues 52, 53, 54, 55, and 58 of the downstream CAP subunit to at least one of residues 593, 596, 597, 599, and 603 of σR4 and vice versa (mutational analysis [58,60]); and proximity of residue 58 of the downstream CAP subunit to residue 596 of σR4 (suppression analysis [59]) (residues numbered as in E. coli RNAP). The models have been deposited in the PDB (PDB **** and ****). Figures were prepared using PyMol [http://www.pymol.org]. The view orientation reflects rotation by −45 on the y-axis relative to the “upstream” view orientation in published models of the RNAP-promoter open complex [39,51**]).
Transcription activation at Class II CAP-dependent promoters
At Class II CAP-dependent promoters, CAP binds at or near position −41.5 and makes three sets of protein-protein interactions with RNAP (Fig 3b [1]). “Activating region 1” of the upstream subunit of CAP (AR1; blue in Fig 3b [34,35,53,54]) interacts with the “287 determinant” of αCTD (yellow in Fig 3b [55]). “Activating region 2” of the downstream subunit of CAP (AR2; dark green in Fig 3b [56]) interacts with residues 162–165 within αNTDI (orange in Fig 3B [1,41,56]). “Activating region 3” of the downstream subunit of CAP (AR3; olive green in Fig 3b [53,54,56,57,58,59]) interacts with residues 593–603 within the module of σ70 responsible for recognition of the promoter −35 element, σ70 region 4 (σR4; pink in Fig 3b [59,60]).
The AR1-αCTD interaction recruits αCTD to the DNA segment immediately upstream of the DNA site for CAP, with αCTD-DNA interaction being mediated by the “265 determinant” of αCTD (red in Fig 3b [1,55]) and the DNA minor groove [61,62,63] and thereby recruits RNAP to the promoter to form the RNAP-promoter closed complex (Fig 3b [56,64]). The AR2-αNTD and AR3-σR4 interactions activate transcription through a post-recruitment mechanism, facilitating isomerization of the RNAP-promoter closed complex to yield the RNAP-promoter open complex [56,58,64].
As noted above, RNAP contains two protomers of αCTD: αCTDI and αCTDII (Fig 3b [29]). At Class II CAP-dependent promoters, one αCTD protomer--either αCTDI or αCTDII, but preferably αCTDI--interacts with CAP [1,41,42]. The other αCTD protomer interacts non-specifically with upstream DNA [1,62,63].
Figure 5b presents a structural model of the intact, full Class II CAP-RNAP-promoter complex at CC(−41.5). The model was constructed in three steps: (i) joining the crystal structures of the CAP-αCTD-DNA complex [44**], the σR4-(−35 element) complex [47**], and the RNAP-DNA complex [49**]--superimposing DNA segments of the three structures onto a single, continuous DNA segment having a site for αCTD, a site for CAP, a −35 element, and a −10 element, spaced as at CC(−41.5); (ii) refining local DNA-helix parameters in the downstream half of the DNA site for CAP (positions −33 to −38), using experimentally defined AR2-αNTDI interactions [41,56], AR3-σR4 interactions [58,59,60], and non-interpenetration as constraints, and using sequence-dependent DNA-deformation energies [50] as restraints; and (iii) modelling downstream DNA segments as in published models of the RNAP-promoter open complex [39,51**]. In step (ii), in order to satisfy the refinement constraints, it was necessary to introduce a relatively large change in local DNA-helix parameters in the downstream half-site of the DNA site for CAP: specifically, it was necessary to replace the primary kink within the downstream half of the DNA site for CAP (~40° roll at the −34/−35 base-pair step) by a smooth bend (~10° roll distributed over the −34/−35, −35/−36, and −36/−37 base-pair steps, yielding a local DNA geometry reminiscent of that observed in two structures of CAP-DNA complexes (see above [22*,24]). The resulting model is consistent with all available experimental information and provides an indispensable structural framework for understanding Class II CAP-dependent transcription. One important feature of the model is the proposed proximity between the DNA minor grooves at positions −73 and −83 and αCTD (Fig 5b). The proposed proximity accounts for results indicating that the αCTD protomer not in contact with CAP interacts with the DNA minor groove at positions −73 and −83 [62,63] and is available in principle to interact with a second activator in the −93, or −103 region [42,62,63]. Another important feature of the model is the proposed requirement for restructuring of DNA local geometry in the downstream half of the DNA site for CAP (conversion of the primary kink to a smooth bend) in order to permit formation of AR2-αNTDI and AR3-σR4 interactions. The proposed requirement for restructuring provides a possible explanation for the observation that AR2-αNTDI and AR3-σR4 interactions do not facilitate binding of RNAP to the promoter to yield the RNAP- promoter closed complex, but do facilitate isomerization of the RNAP-promoter closed complex to yield the RNAP-promoter open complex [56,58,64]: i.e., restructuring, and formation of restructuring-dependent AR2-αNTDI and AR3-σR4 interactions, may occur only during isomerization of the RNAP-promoter closed complex to yield the RNAP-promoter open complex.
Prospect
DNA binding and transcription activation by CAP should be amenable to a complete structural description. Priorities for future work include: (i) determination of high-resolution structures of CAP-DNA complexes having all possible base-pair steps at the primary-kink site (structures relevant to DNA bending and Class II CAP-dependent transcription), (ii) determination of high-resolution structures of the CAP-αCTD-σR4-DNA and CAP-σR4-DNA complexes (structures spanning the upstream-promoter and core-promoter regions of the Class I and Class II CAP-RNAP-promoter complexes), and (iii) determination of low-resolution structural envelopes or high-resolution structures of intact Class I and Class II CAP-RNAP-promoter complexes. Based on recent experience, progress is likely to be rapid.
Acknowledgments
We thank A. Kapanidis, A. Napoli and N. Naryshkin for discussion. Research on CAP in the authors’ laboratories is funded by NIH grants GM21589 (H.M.B. and C.L.L), GM34809 (W. Olson and D.S.), GM41376 (R.H.E.), GM64375 (R. Levy and R.H.E.), and by a Howard Hughes Medical Investigatorship (R.H.E.).
Abbreviations
- CAP
catabolite activator protein
- AR1, AR2, AR3
activating region 1, activating region 2, activating region 3
- RNAP
RNA polymerase holoenzyme (αIαIIββ′ωσ70)
- αNTD
RNAP α subunit N-terminal domain
- αCTD
RNAP α subunit C-terminal domain
- σR2, σR3.1, σR4
σ70 region 2, σ70 region 3.1, σ70 region 4
References
- 1.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 2.Rhodius VA, Busby SJ. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 3.Woychik NA, Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 4.Passner JM, Schultz SC, Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution. J Mol Biol. 2000;304:847–859. doi: 10.1006/jmbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- 5.Chu SY, Tordova M, Gilliland GL, Gorshkova I, Shi Y, Wang S, Schwarz FP. The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding. J Biol Chem. 2001;276:11230–11236. doi: 10.1074/jbc.M010428200. [DOI] [PubMed] [Google Scholar]
- 6.Lanzilotta WN, Schuller DJ, Thorsteinsson MV, Kerby RL, Roberts GP, Poulos TL. Structure of the CO sensing transcription activator CooA. Nat Struct Biol. 2000;7:876–880. doi: 10.1038/82820. [DOI] [PubMed] [Google Scholar]
- 7.Won HS, Yamazaki T, Lee TW, Yoon MK, Park SH, Kyogoku Y, Lee BJ. Structural understanding of the allosteric conformational change of cyclic AMP receptor protein by cyclic AMP binding. Biochemistry. 2000;39:13953–13962. doi: 10.1021/bi000012x. [DOI] [PubMed] [Google Scholar]
- 8.Dong A, Malecki JM, Lee L, Carpenter JF, Lee JC. Ligand-induced conformational and structural dynamics changes in Escherichia coli cyclic AMP receptor protein. Biochemistry. 2002;41:6660–6667. doi: 10.1021/bi020036z. [DOI] [PubMed] [Google Scholar]
- 9.Harman JG. Allosteric regulation of the cAMP receptor protein. Biochim Biophys Acta. 2001;1547:1–17. doi: 10.1016/s0167-4838(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 10.McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- 11.Ebright RH, Ebright YW, Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989;17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 13.Parkinson G, Wilson C, Gunasekera A, Ebright YW, Ebright RE, Berman HM. Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson G, Gunasekera A, Vojtechovsky J, Zhang X, Kunkel TA, Berman H, Ebright RH. Aromatic hydrogen bond in sequence-specific protein DNA recognition. Nat Struct Biol. 1996;3:837–841. doi: 10.1038/nsb1096-837. [DOI] [PubMed] [Google Scholar]
- 15.Gartenberg MR, Crothers DM. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- 16.Dalma-Weiszhausz DD, Gartenberg MR, Crothers DM. Sequence-dependent contribution of distal binding domains to CAP protein-DNA binding affinity. Nucleic Acids Res. 1991;19:611–616. doi: 10.1093/nar/19.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunasekera A, Ebright YW, Ebright RH. DNA sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J Biol Chem. 1992;267:14713–14720. [PubMed] [Google Scholar]
- 18.Ebright RH, Cossart P, Gicquel-Sanzey B, Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984;311:232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- 19.Ebright RH, Kolb A, Buc H, Kunkel TA, Krakow JS, Beckwith J. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc Natl Acad Sci U S A. 1987;84:6083–6087. doi: 10.1073/pnas.84.17.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XP, Ebright RH. Identification of a contact between arginine-180 of the catabolite gene activator protein (CAP) and base pair 5 of the DNA site in the CAP-DNA complex. Proc Natl Acad Sci U S A. 1990;87:4717–4721. doi: 10.1073/pnas.87.12.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci U S A. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Gunasekera A, Zhang X, Kunkel TA, Ebright RH, Berman HM. Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: alteration of DNA binding specificity through alteration of DNA kinking. J Mol Biol. 2001;314:75–82. doi: 10.1006/jmbi.2001.5090. This paper presents crystallographic evidence that: (i) the Glu181→Asp181 substitution of CAP eliminates specificity between the base-pair step T:A/G:C and the base-pair step C:G/G:C at the primary-kink site; and (ii) the Glu181→Asp181 substitution eliminates DNA kinking at the primary-kink site. The results support the proposal [13] that CAP distinguishes between T:A/G:C and C:G/G:C at the primary-kink by an indirect-readout mechanism involving sequence effects on DNA kinking. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Vojtechovsky J, Parkinson GN, Ebright RH, Berman HM. Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: DNA binding specificity based on energetics of DNA kinking. J Mol Biol. 2001;314:63–74. doi: 10.1006/jmbi.2001.5089. This paper compares crystal structures of CAP-DNA complexes containing the consensus base-pair step T:A/G:C or the non-consensus base-pair step C:G/G:C at the primary-kink site. The results show that complexes containing T:A/G:C or C:G/G:C at the primary-kink site exhibit similar overall DNA bend angles and similar local geometries of DNA kinking in each of two crystal lattices. The results imply that indirect readout of T:A/G:C vs. C:G/G:C at the primary-kink site does not involve differences in the geometry of DNA kinking, but, rather, solely, differences in the energetics of DNA kinking. [DOI] [PubMed] [Google Scholar]
- 24.Passner JM, Steitz TA. The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc Natl Acad Sci U S A. 1997;94:2843–2847. doi: 10.1073/pnas.94.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapanidis AN, Ebright YW, Ludescher RD, Chan S, Ebright RH. Mean DNA bend angle and distribution of DNA bend angles in the CAP-DNA complex in solution. J Mol Biol. 2001;312:453–468. doi: 10.1006/jmbi.2001.4976. This paper reports nanosecond time-resolved fluorescence and millisecond time-resolved luminescence measurements defining the mean DNA bend angle and distribution of DNA bend angles in the CAP-DNA complex in solution. The results indicate that the mean DNA bend angle is 77(±3)°--consistent with the mean DNA bend angle observed in crystal structures, 80(±12)°--and that the distribution of DNA bend angles is relatively narrow, with an upper bound of ~100°. The methods described will permit mutational analysis of CAP-induced DNA bending and the role of CAP-induced DNA bending in transcriptional activation (see [27]) [DOI] [PubMed] [Google Scholar]
- 26.Kahn JD, Crothers DM. Measurement of the DNA bend angle induced by the catabolite activator protein using Monte Carlo simulation of cyclization kinetics. J Mol Biol. 1998;276:287–309. doi: 10.1006/jmbi.1997.1515. [DOI] [PubMed] [Google Scholar]
- 27.Kapanidis AN. PhD Thesis. New Brunswick, NJ: Rutgers University; 1999. Fluorescence resonance energy transfer (FRET) studies of DNA bending induced by the Escherichia coli catabolite activator protein (CAP) [Google Scholar]
- 28.Hardwidge PR, Zimmerman JM, Maher LJ., 3rd Charge neutralization and DNA bending by the Escherichia coli catabolite activator protein. Nucleic Acids Res. 2002;30:1879–1885. doi: 10.1093/nar/30.9.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 30.Busby S, Ebright RH. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhang X, Ebright RH. Identification of the activating region of catabolite gene activator protein (CAP): isolation and characterization of mutants of CAP specifically defective in transcription activation. Proc Natl Acad Sci U S A. 1993;90:6081–6085. doi: 10.1073/pnas.90.13.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Busby S, Ebright RH. Identification of the functional subunit of a dimeric transcription activator protein by use of oriented heterodimers. Cell. 1993;73:375–379. doi: 10.1016/0092-8674(93)90236-j. [DOI] [PubMed] [Google Scholar]
- 33.Niu W, Zhou Y, Dong Q, Ebright YW, Ebright RH. Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). I. Saturation and alanine-scanning mutagenesis. J Mol Biol. 1994;243:595–602. doi: 10.1016/0022-2836(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Pendergrast PS, Bell A, Williams R, Busby S, Ebright RH. The functional subunit of a dimeric transcription activator protein depends on promoter architecture. EMBO J. 1994;13:4549–4557. doi: 10.1002/j.1460-2075.1994.tb06776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Merkel TJ, Ebright RH. Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). II. Role at Class I and class II CAP-dependent promoters. J Mol Biol. 1994;243:603–610. doi: 10.1016/0022-2836(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 36.Savery NJ, Lloyd GS, Busby SJ, Thomas MS, Ebright RH, Gourse RL. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J Bacteriol. 2002;184:2273–2280. doi: 10.1128/JB.184.8.2273-2280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Severinov K, Goldfarb A, Fenyo D, Chait B, Ebright RH. Location, structure, and function of the target of a transcriptional activator protein. Genes Dev. 1994;8:3058–3067. doi: 10.1101/gad.8.24.3058. [DOI] [PubMed] [Google Scholar]
- 38.Murakami K, Fujita N, Ishihama A. Transcription factor recognition surface on the RNA polymerase alpha subunit is involved in contact with the DNA enhancer element. EMBO J. 1996;15:4358–4367. [PMC free article] [PubMed] [Google Scholar]
- 39.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 40.Chen H, Tang H, Ebright RH. Functional interaction between RNA polymerase alpha subunit C-terminal domain and sigma70 in UP-element- and activator-dependent transcription. Mol Cell. 2003;11:1621–1633. doi: 10.1016/s1097-2765(03)00201-6. This paper shows that αCTD functionally interacts with σ70 at a subset of UP-element- and activator-dependent promoters [including Class I CAP-dependent promoters where the DNA site for CAP is centered at position -61.5, such as the lac and CC(-61.5) promoters], defines the determinants of αCTD and σ70 required for the interaction, and presents a structural model for the interaction. The αCTD-σ70 interaction spans the upstream promoter and core promoter, thereby linking recognition of upstream-promoter-elements and activators in the upstream promoter with recognition of the −35 element in the core promoter. [DOI] [PubMed] [Google Scholar]
- 41.Niu W. PhD Thesis. New Brunswick, NJ: Rutgers University; 1999. Identification and characterization of interactions between a transcription activator and the general transcription machinery. [Google Scholar]
- 42.Lloyd GS, Niu W, Tebbutt J, Ebright RH, Busby SJ. Requirement for two copies of RNA polymerase alpha subunit C-terminal domain for synergistic transcription activation at complex bacterial promoters. Genes Dev. 2002;16:2557–2565. doi: 10.1101/gad.237502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee DJ, Busby SJ, Lloyd GS. Exploitation of a chemical nuclease to investigate the location and orientation of the Escherichia coli RNA polymerase alpha subunit C-terminal domains at simple promoters that are activated by CRP. J Biol Chem. 2003 doi: 10.1074/jbc.M308300200. [DOI] [PubMed] [Google Scholar]
- 44.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. This paper reports the crystal structure of the CAP-αCTD-DNA complex at a resolution of 3.1 Å. The structure establishes that CAP makes direct protein-protein interactions with αCTD (involving AR1 of CAP and the 287 determinant of αCTD), and that αCTD makes direct protein-DNA interactions with the DNA segment adjacent to the DNA site for CAP (involving the 265 determinant of αCTD and the DNA minor groove adjacent to the DNA site for CAP). The structure further establishes that there are no large-scale conformational changes in CAP and αCTD and that the interface between CAP and αCTD is small--consistent with the proposal that CAP-αCTD interaction contributes to transcription activation through a simple “recruitment” mechanism. The structure in this paper is the first high-resolution structure of a complex between a transcriptional activator and a functional target within the general transcription machinery. [DOI] [PubMed] [Google Scholar]
- 45.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 46.Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 47.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. This paper reports crystal structures of three of four functional regions of Thermus aquaticusσ70:σR2, σR3.1, and σR4. This paper also reports the crystal structure of a σR4-(−35 element) complex. [DOI] [PubMed] [Google Scholar]
- 48.Ross W, Schneider DA, Paul BJ, Mertens A, Gourse RL. An intersubunit contact stimulating transcription initiation by E. coli RNA polymerase: interaction of the alpha C-terminal domain and sigma region 4. Genes Dev. 2003;17:1293–1307. doi: 10.1101/gad.1079403. This paper shows that αCTD functionally interacts with σ70 at a subset of UP-element-dependent promoters, defines the determinants of αCTD and σ70 required for the interaction, and presents a structural model for the interaction. The results and model are substantially consistent with those of [41] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. This paper reports the crystal structure of T. aquaticus RNAP in complex with a DNA “fork junction” (a DNA fragment containing a −35 element, a −35 element/−10 element spacer, and a partial −10 element) [DOI] [PubMed] [Google Scholar]
- 50.Coleman BD, Olson WK, Swigon D. Theory of sequence-dependent DNA elasticity. J Chem Phys. 2003;118:7127–7140. [Google Scholar]
- 51.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. This paper reports use of systematic fluorescence resonance energy transfer and distance-constrained docking to define the three-dimensional structures of RNAP and the RNAP-promoter open complex in solution. [DOI] [PubMed] [Google Scholar]
- 52.Tebbutt J, Rhodius VA, Webster CL, Busby SJ. Architectural requirements for optimal activation by tandem CRP molecules at a class I CRP-dependent promoter. FEMS Microbiol Lett. 2002;210:55–60. doi: 10.1111/j.1574-6968.2002.tb11159.x. [DOI] [PubMed] [Google Scholar]
- 53.Williams R, Bell A, Sims G, Busby S. The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res. 1991;19:6705–6712. doi: 10.1093/nar/19.24.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.West D, Williams R, Rhodius V, Bell A, Sharma N, Zou C, Fujita N, Ishihama A, Busby S. Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at class II promoters. Mol Microbiol. 1993;10:789–797. doi: 10.1111/j.1365-2958.1993.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 55.Savery NJ, Lloyd GS, Kainz M, Gaal T, Ross W, Ebright RH, Gourse RL, Busby SJ. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams RM, Rhodius VA, Bell AI, Kolb A, Busby SJ. Orientation of functional activating regions in the Escherichia coli CRP protein during transcription activation at class II promoters. Nucleic Acids Res. 1996;24:1112–1118. doi: 10.1093/nar/24.6.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhodius VA, Busby SJ. Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J Mol Biol. 2000;299:295–310. doi: 10.1006/jmbi.2000.3736. [DOI] [PubMed] [Google Scholar]
- 59.Rhodius VA, Busby SJ. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase sigma(70) subunit: application of suppression genetics. J Mol Biol. 2000;299:311–324. doi: 10.1006/jmbi.2000.3737. [DOI] [PubMed] [Google Scholar]
- 60.Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 61.Attey A, Belyaeva T, Savery N, Hoggett J, Fujita N, Ishihama A, Busby S. Interactions between the cyclic AMP receptor protein and the alpha subunit of RNA polymerase at the Escherichia coli galactose operon P1 promoter. Nucleic Acids Res. 1994;22:4375–4380. doi: 10.1093/nar/22.21.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murakami K, Owens JT, Belyaeva TA, Meares CF, Busby SJ, Ishihama A. Positioning of two alpha subunit carboxy-terminal domains of RNA polymerase at promoters by two transcription factors. Proc Natl Acad Sci U S A. 1997;94:11274–11278. doi: 10.1073/pnas.94.21.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belyaeva TA, Rhodius VA, Webster CL, Busby SJ. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase alpha subunits. J Mol Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- 64.Rhodius VA, West DM, Webster CL, Busby SJ, Savery NJ. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 1997;25:326–332. doi: 10.1093/nar/25.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang G, Darst SA. Structure of the Escherichia coli RNA polymerase alpha subunit amino-terminal domain. Science. 1998;281:262–266. doi: 10.1126/science.281.5374.262. [DOI] [PubMed] [Google Scholar]