Abstract
We propose a new active mask algorithm for the segmentation of fluorescence microscope images of punctate patterns. It combines the (a) flexibility offered by active-contour methods, (b) speed offered by multiresolution methods, (c) smoothing offered by multiscale methods, and (d) statistical modeling offered by region-growing methods into a fast and accurate segmentation tool. The framework moves from the idea of the “contour” to that of “inside and outside”, or, masks, allowing for easy multidimensional segmentation. It adapts to the topology of the image through the use of multiple masks. The algorithm is almost invariant under initialization, allowing for random initialization, and uses a few easily tunable parameters. Experiments show that the active mask algorithm matches the ground truth well, and outperforms the algorithm widely used in fluorescence microscopy, seeded watershed, both qualitatively as well as quantitatively.
I. INTRODUCTION
Biology has undergone a revolution in the past decade in the way it can examine and analyze processes in live cells, due largely to advances in fluorescence microscopy [2], [3]. Biologists can now collect images across a span of resolutions and modalities, multiple time points as well as multiple channels marking different structures, leading to enormous quantities of image data. As visual processing of such high-dimensional and large data sets is time consuming and cumbersome, automated processing is becoming a necessity. Segmentation is often the first step after acquisition, as it enables biologists to access specific structures of interest (for example, individual cells) within an image.
A. Goal
The acquisition of fluorescence microscope images is done by introducing (into the cells) nontoxic fluorescent probes capable of tagging proteins or molecules of interest. A fluorescent microscope images these cells by illuminating the specimen with light of a specific wavelength, exciting the fluorescent probes to emit light of a longer wavelength; a CCD camera records photon emissions resulting in a digital image. As only some parts of the sample are tagged and the tagging is not uniform, the resulting image looks like a distribution of bright dots on a dark background, a punctate pattern, as in Fig. 1. We focus on images in which such patterns represent individual cells in a multicell specimen. Thus, we aim to:
Fig. 1.
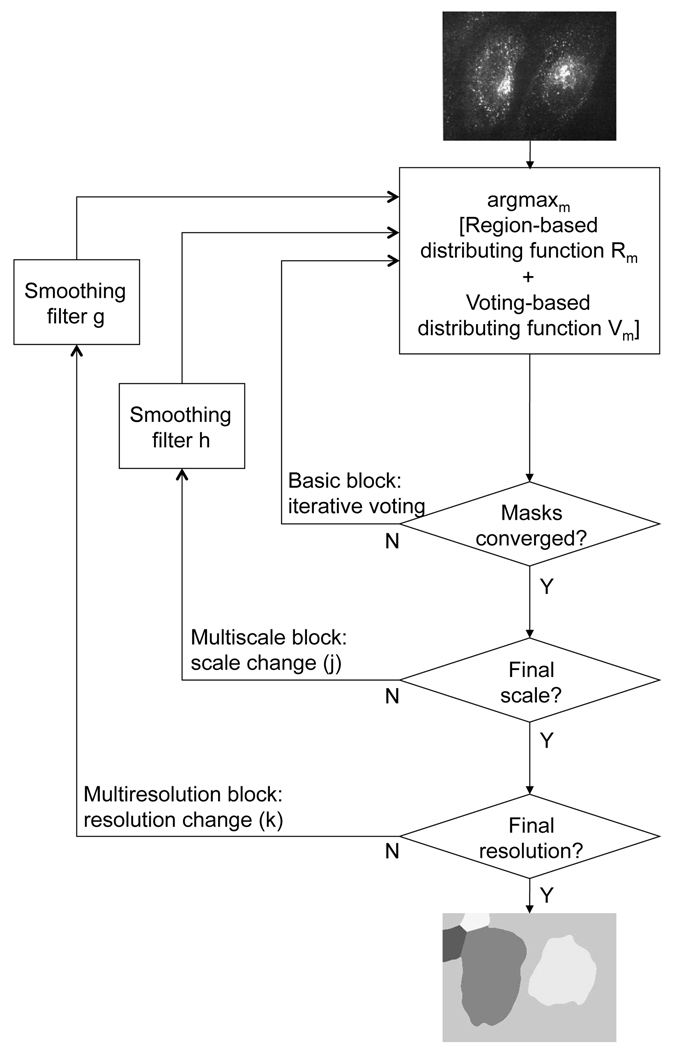
Active mask algorithm. In the basic block, the region-based and voting-based distributing functions work in concert to first separate foreground from background and then iteratively separate masks in the foreground. In the multiscale block, we look at local areas at multiple scales to allow the algorithm to discern cell boundaries and escape local optima. In the multiresolution block, we start at a coarse scale to obtain a fast first segmentation, and then change the scale to refine the result.
Develop an algorithm for the segmentation of fluorescence microscope images of punctate patterns.
B. Segmentation in Fluorescence Microscopy
As different imaging modalities present highly specific challenges, a solution developed for one problem often cannot be applied to another without compromising segmentation accuracy [4]. This specificity has resulted in a vast body of literature; we briefly mention only those works which directly relate to fluorescence microscopy.
In the fluorescence microscope image segmentation community, two algorithms are in wide use: the Voronoi [5], [6] and the seeded watershed algorithms [7], [8]. The watershed algorithm was designed as a “universal segmenter”, and, as it often results in splits or merges, various modifications have been proposed over the years [9], [10]. Its performance is critically dependent on the initial seeds, and, unless the background is seeded, it does not produce tight contours around the cells, thus including a significant portion of the background with the cell (similarly for the Voronoi algorithm).
Region-based segmentation methods could be suited to segment fluorescence microscope images due to the relative homogeneity of statistical properties of the foreground and background in such images. Although there exists a number of region-growing segmentation methods designed by the image processing and computer vision communities [11]–[15], these methods have not been applied directly to fluorescence microscope cell images.
Active contour algorithms are considered state-of-the-art in medical image segmentation and have recently been used for the segmentation of biological images [16]–[18]. Inspired by the segmentation accuracy produced by the active-contour algorithms, we modified an existing image statistics-based active contour algorithm, STACS [19], to fluorescence microscope images. To be precise, we kept STACS’s region-based and curvature-based forces, and imposed topology preservation to keep the number of cells in the image constant. The resulting algorithm, TPSTACS [18], performed extremely well, albeit slowly. Aiming to keep the flexibility of the active-contour based algorithms, which allow for the choice and/or design of forces suited to a given application, we followed it by its multiresolution version, whereby we achieved an order-of-magnitude increase in speed by segmenting the coarse version first, followed by refinements at finer levels. We finally added a multiscale flavor, to allow for both the use of convolution-based processing to increase the speed, as well as for “smoothing to connect the dots” to discern boundaries of cells [20].
C. Idea
In our previous attempts to segment fluorescence microscope images, several issues and questions repeatedly arose: (a) What is the “contour” in a digital image? (b) Updating the level set function in active-contour algorithms is inefficient and slow. (c) How do we reconstruct the level set function in the multiresolution version? (d) Updating in large increments in the multiscale version does not preserve topology. To address these issues, we seek:
An algorithm that combines the:
flexibility offered by active-contour methods,
speed offered by multiresolution methods,
smoothing offered by multiscale methods,
statistical modeling offered by region-growing methods.
We work on fluorescence microscope images of punctate patterns, and assume that: (a) the statistical properties of the foreground (cell) and background are distinct and relatively uniform; (b) the foreground is bright, while the background is dark. The first assumption is crucial, the second not at all; one can easily modify the algorithm should the situation be reversed in another modality (such as brightfield microscopy). Thus, in this paper, we are essentially looking for two different statistical models in the image (foreground and background). We note, however, that the techniques presented here may be generalized to the case of more models (by adding additional region-based distributing functions to the foreground voting, discussed later in the paper).
D. Related Work
Literature in recent years have shown that a combination of techniques (such as edge-based, region-based, shape-based, etc.) is more useful to solve problems that are intractable using any one method alone. In particular, the active contour frame-work is one that is amenable to the combination of multiple techniques. This, together with the encouraging success from our previous efforts, has inspired the present work [20].
A seminal work showed that active contour algorithms can be used to segment images that lack edges [21]. As level-sets adapt gracefully to the image topology, multiphase level-sets were designed to distinguish multiple regions of interest sans tedious topology preservation [22], [23]. Likewise, the combination of parametric active contours and region-based methods has been used effectively for multiband segmentation [24]. Further, methods which use the level-set embedding to keep track of the regions using a discrete grid have demonstrated the efficiency of this class of algorithms [25]–[27]. Multi-scale and multiresolution methods have also been used with segmentation algorithms, and active contours in particular, to improve their accuracy and speed [28]–[31]. While the idea of using numbered masks is prevalent in region-based methods, the PDE literature has also made use of this [32]. Related PDE literature also includes algorithms that evolve regions based on mean curvature [32]–[35].
E. Outline
In Section II, we introduce the basic framework behind the active mask algorithm. We argue for moving from the idea of the “contour” to that of “inside and outside,” that is, to that of masks. We then show how we adapt to the topology of the image by allowing for multiple masks. While active-contour forces act on the contour, we introduce distributing functions, which act on the masks as a whole. We first describe the region-growing distributing functions, which depend on the statistical properties of the image and act to roughly segment the foreground from the background. The voting-based distributing functions that follow aim to separate the foreground into multiple masks. We then explain how these functions work in concert to form the basic block of the active mask algorithm (see basic block in Fig. 1).
In Section III, we add the multiscale block, which iterates on the basic block by increasing/decreasing the “magnification” of the “lens” we use; essentially, we look at local areas at multiple scales to allow the algorithm to discern the cell boundaries and escape local optima (see the multiscale block in Fig. 1). We follow with the multiresolution block; it starts at a coarse version of the image and quickly achieves the initial segmentation by going through the basic and multiscale blocks. It then “lifts” that segmentation result to a finer resolution and repeats the process (see the multiresolution block in Fig. 1). We then illustrate the entire multiresolution-multiscale active mask algorithm.
Finally, in Section IV, we describe our biological testbed on which we demonstrate the performance of the algorithm. We compare the performance of the active mask algorithm with that of the seeded watershed algorithm as well as the ground truth (images hand-segmented by experts), and show that the active mask algorithm outperforms seeded watershed both qualitatively and quantitatively.
II. ACTIVE MASK SEGMENTATION: BASIC BLOCK
Our segmentation task is to both separate foreground (cells) from the background, as well as to separate cells from each other. In Fig. 1, we give a pictorial overview of how we accomplish this task; in this section, we concentrate on the basic block of the active mask algorithm.
A. Framework
We are given a d-dimensional image f of size N1 × … × Nd with (M − 1) cells present. A multidimensional pixel in that image is denoted as n = (n1, … , nd). We choose to depart from the active-contour framework based on contours. Instead, given an image f, our goal is to find an M-valued image ψ of identical size, where ψ(n) = m implies that the pixel n lies inside the mask m attempting to approximate cell m. In short, ψ will be the truth table for whether or not a given pixel lies within the corresponding segmented region, dubbed a mask.
Ideally, we seek an algorithm that iteratively adjusts multiple masks and ultimately results in each cell being perfectly covered by a single mask, with one mask for the background. We now introduce some notation to simplify the expressions in the algorithm that follows. Let be a rectangular subset of the d-dimensional integer lattice ℤd. We regard our image f as a member of ℓ2(Ω), that is, a member of ℓ2(ℤd) that satisfies f(n) = 0 whenever n ∉ Ω. Similarly to [32], we regard a collection of M masks as a function ψ that assigns each pixel n ∈ Ω a value ψ(n) ∈ {1, … , M}. Here, n is an element of the mth mask if ψ(n) = m. That is, a collection of masks is defined to be an element of:
The segmented regions are ψ−1{m} = {n ∈ ℤd|ψ(n) = m}, m = 1, … , M, each having a corresponding mask μm, which is a binary-valued characteristic function:
| (1) |
Fig. 2 illustrates both the collection of all masks ψ as well as representative individual masks μm derived from ψ, for M = 10.
Fig. 2.
An image with nine cells, its corresponding collection of masks ψ, and representative individual binary masks μ1 (background), μ2 and μ4.
Based on the statistics of the image, it is relatively easy to roughly separate foreground/background pixels; this will be the task of the region-based distributing functions Rm described in Section II-B. Given those foreground pixels, the main challenge is to delineate the cells, especially those that touch. As cells almost always have smooth boundaries, we need to partition a homogeneous region (the foreground) into multiple regions with smooth boundaries between them; this will be the task of the voting-based distributing functions Vm described in Section II-C.
B. Region-Based Distributing Functions
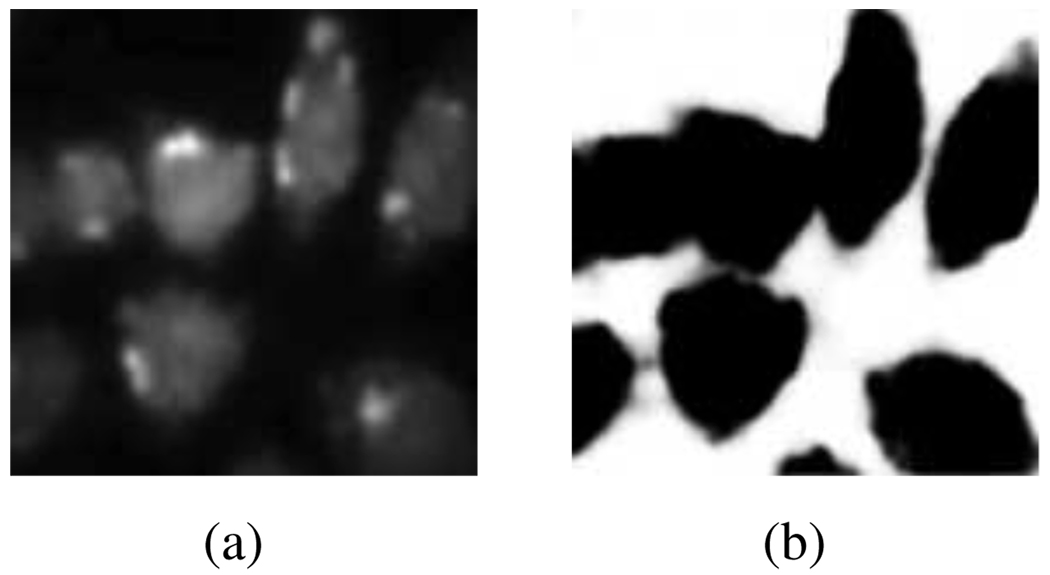
We now describe the action of the region-based distributing functions, which depend on the statistical properties of the image, and which are tasked with separating background from foreground. For the purpose of segmenting fluorescence microscope images, we arbitrarily decide to always let the first mask represent the background, with the remaining (M − 1) masks representing individual cells. In our active mask algorithm below, the mask to which a given pixel belongs is iteratively determined by letting it and its neighboring pixels vote. The role of the region-based distributing function R1 is to skew the voting of points of lower average intensity towards the first mask, and those of higher average intensity away from it. In practice, we construct R1 via a soft-thresholding of a smoothed version of the original image f (see Fig. 3).
Fig. 3.
An illustration of the effect of the region-based distributing function R1 applied to image f from Fig. 2(a) (detail). (a) Smoothed image (f ⋆ h)(n); (b) soft-thresholded image as in (2).
The region-based distributing function R1 is defined to be:
| (2) |
for some region-based lowpass filter h. In (2): sig: ℝ → ℝ is any sigmoid-type function which asymptotically achieves the values ±1 at ±∞, respectively; (f ⋆ h) denotes convolution adjusted for boundary issues as in (4); the skewing factor α ∈ (−1, 0) should be close to −1; β determines the harshness of the threshold; average border intensity γ should be taken as the average intensity of those pixels which lie on the boundary between being inside every cell and being outside them all.
In this work, we choose the following for the region-based lowpass filter h and the sigmoid function:
| (3) |
where a > 0 is called the scale of the region-based lowpass filter. Such an R1 has the properties that:
For a typical pixel n outside a cell, (f ⋆ h)(n) < γ, and so R1(n) ≈ −α ≈ 1. During the voting described in Section II-D, R1(n) then skews the voting so that the background is chosen.
For a typical pixel n inside a cell, (f ⋆ h)(n) > γ, and so R1(n) ≈ α ≈ −1. During the voting described in Section II-D, R1(n) then skews the voting so that any mask but the background one is chosen.
Since the geometry of the cells (smoothness) alone is sufficient to distinguish cells from each other, we took Rm(n) = 0 for all masks but the first one, as we explain in Section II-D.
We designed R1 to depend purely on the density of the underlying fluorescence microscope images; in general, the region-based distributing functions may take into account other statistical properties, edges, textures, and morphological features present in the image. Because of (6), one should scale the Rm’s so as not to exceed 1 in magnitude, as then, their value will completely override the smoothing action of the majority voting introduced later in (9).
C. Voting-Based Distributing Functions
We now describe the action of the voting-based distributing functions, which depend on the geometric properties of the image only, and whose task is to separate individual foreground masks from each other. We draw on the idea of majority voting based on local averages. We do that by computing a convolution (f * g)(n) = ∑ℓ∈ℤd f(n − ℓ)g(ℓ) with some nonnegative voting-based lowpass filter g. Near the boundary of the image, such averages take into account many values of the image that were artificially defined to be zero, and as such, are disproportionately small. We thus compute these averages using the nonstandard, noncommutative convolution:
| (4) |
where χΩ is the characteristic function of Ω. If f ∈ ℓ2(Ω), then (f ⋆ g) ∈ ℓ2(Ω) is well-defined for any nonnegative, nonzero g ∈ ℓ1(ℤd). In practice, we compute both the numerator and denominator of (4) by FFT-implemented circular convolutions of zero-padded versions of f, g and χΩ. In so doing, the segmentation of even large images may be computed with relative efficiency.
For example, in this work, we take g to be
| (5) |
where b is the scale of the voting-based lowpass filter.
The basic step of the voting-based iteration is as follows: Given a collection of multiple masks ψi at iteration i, we begin by forming the masks μm as in (1). Next, we allow each mask to spread its influence by convolving it with g. Because partitions Ω, , and so the values of these convolutions sum to 1:
| (6) |
The voting-based distributing function Vm,i is defined to be:
| (7) |
for n ∈ Ω and mask m at iteration i. The outcome of the voting is then determined to be:
| (8) |
for any n ∈ Ω, and ψi+1(n) = 0 otherwise. Here, Vm,i(n) represents the degree to which pixel n wants to belong to mask m, based on the influence of its neighboring pixels at iteration i. In other words, the mask m producing the largest value of Vm,i(n) is the new mask to which pixel n will belong. We have essentially pitted the masks against each other; each mask tries to conquer any neighboring pixels, and may only be stopped by another mask attempting to do the same.
Similar ideas have appeared in the PDE literature in the context of level sets. In particular, analog versions of our voting-based distributing functions have been used to evolve regions according to mean curvature [32]–[35]. Even in this continuous-domain setting, this method for evolving a contour is “difficult … to analyze theoretically or investigate numerically” [34]. We focus on the discrete version of this idea, which is related to median filtering and the threshold growth of cellular automata [36], a setting in which rigorous proofs of convergence remain elusive.
We illustrate (8) in Fig. 4 as well as with the following simple example:
Fig. 4.
An illustration of the effect of the voting-based distributing functions. (a)–(c) are details from Fig. 5(c)–(e), meant to demonstrate the separation of the foreground region into masks with smooth boundaries.
Example 1: Consider the ith iteration of a 2D image f with the voting-based lowpass filter g being 1 on a 3 × 3 square centered at the origin, and 0 otherwise. Further, suppose that we have M = 3 masks and that the values of ψi in a 3 × 3 neighborhood of some pixel n = (1, 1) are:
 |
Computing Vm,i(1, 1) for m = 1, 2, 3, yields 3/9, 5/9 and 1/9, respectively; thus, in iteration i + 1, the pixel (1, 1) will change its membership from mask 1 to mask 2.
The evolution of the algorithm based on the voting-based distributing functions is given in a pseudocode form in Algorithm 1. The skewing factor in the algorithm is needed for later; to examine just the effect of the voting-based functions, we set Rm = 0 for all m. Experiments reveal that Algorithm 1 will eventually result in masks at equilibrium (that is, further iteration of the voting procedure causes no further change and the algorithm has converged). The minimum thickness of the masks at equilibrium seems to be mostly dependent on the size of the support of the voting-based lowpass filter g. Similarly, the smoothness of the boundaries between distinct masks at equilibrium seems to depend greatly on the smoothness of the voting-based lowpass filter g. If we are looking for smooth boundaries, we should avoid the blocky g described in the example above, and instead choose a g weighted in a more isotropic manner as in (5). In this sense, Algorithm 1 is the active mask version of the smoothing forces traditionally associated with active contour algorithms.
| Algorithm 1 [Active mask voting] |
|
Input: Number of masks M, iteration number i, collection of masks ψi, voting-based lowpass filter g, skewing factors Rm. |
| Output: Collection of masks ψ and final iteration number i. |
| AMVoting(M, i, ψi, g, Rm) |
| while ψi+1 ≠ ψi do |
| Compute the voting-based distributing functions Vm (7) |
| Active mask voting (9) |
| i = i + 1 |
| end while |
| return (ψi, i) |
D. Active Mask Basic Block Evolution
Iteratively applying Algorithm 1 will not successfully segment the image in question, as it does not take the image itself
| Algorithm 2 [Active mask basic block] |
|
Input: Image f, initial number of masks M, initial collection of masks ψ0, scale of the region-based lowpass filter a, scale of the voting-based lowpass filter b, skewing factor α, harshness of the threshold β, average border intensity γ. |
| Output: Collection of masks ψ and final iteration number i. |
| ActiveMasks(f, M, ψ0,a, b, α, β, γ) |
| Compute the region-based lowpass filter h (3) |
| Compute the region-based distributing functions Rm (2) |
| Compute the voting-based lowpass filter g (5) |
| Active mask voting as in Algorithm 1 |
| (ψ, i) = AMVoting(M, 0, ψ0, g, Rm) |
| return (ψ, i) |
into account. As with active contours, we need to combine the purely geometric smoothness forces with image-based forces to push the segmentation in the right direction. In the active mask framework, the region-based distributing functions Rm, given by (2) for m = 1 and taken to be Rm = 0, for m = 2, … , M, will skew the voting of the purely geometric voting-based distributing functions Vm (7).
The active mask basic block is defined to be:
| (9) |
for n ∈ Ω, and ψi+1(n) = 0 otherwise. Thus, as we explained in Section II-B, with α ≈ −1, for a typical pixel n inside a cell, R1 skews the voting so that any mask but the background one is chosen. Similarly, for a typical pixel n outside a cell, R1 skews the voting so that the background is chosen.
To start the iterative process, we initialize the masks ψ randomly. That is, for any given n ∈ Ω, we let ψ0(n) be a number randomly chosen from {1, … , M}, denoted ψ0(n) = rand(M). The strong preference that R1 exhibits for the background will quickly distinguish it from the foreground. With the foreground versus background question settled, the remaining masks compete for supremacy, in a manner similar to the unbiased voting (8). Inevitably, larger masks will consume smaller ones. If at some point ψi(n) ≠ m0 for all n ∈ Ω, then a given mask is no longer present at a given iteration and is eliminated forever, reducing the number of masks M, and consequently, the number of convolutions one must compute in (7). Ultimately, the iterations will converge when the remaining masks that correspond to the cells achieve an equilibrium. This will typically happen when the inherent geometry of the cells causes a stalemate. For example, when two round cells touch, a separate mask grows to dominate each cell. The boundary between the two masks will coincide with the boundary between the two cells, as the narrowing of the cells near their boundary creates a narrow pass which neither side is able to conquer. Thus, the repeated application of Algorithm 1 will often result in a first mask which contains the background, while the remaining nonzero masks each describe a nonzero cell. The steps of the basic block of the active mask algorithm are given in a pseudocode form in Algorithm 2.
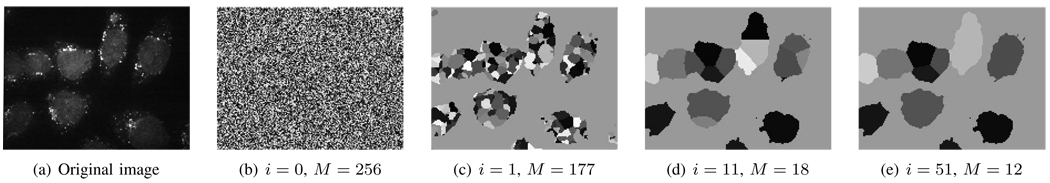
The evolution of the basic block of the active mask algorithm and the successive reduction in the number of masks as the iterations converge are illustrated in Fig. 5. Fig. 5(a) shows a fluorescence microscope image of HeLa cells with six whole cells visible and three partially visible. Fig. 5(b) shows the initial random state of ψ with M = 256 masks (we call this iteration i = 0). In the first iteration, R1 coarsely separates the background from the foreground, discarding empty masks and thus drastically reducing the number of masks to M = 177 as shown in Fig. 5(c). Subsequently, the masks in the foreground region compete with each other, with the geometry of the cells prevailing. Thus, in Fig. 5(d), at i = 11, we see the masks showing an increased correspondence to the cells while the number of masks is steadily decreasing, with convergence achieved in iteration i = 51.
Fig. 5.
An illustration of the evolution of the basic block of the active mask algorithm with random initialization. (a) A COPII image (enhanced for visibility). (b) Random initialization with M = 256 initial masks. (c) In iteration i = 1, the the number of masks reduces to M = 177. (d)–(e) Further iterations resulting in convergence at iteration i = 51, with a final number of masks M = 12.
The active mask algorithm works easily with higher-dimensional images; in fact, the cells we seek to segment are 3D; we give an example of 3D segmentation in Fig. 6. The blobs in Fig. 6 are segmented cells; a 2D slice along the z-axis of this volume represents the segmentation outcome for the corresponding 2D image.
Fig. 6.
3D active mask segmentation of a z-stack. The blobs are segmented cells.
No algorithm is perfect, and occasionally splits and merges of cells will occur, particularly during 2D segmentation (for instance, a split is visible in Fig. 5(e) in what should be mask μ4 (see Fig. 2(e))). Generally, these anomalies depend on the design of the voting-based and region-based distributing functions, and how sensitive the filters are to the features that distinguish the objects of interest. In particular, for the segmentation of fluorescence microscope cell images with the functions V and R described above, if the region-based function has not been smoothed adequately, the masks during the local majority voting may split a single cell into two or more regions based on the kinks on the surface of a single cell. Alternatively, if the scale of the voting-based lowpass filter is too small, the function may be too sensitive to small indentations in the foreground, causing spurious splits. Likewise, if the scale parameters of the filters are too large, cusps between touching cells may be smoothed out, resulting in spurious merges. We address these problems in the following section.
III. ACTIVE MASK SEGMENTATION: MULTIRESOLUTION AND MULTISCALE BLOCKS
In the previous section, we described the basic block of the active mask segmentation algorithm. We also noted instances when splits occur (see Fig. 5(e)). We now address the issue of splits by allowing the scale parameters to change; we address the issue of speed by allowing resolution to change.
A. Multiscale Block
To overcome the problem of spurious splits noted in Section II-D, we could slowly change the scale a > 0 of the region-based lowpass filter h from which the region-based distributing function (2) is computed. For example, with h as in (3), choosing a to be large results in a quickly converging algorithm but one whose ultimate masks overestimate the size of the cells in addition to oversmoothing their boundaries, while choosing a to be small yields better masks but slower convergence.
For the best of both worlds, a is initially taken to be large to rapidly obtain a coarse segmentation. Then, after (9) has converged for this specific a, these resulting masks form the initial guess for the rerunning of (9) with a slightly smaller a. When the change in a is slight, the second iteration will converge in a few steps. This process is then repeated, gradually pulling a down to a point where experimentation reveals the ultimate segmentation as being a good match to the ground truth.
B. Multiresolution Block
In addition to simplifying one’s perspective over discrete domains, a mask-based formulation of segmentation permits an easy implementation of multiresolution schemes to speed the algorithm’s performance, by obtaining a fast and coarse segmentation of the image at a low resolution and then successively refining the result at higher resolutions [37]. While one could use any lowpass filter to obtain a coarse version of the original image, our choice, Haar, was guided by two principles: simplicity and the ability to model the signal readout behavior of the CCD camera of a fluorescence microscope [38].
1) From Fine to Coarse
Let H be the Haar smoothed and downsampled (coarse) version of the original image f:
| (10) |
For example, for d = 1, the output H f is half the size of f and is given by H f(n) = [f(2n) + f(2n + 1)]/2.
Thus, we consider a multiresolution segmentation algorithm in which we first segment a coarse version of f, HKf (H applied to f K times), which is smaller than f by a factor of 2K in each dimension. Once Algorithm 2 has iteratively produced segmentation masks ψ(K) for HK f, we use this information as a starting point for the segmentation of the slightly more detailed image HK−1f.
2) From Coarse to Fine
The multiresolution block at iteration k−1 will begin with the lifted version of the coarser mask ψ(k). The lifted version ψ(k−1) is obtained by copying each binary value of ψ(k) into 2d values of ψk−1:
| (11) |
where the flooring operation is performed coordinate-wise (equivalent to upsampling and repetition interpolation). For example, for d = 1 and if ψ(k) = 1101, ψ(k−1) = 11110011. Here, a contour-based implementation of this same idea would be unnecessarily complicated: one would need to first determine the inside of a given contour to find a mask, then apply (11) to this mask to obtain a new one of twice its size, and finally take the boundary of this resulting mask to find the new contour. In short, multiresolution segmentation lends itself more naturally to mask-based, rather than contour-based, segmentation algorithms.
C. Multiresolution Multiscale Active Mask Evolution
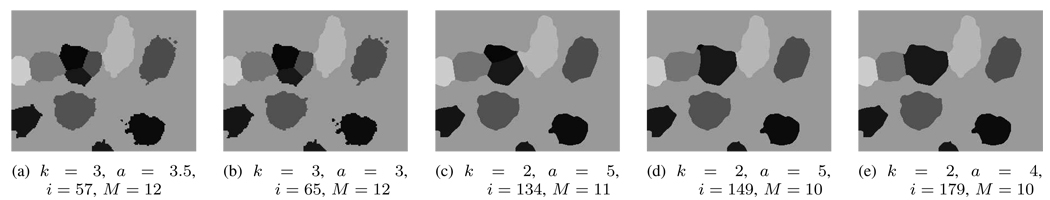
The complete multiresolution multiscale algorithm, beginning with some randomly chosen is given in a pseudocode form in Algorithm 3 and shown in Fig. 1, with index k referring to the multiresolution block, and index j to the multiscale block. We illustrate the evolutionary behavior of the algorithm in Fig. 7. For any fixed k and j, we iteratively apply (9) until the masks are in equilibrium. We resume the process shown in Fig. 5(e), at k = 3 and aj = 4, and use those masks as the starting point for the next step in the scale change. When the change of scales for a given resolution is completed (see Fig. 7(a)–(b)), we lift the resulting masks to a higher resolution, k = 2, and begin the process again (see Fig. 7(c)–(e)). The final masks are given by the function where K0 = 2 in the example. Note how the splits from Fig. 5(e) are no longer present in Fig. 7(e). We could change the scale parameter and pull the resolution back to the original resolution; however, as satisfactory segmentation has already been achieved at this stage, we choose k = 2 to stop. When k = 0, the masks given by the function provide a full-resolution, multiscale active mask segmentation of the
| Algorithm 3 [Multiresolution multiscale active masks] |
|
Input: Image f, initial number of masks M, initial resolution level K, scale of the region-based lowpass filter a, scale of the voting-based lowpass filter b, skewing factor α, harshness of the threshold β, average border intensity γ. |
| Output: Collection of masks ψ and final iteration number i. |
| MRMSActiveMasks(f, M, K, a, b, α, β, γ) |
| Initialization |
| i = 0, k = K, j = 1, , I = f |
| Multiresolution block |
| while k ≥ 0 do |
| Compute coarse version of the image |
| f(n) = Hk I (n) |
| Multiscale block |
| while j ≤ Jk do |
| Basic block |
| Change of scale parameter at resolution k |
| j = j + 1, i = i + p |
| end while |
| Lift mask ψ to the next higher resolution k − 1 |
| k = k − 1 |
| end while |
| return (ψ, i) |
original image. The scale b of the voting-based lowpass filter g used in the voting-based distributing function, chosen to be a fixed constant here, can also be changed. The smaller the scale b, the longer it takes for ψ(k,jk) to converge, but the boundaries between the foreground masks are more accurate. The parameters α, β and γ are fixed constants which are experimentally determined for optimal performance. The choice of all the parameters as well as the performance of the algorithm, both in terms of speed and accuracy, are given in the following section.
Fig. 7.
An illustration of the evolution of the multiresolution multiscale active mask algorithm starting from Fig. 5(e) which used resolution level k = 3 and scale of the region-based lowpass filter a = 4 to Fig. 7 at resolution level k = 2 and scale a = 4, with the final number of masks M = 10. This final result is also shown in pseudo color in Fig. 11(c). (Images at different resolutions have been scaled to the same size for display purposes.)
IV. EXPERIMENTAL RESULTS
There are many biological problems for which punctate markers of the cell periphery are used. We focus on one such problem where manual segmentation presents a critical obstacle.
A. Biological Testbed: Influence of Golgi Protein Expression on the Size of the Golgi Body and Cell
The Golgi body is a membrane-bound organelle in eukaryotic cells whose cytoplasmic surface is a site for a number of important signaling pathways [39]. Further, the Golgi body mediates the processing and sorting of proteins and lipids in the final stages of their biosynthesis. Defects in the biosynthetic pathway are responsible for many human diseases [40]. An understanding of the molecular basis of these defects is paving the way to future effective therapeutics [41].
The Linstedt lab seeks to understand how the Golgi body’s underlying structures are established and maintained, how they are regulated in stress as well as the purpose of each structural feature, that is, they seek a structure/function analysis from the underlying components of the organelle. The current hypothesis being tested is that the affinity of interaction between vesicle coat complexes and SNARE molecules (and other Golgi proteins) establishes and controls the size of Golgi compartments [42]. These tests depend on a quantitative ratiometric assay comparing Golgi size to cell size. The assay is fluorescence microscopy-based and accurate segmentation is a critical step in determining cell volume. To date, the experimental tests of this hypothesis have resulted in the discovery that direct interactions between the cytoplasmic domains of Golgi proteins exiting the endoplasmic reticulum (another membrane-bound organelle in eukaryotic cells) and the COPII (a vesicle that transports proteins from the endoplasmic reticulum to the Golgi body) component Sar1p regulate COPII assembly, providing a variable exit rate mechanism that influences Golgi size [43].
1) The Need for Automated Segmentation
Hand segmentation of cell boundaries was performed based on the diffuse hazy background staining of the COPII subunit Sec13 in each image. Hand segmentation limits the analysis to cells with a flat morphology (fewer slices in a z-stack) and to a small number of cells. To extend the findings, the Linstedt lab needs to analyze a developmental time course over a large number of cells and, as the cell types involved are not flat, it will be necessary to have a larger number of slices per cell.
The segmentation challenge posed by these images is that the COPII subunit Sec13 exhibits not only a bright punctate pattern in the cytoplasm, but also a hazy cytoplasmic pattern (see Fig. 9(a) for an example). Consequently, the signal is not uniform and not always strong, which makes it hard to visually determine the boundary of a cell. Further, as these experiments do not require the nuclear protein to be imaged, there is no parallel channel from which to obtain accurate seeds as initialization for the seeded watershed or Voronoi algorithms. As the computation of cell area/volume is critical in this application, algorithms that do not yield tight contours are not suitable. Using rule-based methods to obtain tight contours or approaches, such as active contours that require involved tuning for each image, make the segmentation semi-automated at best, and would require investing a considerable effort to obtain good segmentation results. Thus, we look to the active mask algorithm to segment these images.
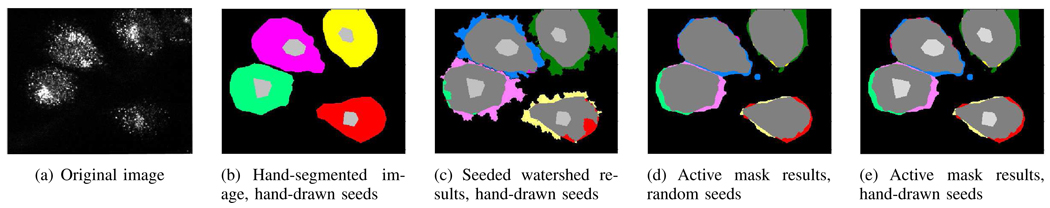
Fig. 9.
(a) An image of HeLa cells marked with COPII at one-fourth the original resolution. (b) Hand-segmented masks (green, pink, yellow and red), together with hand-drawn seeds (light gray). (c) Seeded watershed algorithm initialized with hand-drawn seeds and overlaid on the hand-segmented masks for comparison. (The excess areas marked by seeded watershed algorithm are indicated in light pink, blue, green and yellow.) Area similarity is AS=82.48%. (d) Active mask algorithm initialized with random seeds overlaid on the hand-segmented masks. Area similarity is AS=91.80%. (e) Active mask algorithm initialized with hand-drawn seeds overlaid on hand-segmented masks. Area similarity is AS=91.80%.
B. Data Sets
The data was generated in two experiments.
1) DS1
The first data set consists of 15 z-stacks containing 3–8 cells each. Each stack contains 40 slices of size 1024 × 1344. The HeLa cells were double labeled with the COPII subunit Sec13 (a cytosolic protein peripherally associated with the membrane) and the Golgi marker protein, giantin, in two parallel channels (see Fig. 11(a)–(b)). Sec13 staining has a diffuse cytoplasmic background used to mark the boundary of the cell. The pixel size was 0.05 μm in x/y directions and 0.3 μm in z direction. The findings based on hand segmentation of these images were reported in [43]. We developed the cell-volume computation and Golgi-body segmentation algorithm based on two of the stacks and tested it on others.
Fig. 11.
(a) Original HeLa cell image (COPII, also shown in Fig. 5(a)). (b) Original HeLa Golgi image (giantin). (c) Active mask segmentation of the parallel cell image. (d) Active mask segmentation of the corresponding Golgi image. Using cell segmentation to initialize Golgi segmentation helps associate the different fragments of the Golgi-body in a 2D slice of a cell to their corresponding cells.
2) DS2
The second data set consists of 5 z-stacks containing 2–4 cells each. Each stack contains 20 slices of size 1024 × 1344. The HeLa cells were labeled with Sec13. While the cells imaged in both DS1 and DS2 belong to the same cell line (HeLa), the ones in DS2 are thinner, and thus we have fewer slices per stack. The pixel size was the same as for DS1.
3) Hand-Segmented DS2
To assess the performance of the algorithm, we used the hand-segmented version of the DS2 set as ground truth. The images were hand segmented by two experts in three separate sessions. This was done to minimize the effect of an individual’s bias on a given day as well as ensuring sufficient respite to segment as accurately and precisely as possible (see Fig. 9(a)–(b)).
C. Parameter Selection
We now discuss the selection of parameters as input to Algorithm 3; for convenience, we summarize them in Table I.
TABLE I.
Parameters Used In Experiments.
| Param. | Description | Ref. | Value | Res. k |
|---|---|---|---|---|
| M | Initial number of masks | 256 | ||
| K/K0 | Initial/final resolution levels | 3/2 | 3/2 | |
| a | Scale of the | (3) | (4, 3.5, 3) | 3 |
| region-based lowpass filter | 5 | 2 | ||
| b | Scale of the | (5) | 8 | 3 |
| voting-based lowpass filter | 16 | 2 | ||
| α | Skewing factor | (2) | −0.9 | |
| β | Harshness of the threshold | (2), (12) | 1.5 – 2.0 | |
| γ | Average border intensity | (2), (12) | 3.0 – 4.2 |
1) Initial Number of Masks M
We initialized the algorithm with M = 256 random masks. The larger the M, the lower the possibility of unwanted merges.
2) Initial Resolution Level K
In this work, we decomposed the original images to K = 3 levels (one-eighth the original resolution), lifting the result as per (11), to k = 2 (one-fourth the original resolution). We did not refine any further, as we obtained a satisfactory result at k = 2.
3) Scale factor a of the Region-Based Lowpass Filter
For the multiscale evolution phase, we did not overly tune the parameters. We determined the scale parameter aK,1 for the region-based lowpass filter h in Algorithm 3 based on the resolution k as well as the approximate size of a cell at the coarsest resolution, k = K. We began with a3,1 = 4 at one-eighth the original resolution and pulled back to a3,3 = 3 in decrements of 0:5 (the numbers in parentheses in Table I). At k = K −1 = 2, that is, one-fourth the original resolution, we used just one value, a2,1 = 5. In practice, the more gradual the scale change, the faster the convergence for different scales j at a fixed k. Further, while it is not necessary to decrement the values during the scale change process (they can also be incremented), larger a allows us to obtain a fast coarse segmentation, whereas smaller a takes longer to compute but provides a finer segmentation. We used the same values of a on all the images, without tuning them per image or stack.
4) Scale Factor b of the Voting-Based Lowpass Filter
We changed the scale for the voting-based lowpass filter g in (5) based on k; for k = 3, b = 8, for k = 2, b = 16. In selecting the values, we were guided by the twin goals of expediting convergence while not compromising on the quality of the boundary.
5) Skewing Factor α
We chose α = −0:9 as the weight of R1 given by (2).
6) Harshness of the Threshold β and Average Border Intensity γ
To facilitate and broaden the use by biologists, we have made an effort to use those parameters deemed intuitive; thus, to determine β and γ, we use the average foreground and background value, denoted μfore and μback, respectively. We then set
| (12) |
While the tuning was minimized to a large extent through scale change, it was not entirely eliminated. We determined β and γ based on one image from each stack and used the same numbers for all the other images from that stack. These coarse adjustments are possible because the performance of the algorithm is robust within a small range of the parameter values. While it may be possible to obtain better results than we report with a more involved tuning procedure, it is not practical to expect an involved tuning of the parameters in high-throughput applications, and thus, we did not resort to such a tuning. The range of parameter values is given in Table I, while values used for each stack as well as the scripts used to obtain the results in this section are provided with the reproducible research compendium at [44].
D. Reference Algorithm: Seeded Watershed
Our choice of the reference algorithm was entirely guided by our general goal, that of segmenting fluorescence microscope images of punctate patterns. As the seeded watershed algorithm is widely used for such a task, it was one of our candidates. Following reviewers’ comments, we also considered algorithms from [13]–[15] as well as general region-growing from a single seed point (available in Matlab). Of these, sufficient information/source code with instructions was available only for the seeded watershed algorithm and the general region-growing algorithm available in Matlab. Since the gray levels inside and outside the cells are not uniform for the class of images we consider, the general region-growing algorithm did not perform well. We thus chose to compare the performance of our active mask algorithm to that of the seeded watershed algorithm, as well as the hand-segmented masks.
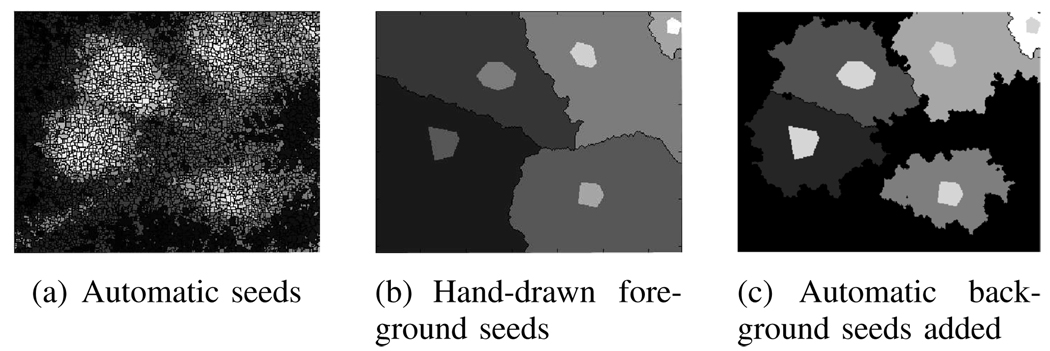
When we use the watershed algorithm described in [7], the algorithm bins regions in the image based on their grayscale values. We obtain a large number of regions (spurious splits) despite smoothing the fluorescence microscope images to reduce the effect of shot noise (see Fig. 8(a)). This result requires a postprocessing step defining rules to merge the highly fragmented regions into a reasonable number of regions. We also used hand-drawn seeds to initialize the algorithm and used the version described in [8] to obtain as many regions as the initial seeds. Even this proved insufficient as the entire image was assigned to one region or another (see Fig. 8(b)). The resulting cell masks were not tight and included a significant portion of the background. We then further tuned the seeding procedure to seed regions in the background to prevent the foreground regions from expanding to the image borders. This result produced tighter masks (see Fig. 9(c)) which we used for comparison with results produced by the active mask algorithm.
Fig. 8.
Seeded watershed algorithm results: (a) With automatic seeds, the algorithm results in 5179 regions that need to be merged based on rules [7]. (b) With hand-drawn foreground seeds (light gray) but no background seeds, the algorithm results in regions that include a significant portion of the background with each cell. (c) Adding background seeds, the algorithm results in reasonable segmentation (background seeds are automatically chosen as every minimum in the smoothed cell image).
E. Performance Evaluation
We now discuss the performance of the algorithm both qualitatively (visual inspection) as well as quantitatively. We also report the runtime.
1) Qualitative Evaluation
We compared the results of the active mask algorithm with the hand-segmented masks, shown in Fig. 9. As we noted before, the seeded watershed with automatic seeds performs poorly without extensive post-processing. Both the seeded watershed algorithm with hand-drawn foreground and automatic background seeds as well as the active mask algorithm with either random or hand-drawn seeds, seem to include a slightly larger area than that marked by hand segmentation. The results of active mask algorithm seem to match the area marked by the experts better than the masks produced by seeded watershed algorithm (see Fig. 9(c)–(e)).
Further, it appears that the active mask algorithm initialized with random seeds performs almost as well as when initialized with hand-drawn seeds. Although this is the case for a majority of the images, due to a random initial configuration, there are some anomalous results as well, such as the one shown in Fig. 10(a). If indeed the problem was only that of an unfavorable initial configuration, often just rerunning the algorithm without changing parameters produces a better result. If the problem resulted from insufficient information or a choice of parameters that induced the masks to split a cell at a particular scale, changing the scale parameters of the filter(s) in the distributing function(s) eliminates spurious splits during the course of the evolution itself (see Fig. 5(b)–(e)). Introducing details available in a higher-resolution version of the image can also help recover from such spurious splits (see Fig. 7). These techniques are already built into the mask evolution phase. However, they do not help in recovering from a spurious merge during the course of the evolution. As it is not always possible to rerun the algorithm, if seeds are available, for example, in the form of reliable information from a parallel channel, they do help in limiting anomalies resulting from the initial randomness (see Fig. 10(b)).
Fig. 10.
Effect of initialization on Algorithm 3: (a) A merge occurs, two cells are covered by the same mask. (b) Given reasonable seeds, the anomaly from (a) disappears.
2) Quantitative Evaluation
To quantify the performance of the algorithm, we used the standard performance measure, area similarity (AS), for each cell in each of 80 different 2D slices. AS normalizes twice the area that is common to the masks by the sum of the areas of hand segmentation and the algorithm, and thus penalizes the algorithm that produces larger regions. Performance of AS ≥ 70% generally implies a good agreement of the algorithm’s result with the ground truth [45]. The area similarity between the mask j in the active mask algorithm (or seeded watershed algorithm) and the mask i in the hand-segmented data set (where n(AMj) is the number of nonzero values in the mask AMj) and is given by,
For partial cells (cells on the periphery), hand-segmented masks were not provided. While the active mask algorithm segmented every cell in the image, whether it was at the periphery or at the center, the seeded watershed algorithm tended to include peripheral cells in the same region as a neighboring central cell. For the seeded watershed algorithm, this required seeding the incomplete cells to avoid leakage of watershed regions of valid whole cells in the image. To compute the numbers, we considered only those cells that were chosen in the hand-segmented data set. As the density of cells varies per image, and to account for the influence of neighboring cells on the performance of the algorithm for a cell, we averaged the performance measures per image before computing the overall average for the data set.
A summary of the results is presented in Table II. For the seeded watershed algorithm, AS=72.94% with σ=13.90 for hand-drawn seeding, while for the active mask algorithm, AS=84.73% with σ=7.90 and AS=86.06% with σ=7.15 for random seeding and hand-drawn seeding, respectively. 1 AS for the seeded watershed with automatic seeding is very low. Whereas in practice the seeded watershed is not initialized with automatic seeding as such, we have provided this measure for completeness. As mentioned in Section IV-D, there are various flavors of the algorithm; perhaps a more sophisticated version, such as in combination with active contours, would perform better [46]. We do not comment further on the seeded watershed with automatic seeding. In what follows, we use only the seeded watershed algorithm with perfect seeding for comparison.2 The active mask algorithm outperforms the seeded watershed algorithm in both cases. Moreover, the active mask algorithm has a lesser chance of detecting false positives in the image. As we mentioned earlier, the performance of active mask algorithm with random seeding is not significantly different from that with hand-drawn seeds. These numbers underscore that the algorithm almost always performs just as well with random seeds as it does with hand-drawn seeds.
TABLE II.
Quantitative Assessment. Area Similarity Measure Computed For The Active mask Algorithm And The reference For Evaluation, Seeded Watershed Algorithm.
| Area similarity [%] | ||
|---|---|---|
| Automatic seeds | Hand-drawn seeds | |
| Seeded watershed | 0.14 | 72.94 |
| Active masks | 84.73 | 86.06 |
3) Runtime
We implemented the active mask algorithm in MATLAB (version 2007a) for flexible design and prototyping [47]. Due to the random initial configuration, runtime ranged between 30 sec to 2 min on an Intel Pentium M 1.6 GHz processor with 1.5 GB memory for the results above. We have also created an ImageJ plugin [48], to facilitate wide distribution among biologists. Apart from algorithmic optimization (such as not waiting for absolute convergence at each scale before changing the scale parameter, and using the segmentation of one image to initialize the segmentation of the succeeding image in a stack) to expedite convergence, optimizing the code in compiled languages such as C may yield anywhere between a 10× to a 1000× speedup, if the problem is not memory bound (that is, a lot of data and very few computations) [49].
F. Application-Specific Processing
In Section IV-A, we introduced our biological testbed, the application that motivated the design of an automated segmentation algorithm for fluorescence microscope images of punctate patterns. As examples of application-specific postprocessing modules, we show how cell-volume computation and Golgi-body segmentation were performed, given the segmentation outcome of the active mask algorithm.
1) Cell-Volume Computation
Given the active mask segmentation outcome, each μm in (1) represents a distinct region. If we set d = 2 and segment the images of a stack in 2D to obtain an approximation of the cell volume, we can simply sum the areas of the 2D masks, as it is done when hand segmentation of the 2D images is used to process the images. To ensure the cells along a stack are assigned the same mask number, we may initialize one of the slices in a stack where the cells are fairly discernible with an initial M ≫ L masks, where L is the expected number of cells. The segmentation outcome of this slice can be used to initialize the neighboring masks and so on. Postprocessing to match a region in one mask with the most overlapping region in a successive mask may also be used in computing the volume from a 2D segmentation procedure [1]. This pseudo-3D segmentation (as the 2D segmentation benefits from the information in 3D) also speeds up segmentation of a stack over segmenting each image in the stack independently.
We can also segment in 3D directly. Such a segmentation affords an elegant way of visualizing the cells in the z-stack and the segmentation outcome is not hampered when a cell is occluded by another cell in a 2D section (see Fig. 6). Further, the volume of each cell can be computed in a straightforward manner from such a result.
2) Golgi-Body Segmentation
The Golgi-body is a double-membraned organelle consisting of cisternae stacked to increase its surface area to facilitate secretion. Thus, though there is only one Golgi-body in each cell, in a 2D section, a Golgi-body may appear as multiple fragments (see Fig. 11(b)). If the cells are close, then by using the Golgi channel alone it is not easy to associate the different pieces of the organelle in 2D with others that belong to the same cell. Given the active mask segmentation of cells for the COPII channel of an image, we may use the resulting masks to initialize the segmentation of the corresponding Golgi channel. The advantage is that multiple pieces of the Golgi body belonging to a cell are now marked by the same mask (see Fig. 11(d)). Moreover, such a Golgi mask matches the cell to which the different Golgi fragments belong (see Fig. 11(c)). This facilitates the computation of the Golgi-volume and, subsequently, the ratio of the Golgi volume to the cell volume as required by the application.
V. CONCLUSIONS
We have proposed a new algorithm termed active mask segmentation, designed for segmentation of fluorescence microscope images of punctate patterns, a large class of data. The framework brings together flexibility offered by active-contour methods, speed offered by multiresolution methods, smoothing offered by multiscale methods, and statistical modeling offered by region-growing methods, to construct an accurate and fast segmentation tool. It departs from the idea of the contour and instead uses that of a mask, as well as multiple masks. The algorithm easily performs multidimensional segmentation, can be initialized with random seeds, and uses a few easily tunable parameters.
We compared the active mask algorithm to the seeded watershed one, and showed active masks to be highly competitive, both qualitatively and quantitatively. We have also compared the algorithm to the ground truth, hand-segmented masks provided by experts, and have found good agreement. We have shown how the segmentation results may be used with application-specific postprocessing modules using the example of cell-volume computation and Golgi-body segmentation for the study of the influence of Golgi protein expression on the cell size. Thus, we have demonstrated that, for segmentation of fluorescence microscope images of punctate patterns, active mask segmentation is a viable alternative to hand segmentation or any standard algorithm (such as seeded watershed or level-set based methods).
We are currently expanding the repertoire of distributing functions that can be used to segment a wider range of images such as those of tissues as well as images that possess features such as edges. Moreover, though even simple, continuous-domain versions of our algorithm are difficult to analyze, we are currently seeking proofs of convergence of (9).
REPRODUCIBLE RESEARCH
To facilitate sharing the method with end users as well as developers, we provide the code and information necessary to reproduce the results in this paper at [44]. The code is given both as developed in Matlab, as well as the ImageJ plugin.
ACKNOWLEDGMENTS
We thank the four anonymous reviewers as well as the Associate Editor, Dr. Dimitri Van De Ville, for their insightful and thorough reviews. We would like to thank Justin Newberg for adapting the seeded watershed algorithm to our data and Warren Ruder for his helpful inputs on various markers for cells. Our thanks go to Manuel Gonzales-Rivero and Sarah Hsieh for implementing the volume computation and Golgi-body segmentation modules. We thank Dr. Amina Chebira and Doru-Cristian Balcan for insightful discussions. We are grateful to Prof. Robert Murphy and Prof. José Moura for their valuable suggestions regarding performance evaluation. We thank Gunhee Kim, Kun Qian, Dr. Amina Chebira and Dr. Pablo Hennings Yeomans for their work on the ImageJ plugin.
This work was supported in part by NSF through awards DMS-0405376, ITR-EF-0331657, by NIH through awards R03-EB008870, R01-GM-56779, and the PA State Tobacco Settlement, Kamlet-Smith Bioinformatics Grant.
Footnotes
A preliminary version of this algorithm was presented in [1].
This is with all the preprocessing described in Section IV-D to make the comparison as fair as possible to the seeded watershed algorithm.
Perfect seeding means hand-drawn seeds for the foreground and automatic for the background, as explained earlier in Section IV-D.
REFERENCES
- 1.Srinivasa G, Fickus MC, Gonzales-Rivero MN, Hsieh SY, Guo Y, Linstedt AD, Kovačević J. Active mask segmentation for the cell-volume computation and Golgi-body segmentation of HeLa cell images; Proc. IEEE Int. Symp. Biomed. Imaging; Paris, France. 2008. May, pp. 348–351. [Google Scholar]
- 2.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Bio. 2002;vol. 3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 3.Michalet X, Kapanidis AN, Laurence T, Pinaud F, Doose S, Pflughoefft M, Weiss S. The power and prospects of fluorescence microscopies and spectroscopies. Ann. Rev. Biophys. and Biomol. Str. 2003;vol. 32:161–182. doi: 10.1146/annurev.biophys.32.110601.142525. [DOI] [PubMed] [Google Scholar]
- 4.Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Ann. Rev. Biomed. Eng. 2001;vol. 2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- 5.Morelock M, Hunter E, Moran T, Heynen S, Laris C, Thieleking M, Akong M, Mikic I, Callaway S, DeLeon R, Goodacre A, Zacharias D, Price J. Statistics of assay validation in high throughput cell imaging of nuclear Factor κB nuclear translocation. ASSAY Drug Dev. Tech. 2005;vol. 3:483–499. doi: 10.1089/adt.2005.3.483. [DOI] [PubMed] [Google Scholar]
- 6.Jones TR, Carpenter AE, Golland P. Voronoi-based segmentation of cells on image manifolds. Lecture Notes in Computer Science. 2005:535–543. [Google Scholar]
- 7.Vincent L, Soille P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Patt. Anal. and Mach. Intelligence. 1991 Jun.vol. 13(no 6):583–598. [Google Scholar]
- 8.Meyer F. Topographic distance and watershed lines. Signal Proc. 1994;vol. 38(no 1):113–125. [Google Scholar]
- 9.Krtolica A, Solorzano CO, Lockett S, Campisi J. Quantification of epithelial cells in coculture with fibroblasts by fluorescence image analysis. Cytometry. 2002;vol. 49:73–82. doi: 10.1002/cyto.10149. [DOI] [PubMed] [Google Scholar]
- 10.Wählby C. Ph.D. dissertation. Uppsala Univ.; 2003. Algorithms for applied digital image cytometry. [Google Scholar]
- 11.Reed TR, du Buf JMH. A review of recent texture segmentation and feature extraction techniques. CVGIP: Image Understanding. 1993;vol. 57(no 3):359–372. [Google Scholar]
- 12.Koepfler G, Lopez C, Morel JM. A multiscale algorithm for image segmentation by variational method. SIAM Journ. Num.. Anal. 1994;vol. 31(no 1):282–299. [Google Scholar]
- 13.Nock R, Nielsen F. Statistical region merging. IEEE Trans. Patt. Anal. and Mach. Intelligence. 2004;vol. 26(no 11):1452–1458. doi: 10.1109/TPAMI.2004.110. [DOI] [PubMed] [Google Scholar]
- 14.Ballester C, Caselles C, Gonzales M. Affine invariant segmentation by variational method. SIAM Journ. Appl. Math. 1996;vol. 56(no 1):294–325. [Google Scholar]
- 15.Pavlidis T. Segmentation of pictures and maps through functional approximation. Comp. Graphics and Image Proc. 1972;vol. 1:360–372. [Google Scholar]
- 16.Sarti A, Solorzano CO, Lockett S, Malladi R. A geometric model for 3-D confocal image analysis. IEEE Trans. Biomed. Eng. 2000;vol. 47(no 12):1600–1609. doi: 10.1109/10.887941. [DOI] [PubMed] [Google Scholar]
- 17.Appleton B, Talbot H. Globally optimal geodesic active contours. Journ. Math. Imag. Vis. 2005;vol. 23(no 1):67–86. [Google Scholar]
- 18.Coulot L, Kirschner HE, Chebira A, Moura JMF, Kovčević J, Osuna EG, Murphy RF. Topology preserving STACS segmentation of protein subcellular location images; Proc. IEEE Int. Symp. Biomed. Imaging; Arlington, VA. 2006. Apr., pp. 566–569. [Google Scholar]
- 19.Pluempitiwiriyawej C, Moura JMF, Wu Y, Ho C. STACS: A new active contour scheme for cardiac MR image segmentation. IEEE Trans. Med. Imag. 2005 May;vol. 24(no 5):593–603. doi: 10.1109/TMI.2005.843740. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasa G, Fickus MC, Kovčević J. Multiscale active contour transformations for the segmentation of fluorescence microscope images; Proc. SPIE Conf. Wavelet Appl. in Signal and Image Proc; San Diego, CA. 2007. Aug., pp. 1–15. [Google Scholar]
- 21.Chan T, Vese LA. Active contours without edges. IEEE Trans Image Proc. 2001 Feb.vol. 10(no 2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 22.Zhao H-K, Chan T, Merriman B, Osher S. A variational level set approach to multiphase motion. Journ. Comp. Phys. 1996;vol. 127:179–195. [Google Scholar]
- 23.Vese LA, Chan TF. A multiphase level set framework for image segmentation using the Mumford and Shah model. Int. Journ. Comp. Vis. 2002;vol. 50(no 3):271–293. [Google Scholar]
- 24.Zhu SC, Yuille A. Region competition: unifying snakes, region growing, and Bayes/MDL for multiband image segmentation. IEEE Trans. Patt. Anal. and Mach. Intelligence. 1996;vol. 18:884–900. [Google Scholar]
- 25.Song B, Chan T. Fast algorithm for level set based optimization. Univ. California Los Angeles; CAM Report 2. 2002 http://citeseer.ist.psu.edu/621225.html. [Online]. Available: http://citeseer.ist.psu.edu/621225.html.
- 26.Gibou F, Fedkiw R. A fast hybrid k-means level set algorithm; Proc. Int. Conf. on Stat., Math. and Related Fields; 2005. Nov., [Google Scholar]
- 27.Papandreou G, Maragos P. Multigrid geometric active contour models. IEEE Trans. Image Proc. 2007 Jan.vol. 16(no 1):229–240. doi: 10.1109/tip.2006.884952. [DOI] [PubMed] [Google Scholar]
- 28.Unser M, Aldroubi A. A review of wavelets in biomedical applications. Proc. IEEE. 1996;vol. 84(no 4):626–638. [Google Scholar]
- 29.Laine AF. Wavelets in temporal and spatial processing of biomedical images. Ann. Rev. Biomed. Eng. 2000;vol. 2:511–550. doi: 10.1146/annurev.bioeng.2.1.511. [DOI] [PubMed] [Google Scholar]
- 30.Liò P. Wavelets in bioinformatics and computational biology: State of art and perspectives. Bioinformatics. 2003;vol. 19(no 1):2–9. doi: 10.1093/bioinformatics/19.1.2. [DOI] [PubMed] [Google Scholar]
- 31.Bresson X, Vandergheynst P, Thiran J-P. Multiscale active contours. Int. Journ. Comp. Vis. 2006;vol. 70(no 3):197–211. [Google Scholar]
- 32.Merriman B, Bence JK, Osher SJ. Motion of multiple junctions: A level set approach. Journ. Comp. Phys. 1994;vol. 112:334–363. [Google Scholar]
- 33.Ruuth SJ. Efficient algorithms for diffusion-generated motion by mean curvature. Journ. Comp. Phys. 1998;vol. 144:603–625. [Google Scholar]
- 34.Ruuth SJ, Merriman B. Convolution-thresholding methods for interface motion. Journ. Comp. Phys. 2001;vol. 169:678–707. [Google Scholar]
- 35.Cao F. Geometric curve evolution and image processing. vol. 1805. Springer: ser. Lecture Notes on Math; 2003. [Google Scholar]
- 36.Gravner J, Griffeath D. Cellular automaton growth on ℤ2: Theorems, examples and problems. Adv. Appl. Math. 1998;vol. 21:241–304. [Google Scholar]
- 37.Leroy B, Herlin IL, Cohen LD. In: Multi-resolution algorithms for active contour models. Sporring J, Nielsen M, Florack LMJ, Johansen P, editors. vol. 219. Berlin, Germany: Springer: ser. Lecture Notes in Control and Inform. Sci.; 1996. [Google Scholar]
- 38.Webb RH, Dorey KC. In: The pixilated image. 2nd ed. Pawley JB, editor. New York, NY: Springer: ser. Handbook of Biological Confocal Microscopy; 1995. [Google Scholar]
- 39.Donaldson J, Lippincott-Schwartz J. Sorting and signaling at the Golgi complex. Cell. 2000;vol. 101(no 7):693–696. doi: 10.1016/s0092-8674(00)80881-8. [DOI] [PubMed] [Google Scholar]
- 40.Aridor M, Hannan LA. Traffic jam: A compendium of human diseases that affect intracellular transport processes. Traffic. 2000;vol. 1(no 11):836–851. doi: 10.1034/j.1600-0854.2000.011104.x. [DOI] [PubMed] [Google Scholar]
- 41.Aridor M, Hannan LA. Traffic jams II: An update of diseases of intracellular transport. Traffic. 2002;vol. 3(no 11):781–790. doi: 10.1034/j.1600-0854.2002.31103.x. [DOI] [PubMed] [Google Scholar]
- 42.Puthenveedu M, Linstedt AD. Subcompartmentalizing the Golgi apparatus. Curr. Opin. Cell Biol. 2005;vol. 17(no 4):369–375. doi: 10.1016/j.ceb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Linstedt AD. COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. Journ. Cell Biol. 2006;vol. 174(no 1):53–56. doi: 10.1083/jcb.200604058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasa G, Fickus MC, Guo Y, Linstedt AD, Kovačević J. Active mask segmentation of fluorescence microscope images. doi: 10.1109/TIP.2009.2021081. [Online]. Available: http://www.andrew.cmu.edu/user/jelenak/Repository/08_SrinivasaFGLK/08_SrinivasaFGLK.html. [DOI] [PMC free article] [PubMed]
- 45.Bartko JJ. Measurement and reliability: Statistical thinking considerations. Schiz. Bullet. 1991;vol. 17(no 3):483–489. doi: 10.1093/schbul/17.3.483. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen HT, Worring M, Boomgaard R. Watersnakes: Energy-driven watershed segmentation. IEEE Trans. Patt. Anal. and Mach. Intelligence. 2003;vol. 25(no 3):330–342. [Google Scholar]
- 47.Kennedy K, Koelbel C, Schreiber R. Defining and measuring the productivity of programming languages. Int. Journ. High Perf. Comp. Appl. 2004;vol. 18(no 4):441–448. [Google Scholar]
- 48.Image: Image processing and analysis in Java. http://rsbweb.nih.gov/ij/. [Online]. Available: http://rsbweb.nih.gov/ij/
- 49.DeRose L, Padua D. Techniques for the translation of MATLAB programs into FORTRAN 90. ACM Trans. Prog. Lang. and Sys. 1999;vol. 21(no 2):286–323. [Google Scholar]