Abstract
Although hibernating mammals wake occasionally to eat during torpor, this period represents a state of fasting. Fasting is known to alter the gut microbiota in nonhibernating mammals; therefore, hibernation may also affect the gut microbiota. However, there are few reports of gut microbiota in hibernating mammals. The present study aimed to compare the gut microbiota in hibernating torpid Syrian hamsters with that in active counterparts by using culture-independent analyses. Hamsters were allocated to either torpid, fed active, or fasted active groups. Hibernation was successfully induced by maintaining darkness at 4°C. Flow cytometry analysis of cecal bacteria showed that 96-h fasting reduced the total gut bacteria. This period of fasting also reduced the concentrations of short chain fatty acids in the cecal contents. In contrast, total bacterial numbers and concentrations of short chain fatty acids were unaffected by hibernation. Denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA gene fragments indicated that fasting and hibernation modulated the cecal microbiota. Analysis of 16S rRNA clone library and species-specific real-time quantitative PCR showed that the class Clostridia predominated in both active and torpid hamsters and that populations of Akkermansia muciniphila, a mucin degrader, were increased by fasting but not by hibernation. From these results, we conclude that the gut microbiota responds differently to fasting and hibernation in Syrian hamsters.
Some mammalian species have evolved with the physiological phenomenon of hibernation to survive unfavorable winter environments (9). Hibernation is realized by entering torpor in order to eliminate the need to maintain a constant, high body temperature. During torpor, typical hibernating mammals, such as hamsters and ground squirrels, lower their body temperature to only a few degrees above ambient temperatures to reduce energy expenditure. Torpor is interrupted by periods of intense metabolic activity. During these interbout arousals, physiological parameters are restored rapidly to near-normal levels. Thus, hibernators alternate between hypothermic and euthermic states during hibernation.
Some hibernating mammals awake to forage during torpor, while food-storing hibernators such as hamsters eat cached food during interbout arousals. However, hibernation essentially involves periods of fasting. Fasting is known to affect the gut microbiota in nonhibernating mammals such as mice (12); therefore, it is possible that hibernation also influences the gut microbiota. Given that the gut microbiota plays important roles in mammalian tissue development and homeostasis (28), it was of interest to investigate the changes in the gut microbiota that may take place during hibernation. To date, this issue has received little attention; to our knowledge, there are only two reports on the gut microbiota in hibernating mammals. Schmidt et al. showed that although the total counts of coliforms, streptococci, and psychrophilic organisms in the feces of arctic ground squirrels held in a cold room at 3°C remained constant the composition changed, with a decrease in coliform count and a 1,000-fold increase in the number of aerobic psychrophilic gram-negative bacteria (31). Barnes and Burton reported that although there was some reduction in total numbers of viable bacteria in the cecum during hibernation, composition of the microbiota remained stable (6). In terms of amphibians, Banas et al. and Gossling et al. reported a reduction and compositional changes of the gut microbiota in hibernating leopard frogs (4, 5, 18, 19).
Only 20 to 40% of bacterial species from the mammalian intestinal tract can be cultured and identified using classical culture methods (22, 34, 36). In contrast, culture-independent methods based on the amplification of bacterial 16S rRNA genes by PCR have revealed a great diversity of microbiota in environmental samples (3, 37). The present study compared the gut microbiota in hibernating torpid Syrian hamsters with that in active counterparts by using culture-independent analyses.
MATERIALS AND METHODS
Animals and diets.
The following study was approved by the Hokkaido University Animal Use Committee, and the animals were maintained in accordance with the Hokkaido University guidelines for the care and use of laboratory animals. Male Syrian hamsters (age 10 weeks) were purchased from Japan SLC (Hamamatsu, Japan) and housed individually in standard plastic cages with nesting material in a temperature-controlled (23 ± 2°C) room with a dark period from 20:00 to 8:00 h. They were allowed free access to chow (Labo H Standard; Nosan Corporation, Yokohama, Japan) and tap water. Hamsters were kept under these conditions until 12 weeks of age.
Experimental design.
Hamsters were divided into a fed active, nonhibernating group (n = 6), a fasted active, nonhibernating group (n = 6), and a hibernating group (n = 6). The two active groups continued to be housed under the conditions described above, while the latter was housed individually in constant darkness at 4°C in order to induce hibernation (27). All hamsters were allowed free access to chow and tap water. In the hibernating group, a temperature sensor (coupled to a data logger; RTR-52A; T & D Corporation, Nagano, Japan) was attached to each cage to monitor the duration of each hibernation bout. Of the hibernating group, four of the six hamsters experienced 9 to 10 hibernation cycles and were then killed by exsanguination from the abdominal aorta. Six hamsters from the fasted active group were killed by exsanguination from the abdominal aorta while under anesthesia by inhalation with diethyl ether after fasting for 96 h, and the remaining six fed active hamsters were killed without fasting. The cecal contents of all animals were excised and subjected to analyses of microbiota.
Flow cytometry analysis of viable, injured, and dead bacterial cells in cecal contents.
Population and viability of bacteria were analyzed by flow cytometry according to the methods reported by Ben-Amor et al. (7). In brief, a portion (≈100 mg) of the cecal contents was suspended in 1 ml of anaerobic phosphate-buffered saline (PBS) containing 1 mM dithiothreitol and 0.01% (wt/vol) Tween 20 and homogenized by vortexing for 3 min. After centrifugation at 700 × g for 1 min, the supernatant was carefully recovered and centrifuged at 6,000 × g for 3 min. The pellet was washed twice, resuspended in anaerobic PBS, and then serially diluted. Thereafter, the diluted samples were incubated for 15 min at room temperature in anaerobic PBS supplemented with 104 particles/ml fluorospheres (Flow-Check fluorospheres; Beckman Coulter, Tokyo, Japan), 1 μg/ml propidium iodide (Wako Pure Chemical Industries, Osaka, Japan), and 5 nM SYTO-BC (Molecular Probes, Eugene, OR). Samples were analyzed by flow cytometry (Epics XL; Beckman Coulter).
Profile analysis of cecal microbiota by PCR-denaturing gradient gel electrophoresis.
DNA was extracted from cecal contents using a fecal DNA isolation kit (MO BIO Laboratories, Carlsbad, CA) according to the manufacturer's instructions. DNA samples were used as a template to amplify the fragments of the 16S rRNA gene with the universal primers U968-GC (CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC) and L1401 (CGG TGT GTA CAA GAC CC) (37), and denaturing gradient gel electrophoresis (DGGE) analysis of the amplicon was carried out as previously described (15). Quantity One software (version 4.6.0; Bio-Rad, Hercules, CA) was used for band identification and normalization of band patterns from DGGE gels. A dendrogram showing the similarity of band patterns was constructed using the unweighted pair-group method with arithmetic mean clustering method in the Quantity One software as previously described (15).
Analysis of the 16S rRNA gene sequences in cecal bacteria.
Cecal DNA samples were pooled in each group and used as templates to amplify the fragments of the 16S rRNA gene with universal primers U968 (AAC GCG AAG AAC CTT AC) and L1401. PCR was performed in a reaction volume of 25 μl that contained 500 nM of each primer, 1× PCR buffer, 0.2 mM of each deoxynucleoside triphosphate, and 1.25 U of Taq-HS polymerase (Takara, Ohtsu, Japan). The reaction conditions were 94°C for 5 min, followed by 20 cycles of 94°C for 30 s, 56°C for 20 s, and 68°C for 40 s, and a final extension at 68°C for 7 min. The amplicons were purified by using a QIAquick PCR purification kit (Qiagen, Tokyo, Japan) and cloned into pGEM-Easy T vectors (Promega, Madison, WI). Transformation was performed with competent Escherichia coli XL-1 Blue cells, and the transformants were spread on Luria-Bertani agar plates supplemented with 25 μg/ml ampicillin, 30 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and 20 μg/ml isopropyl-β-d-thiogalactopyranoside and incubated overnight at 37°C. White colonies were randomly picked from each sample and grown on Luria-Bertani agar. Clones carrying inserts were identified by colony PCR using a Colony PCR M13 set (Nippongene, Tokyo, Japan). Plasmid DNAs in the positive clones were amplified for sequencing with an Illustra TempliPhi DNA amplification kit (GE Healthcare Bioscience, Tokyo, Japan) according to the manufacturer's instructions. Resultant amplicons were sequenced by using an ABI 3730XL or ABI 3730 apparatus (Applied Biosystems, Carlsbad, CA) with M13-F (GTT TTC CCA GTC ACG ACG TT) as a sequencing primer. Artifact chimeric sequences were detected using the CHECK_ CHIMERA program of the Ribosomal Database Project (RDPII) (10) and omitted from the following analysis. Sequence data were aligned with the Clustal X multiple sequence alignment program (35) and corrected by manual inspection. The sequences were compared to the database of RDPII. Naïve Bayesian rRNA classifier version 2.0 from RDPII was used to assign 16S rRNA gene sequences to the taxonomical hierarchy proposed in Bergey's Manual of Systematic Bacteriology, release 6.0 (10), with a setting threshold value of 90%. The sequences were assigned to individual operational taxonomy units (OTUs) based on a sequence similarity of at least 98% (33). A phylogenetic tree was constructed by the neighbor-joining method (29) in the Clustal X program with 1,000 bootstrap replicates. Analysis of the 16S rRNA gene sequences was also performed using the DDBJ/EMBL/GenBank DNA database and the BLAST algorithm.
Real-time quantitative PCR for Akkermansia muciniphila in cecal contents.
Populations of A. muciniphila were determined by real-time quantitative PCR (RT-qPCR) according to the methods of Collado et al. (11). In brief, amplification and detection of cecal DNA were performed with the thermal cycler Dice real-time system (Takara). An A. muciniphila species-specific primer pair (AM1, CAG CAC GTG AAG GTG GGG AC; AM2, CCT TGC GGT TGG CTT CAG AT) was used. RT-qPCR was performed in a reaction volume of 25 μl containing 12.5 μl of SYBR Premix Ex Taq (Takara), 200 nM each of the forward and reverse primers, and 1 μl of cecal DNA samples. The reaction conditions were 95°C for 5 min, followed by 40 cycles at 95°C for 15 s, 60°C for 40 s, and 72°C for 30 s, and a final extension at 72°C for 5 min. The fluorescent products were detected at the last step of each cycle. A melting curve analysis was made after amplification to distinguish the targeted PCR product from the nontargeted PCR product. All samples were analyzed in duplicate.
Using a cecal DNA sample from a fasted active hamster as a template, a fragment of 16S rRNA gene was amplified by PCR with the A. muciniphila species-specific primer pair (AM1 and AM2, as described above). The size of the amplicon, estimated by agarose gel electrophoresis, was identical to the expected size (327 bp) (11), and the sequence was completely identical to A. muciniphila (data not shown). The amplicon was purified and subcloned into a bacterial plasmid as described above. The plasmid DNA was extracted with QIAprep spin miniprep kit (Qiagen) and used as a standard for RT-qPCR. Additionally, the plasmid DNA was added to each cecal DNA sample in order to check for the presence of PCR inhibitors in the samples, and we confirmed no PCR inhibitors in any samples.
Measurement of short chain fatty acid concentrations in cecal contents.
Concentrations of short chain fatty acids (SCFA) in the cecal contents of hamsters were determined using high-performance liquid chromatography by the internal standard method (20). A portion of the cecal contents (≈100 mg) was homogenized in 2 ml of 10 mM sodium hydroxide solution containing 0.5 g/liter crotonic acid (Wako Pure Chemical Industries) as an internal standard and then centrifuged at 10,000 × g for 15 min. Concentrations of individual SCFA (acetate, propionate, i-butyrate, n-butyrate, i-valerate, n-valerate, succinate, and lactate) in the supernatant were measured using a high-performance liquid chromatography system (LC-6A; Shimadzu, Kyoto, Japan) equipped with a Shim-pack SCR-102H column (inner diameter, 8 mm; length, 30 cm; Shimadzu) and a CDD-6A electroconductivity detector (Shimadzu).
Statistical analyses.
Results are presented as means ± standard errors of the means (SEM) or as individual values for each hamster. Tukey-Kramer's test following one-way analysis of variance (ANOVA) was used to analyze the differences among the means of the experimental groups. StatView for Macintosh (version 5.0; SAS Institute Inc., Cary, NC) was used for the analysis. Differences were considered significant at a P level of <0.05.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained from cloning experiments have been deposited in DDBJ with accession numbers AB511067 to AB511276.
RESULTS AND DISCUSSION
Hibernation in mature hamsters can be induced at any time of the year by subjecting them to a reduced ambient temperature of 4°C and constant darkness for about 45 days (27). In the present study, hibernation was successfully initiated in four out of six hamsters. A representative time course of housing temperature (see Fig. S1 in the supplemental material) exhibits a clear hibernation rhythm with individual hibernation bout and interbout arousal. Thus, monitoring the housing temperature alerted us to the time course of hibernation rhythms for each hamster. Four torpid hamsters that had experienced 9 to 10 hibernation cycles were killed just prior to the start of the next interbout arousal. The duration of the average individual hibernation bout in four hibernating hamsters was 92.3 ± 7.1 h.
Flow cytometry analysis was performed to determine total population and viability of bacteria in cecal contents (see Fig. S2 in the supplemental material), and the results are summarized in Table 1. Wet weights of cecal contents were significantly higher in torpid hamsters than in active hamsters, and there was no significant difference between fed and fasted active hamsters. The cecal contents in fasted active hamsters appeared much more fluidal compared to fed active hamsters and torpid hamsters. Total bacterial populations were significantly lower in fasted active hamsters than in fed active and torpid hamsters, and there was no significant difference between fed active and torpid hamsters. Viabilities of cecal bacteria were significantly higher in torpid hamsters than in active hamsters, although there was no significant difference between fed and fasted hamsters. The proportions of injured bacterial cells were significantly higher in fasted active hamsters than in fed active hamsters and torpid hamsters. Consequently, the proportions of dead bacterial cells were significantly higher in fed active hamsters than in fasted active hamsters and torpid hamsters and tended to be lower in torpid hamsters than in fasted active hamsters. Additionally, the concentrations of SCFA, predominant fermentation products of bacteria, in the cecum are shown in Table 2. The concentrations of total SCFA and acetic acid were significantly lower in fasted active hamsters than in fed active hamsters and torpid hamsters. A similar tendency was shown in the concentrations of propionic, succinic, and lactic acids, although there were no significant differences among the groups. Fasted active hamsters showed no detectable levels of n-butyric acid. As the concentrations of SCFA represent the difference between production and absorption in the cecum, the lower concentrations of cecal SCFA in fasted active hamsters appear to reflect a decrease in the production of SCFA caused by a reduction in bacterial populations and viabilities. Regarding SCFA in the hibernating animals, Hume et al. reported that the concentrations in the cecal contents of free-living alpine marmots were significantly lower in emergence from hibernation in April compared to active animals in summer (21). Those authors described that the lower concentrations of cecal SCFA in emergence might reflect the resting state of the gut microbiota, due to low body temperatures in hibernating animals (21). Although the reduced metabolic activity of gut microbiota due to low temperatures is also presumably the case in the present study, no reductions in the concentrations of SCFA were observed in the cecal contents of torpid hamsters. This might reflect a lower ability to absorb SCFA in the cecum due to reduced metabolic activity during hibernation bouts. Additionally, periodic consumption of cached food during interbout arousal might supply a substantial amount of substrate for fermentation in the cecum of hibernating hamsters.
TABLE 1.
Flow cytometry results for cecal bacteria in Syrian hamstersa
| Treatment group | Cecal contents (g) | Total bacterial no.
|
Viability of cecal bacteria
|
|||
|---|---|---|---|---|---|---|
| Log count/cecum | Log count/g | % Viable | % Injured | % Dead | ||
| Fed | 3.8 ± 0.5 a | 10.6 ± 0.1 a | 10.0 ± 0.1 a | 81.3 ± 1.7 a | 9.2 ± 0.6 a | 9.5 ± 1.3 a |
| Fasted | 3.3 ± 0.3 a | 9.4 ± 0.1 b | 8.8 ± 0.1 b | 77.7 ± 3.1 a | 15.9 ± 2.6 b | 6.4 ± 1.0 b |
| Torpid | 7.6 ± 0.4 b | 10.9 ± 0.1 a | 10.0 ± 0.1 a | 90.9 ± 0.9 b | 5.7 ± 0.7 a | 3.4 ± 0.2 b |
Data are means ± SEM from six, six, and four hamsters in the fed, fasted, and torpid groups, respectively. Mean values with different letters were significantly different (P < 0.05) as estimated by Tukey-Kramer's test following a one-way analysis of variance.
TABLE 2.
Short chain fatty acids in cecal contents of Syrian hamstersa
| SCFA | Cecal contents (μmol/g) in treatment group
|
||
|---|---|---|---|
| Fed | Fasted | Torpid | |
| Acetic acid | 71.8 ± 11.1 a | 18.5 ± 1.9 b | 56.1 ± 6.8 a |
| Propionic acid | 15.6 ± 5.6 | 3.5 ± 0.3 | 9.0 ± 1.1 |
| n-Butyric acid | 21.1 ± 5.8 a | ND b | 11.4 ± 0.8 ab |
| Succinic acid | 3.26 ± 1.69 | 0.22 ± 0.07 | 1.66 ± 0.38 |
| Lactic acid | 4.38 ± 3.01 | 0.16 ± 0.04 | 0.34 ± 0.18 |
| Total SCFA | 116.2 ± 22.9 a | 22.4 ± 2.2 b | 78.5 ± 8.6 a |
Data are means ± SEM of six, six, and four hamsters in the fed, fasted, and torpid groups, respectively. Mean values with different letters were significantly different (P < 0.05) as estimated by Tukey-Kramer's test following a one-way ANOVA. Isobutyric, n-valeric, and i-valeric acids were not detected (ND).
The 16S rRNA gene profiles of the bacterial collections in the cecal samples were generated by PCR coupled with DGGE. The DGGE band profile is shown in Fig. 1A. The intensity and position of detected bands were subjected to cluster analysis. The dendrogram shows three large clusters of fed active hamsters, fasted active hamsters, and torpid hamsters, except for one fed active hamster belonging to the cluster of torpid hamsters (Fig. 1B). The results suggest that the gut microbiota is modulated by fasting and by hibernation in hamsters. To further investigate the difference, 16S rRNA clone libraries were constructed from DNA samples pooled in each group. The results of the phylogenetic analysis are shown in Table 3. For fed active, fasted active, and torpid hamsters, 95, 93, and 91 sequences were analyzed. The class Clostridia in the phylum Firmicutes was the most abundant taxonomic group in all treatment groups. However, the frequency of class Clostridia organisms in fasted active hamsters (37.6%) tended to be lower than that in fed active hamsters and torpid hamsters (82.1 and 81.3%, respectively). By comparison, in fasted active hamsters, the phylum Verrucomicrobia was the second most abundant taxonomic group (24.7%), and the third was the phylum Proteobacteria (20.4%). Based on the 98% sequence similarity criterion (33), 56, 49, and 44 OTUs were found in the libraries from fed active, fasted active, and torpid hamsters, respectively. According to Good's method (17), coverage of the diversity in the 16S rRNA gene sequences was 41, 47, and 52% in fed active, fasted active, and torpid hamsters, respectively. A phylogenetic reconstruction of the relationships between the OTUs is presented in Fig. S3 of the supplemental mateiral. The phylogenetic tree shows six distinctive clusters of the phylum Bacteroidetes, the phylum Verrucomicrobia, the class Bacilli, the class Alphaproteobacteria, the class Deltaproteobacteria, the family Ruminococcaceae, and other families of Clostridia. Although the DGGE band profile shows three distinct clusters of fed active, fasted active, and torpid hamsters (Fig. 1), the phylogenetic tree shows no distinct clusters in association with these animal groups.
FIG. 1.
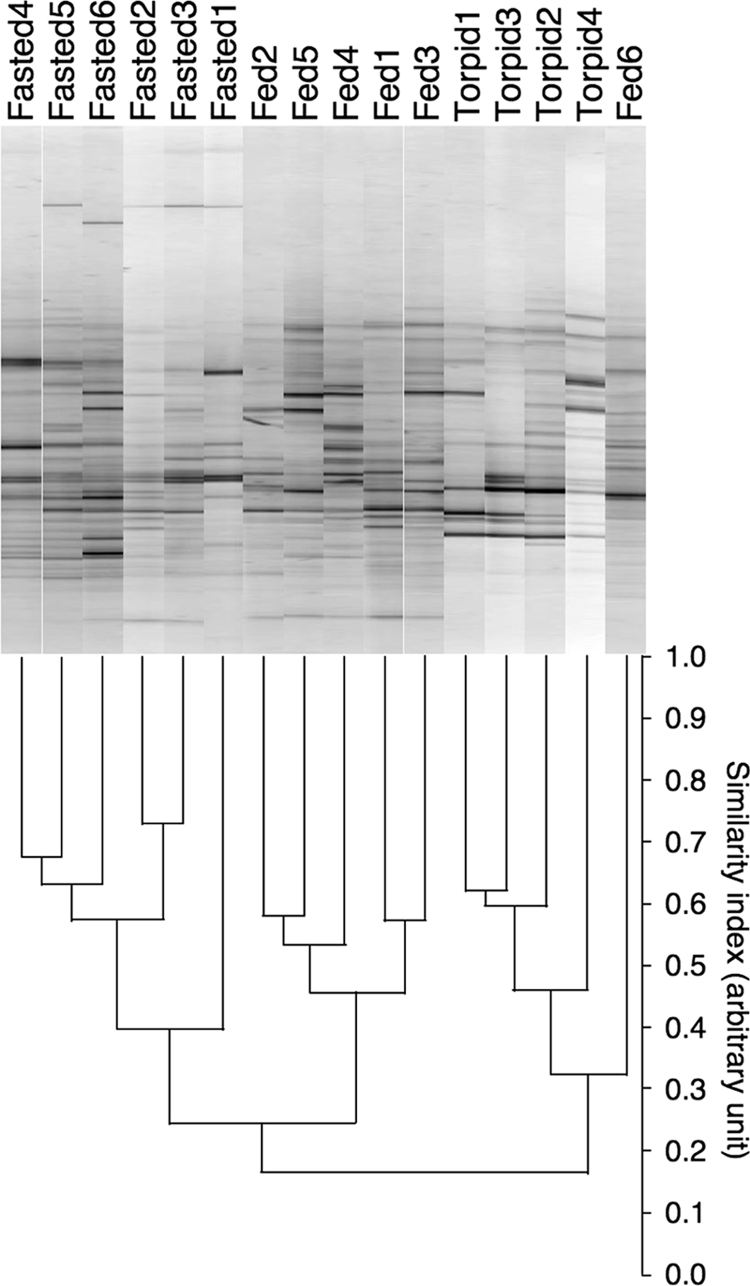
PCR-DGGE analysis of the cecal microbiota based on 16S rRNA gene sequences in hamsters. (Top) DGGE gel image stained with SYBR green. (Bottom) Similarities among DGGE band profiles of cecal bacteria of hamsters were calculated based on the positions and intensities of bands, and the dendrogram of DGGE band profiles was constructed by the unweighted pair-group method with arithmetic mean clustering method. Each lane in the gel and each line in the dendrogram represent individual hamsters. Distances were measured in arbitrary units.
TABLE 3.
Taxonomic distribution of 16S rRNA gene sequences in cecal contents of Syrian hamsters
| Taxonomy | No. (%) of sequences found in treatment group
|
||
|---|---|---|---|
| Fed | Fasted | Torpid | |
| Phylum Bacteroidetes | 0 (0.0) | 1 (1.1) | 2 (2.2) |
| Class Bacteroidetes | 0 (0.0) | 1 (1.1) | 2 (2.2) |
| Order Bacteroidales | 0 (0.0) | 1 (1.1) | 2 (2.2) |
| Unclassified Bacteroidales | 0 (0.0) | 1 (1.1) | 2 (2.2) |
| Phylum Firmicutes | 86 (90.5) | 37 (39.8) | 81 (89.0) |
| Class Bacilli | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Order Lactobacillales | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Family Lactobacillaceae | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Class Clostridia | 78 (82.1) | 35 (37.6) | 74 (81.3) |
| Order Clostridiales | 76 (80.0) | 34 (37.6) | 74 (81.3) |
| Family Veillonellaceae | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Family Lachnospiraceae | 50 (52.6) | 13 (14.0) | 55 (60.4) |
| Family Ruminococcaceae | 21 (22.1) | 18 (19.4) | 13 (14.3) |
| Unclassified Clostridiales | 5 (5.3) | 3 (3.2) | 5 (5.4) |
| Unclassified Clostridia | 2 (2.2) | 1 (1.1) | 0 (0.0) |
| Unclassified Firmicutes | 8 (8.4) | 2 (2.2) | 6 (6.6) |
| Phylum Proteobacteria | 2 (2.2) | 19 (20.4) | 1 (1.1) |
| Class Alphaproteobacteria | 1 (1.1) | 2 (2.2) | 0 (0.0) |
| Unclassified Alphaproteobacteria | 1 (1.1) | 2 (2.2) | 0 (0.0) |
| Class Deltaproteobacteria | 1 (1.1) | 12 (12.8) | 1 (1.1) |
| Order Desulfovibrionales | 1 (1.1) | 12 (12.8) | 1 (1.1) |
| Family Desulfovibrionaceae | 1 (1.1) | 12 (12.8) | 1 (1.1) |
| Unclassified Proteobacteria | 0 (0.0) | 5 (5.4) | 0 (0.0) |
| Phylum Verrucomicrobia | 0 (0.0) | 23 (24.7) | 0 (0.0) |
| Class Verrucomicrobiae | 0 (0.0) | 23 (24.7) | 0 (0.0) |
| Order Verrucomicrobiales | 0 (0.0) | 23 (24.7) | 0 (0.0) |
| Family Verrucomicrobiaceae | 0 (0.0) | 23 (24.7) | 0 (0.0) |
| Unclassified Bacteria | 7 (7.3) | 13 (14.0) | 7 (7.7) |
| Total | 95 (100.0) | 93 (100.0) | 91 (100.0) |
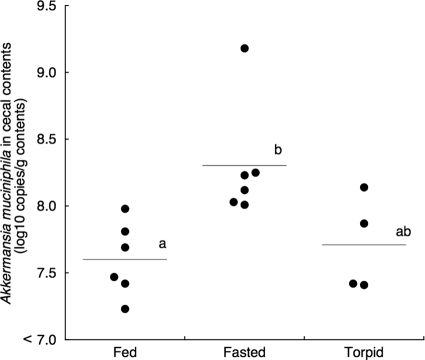
It is noteworthy that all sequences in the phylum Verrucomicrobia detected in fasted active hamsters were classified to the genus Akkermansia (similarity 100%). Derrien et al. (14) isolated Akkermansia muciniphila as a mucin-degrading bacterium in the human intestine. RT-qPCR using a species-specific primer pair for A. muciniphila revealed that the bacterial populations corresponding to A. muciniphila were significantly higher in fasted active hamsters than in fed active hamsters, and those in torpid hamsters were intermediate (Fig. 2). Mucin-degrading bacteria have a competitive advantage during nutrient deprivation, such as during malnutrition, or total parenteral nutrition (TPN) (13, 25). Therefore, increased populations of A. muciniphila in the cecal contents of fasted active hamsters suggest that a lack of food-derived enteral nutrients encourages the growth of mucin-degrading bacteria, such as A. muciniphila. However, the populations of A. muciniphila in the cecal contents of torpid hamsters were comparable to those of fed active hamsters, despite fasting during hibernation bouts. Because the growth of A. muciniphila reportedly occurs at 20 to 40°C, with optimum growth at 37°C (14), lower temperatures might prevent the growth of A. muciniphila in the cecum of torpid hamsters.
FIG. 2.
Populations of Akkermansia muciniphila in cecal contents of hamsters. RT-qPCR with a species-specific primer pair was performed to estimate the populations of A. muciniphila. Each closed circle represents a value for an individual hamster, and horizontal bars represent mean values. Mean values with different letters were significantly different (P < 0.05) as estimated by Tukey-Kramer's test following a one-way analysis of variance.
Because mucins are highly sulfated acidic mucopolysaccharides, degradation of mucins by intestinal bacteria releases sulfate, which in turn stimulates sulfate-reducing bacteria, such as Desulfovibrio spp. (16). In the present study, the family Desulfovibrionaceae was the most common family in the phylum Proteobacteria, the third most abundant taxonomic group in fasted active hamsters, and the frequency of the family Desulfovibrionaceae was higher in fasted active hamsters than in fed active and torpid hamsters (Table 3). These findings suggest that fasting stimulates the growth of sulfate-reducing bacteria, such as Desulfovibrio spp., through increased degradation of mucins by A. muciniphila in the cecum of active hamsters. Considering the toxicity of hydrogen sulfide produced from reduction of sulfate by sulfate-reducing bacteria, abundance of the family Desulfovibrionaceae might be associated with higher proportions of injured bacteria in the cecum of fasted active hamsters.
Mucins are the main constituents of the mucus covering the epithelial surface of the gastrointestinal tract, and thus they contribute to the epithelial protective barrier against pathogenic microorganisms as well as chemical, physical, or enzymatic damage. Therefore, the degradation of intestinal mucins possibly results in reduced barrier function, which in turn becomes pathological. As mucin-degrading bacteria have a competitive advantage during nutrient deprivation (13, 25), the gut mucosal barrier may be disrupted by fasting and/or TPN. Indeed, TPN reportedly reduced the absolute quantity of mucus gel in rat ileum (30) and also induced intestinal bacterial translocation (BT) in rats (1, 2, 24, 32). In contrast, BT was not induced by up to 72 h of fasting in rats (8, 12, 26), although increased mucosal permeability was reported in fasted rats (8). Nevertheless, fasting can exacerbate the risk of morbidity and mortality for BT (23). The present study showed that prolonged fasting stimulated the growth of A. muciniphila in the cecum of active hamsters. In this context, A. muciniphila may be involved in disruption of the gut mucosal barrier, possibly followed by BT under certain circumstances, during deprivation of enteral nutrients.
To our knowledge, the present study is the first to investigate the gut microbiota in hibernating animals by culture-independent methods. The results suggest that the gut microbiota respond differently to fasting and hibernation in Syrian hamsters. Fasting, in particular, stimulated the growth of the mucin degrader A. muciniphila. However, coverage of the diversity in the 16S rRNA gene sequences of gut microbiota was about 50% in the present study. Additionally, the present study made no attempt to assess the dominant species of the gut microbiota by identification of bacterial origins of 16S rRNA gene sequences obtained from DGGE gels and to compare to those from the 16S rRNA gene clone library. Therefore, further studies are required to illustrate more precisely the gut microbiota alterations in response to fasting and hibernation.
Supplementary Material
Acknowledgments
We are thankful to Masaaki Hashimoto of Asahikawa Medical College and Satoru Fukiya and Michiko Tanaka of Hokkaido University for their helpful suggestions.
This study was partly supported by a Grant-in-Aid for Scientific Research from The Ministry of Education, Science, Sports and Culture of Japan.
We declare that no authors (Kei Sonoyama, Reiko Fujiwara, Naoki Takemura, Toru Ogasawara, Jun Watanabe, Hiroyuki Ito, and Tatsuya Morita) have conflicts of interest.
Footnotes
Published ahead of print on 21 August 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alexander, J. W. 1990. Nutrition and translocation. J. Parenter. Enteral Nutr. 14:170S-174S. [DOI] [PubMed] [Google Scholar]
- 2.Alverdy, J. C., E. Aoys, and G. S. Moss. 1988. Total parenteral nutrition promotes bacterial translocation from the gut. Surgery 104:185-190. [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banas, J. A., W. J. Loesche, and G. W. Nace. 1988. Classification and distribution of large intestinal bacteria in nonhibernating and hibernating leopard frogs (Rana pipiens). Appl. Environ. Microbiol. 54:2305-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banas, J. A., W. J. Loesche, and G. W. Nace. 1988. Possible mechanisms responsible for the reduced intestinal flora in hibernating leopard frogs (Rana pipiens). Appl. Environ. Microbiol. 54:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes, E. M., and G. C. Burton. 1970. The effect of hibernation on the caecal flora of the thirteen-lined ground squirrel (Citellus tridecemlineatus). J. Appl. Bacteriol. 33:505-514. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Amor, K., H. Heilig, H. Smidt, E. E. Vaughan, T. Abee, and W. M. de Vos. 2005. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl. Environ. Microbiol. 71:4679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boza, J. J., D. Möennoz, J. Vuichoud, A. R. Jarret, D. Gaudard-de-Weck, R. Fritsché, A. Donnet, E. J. Schiffrin, G. Perruisseau, and O. Ballévre. 1999. Food deprivation and refeeding influence growth, nutrient retention and functional recovery of rats. J. Nutr. 129:1340-1346. [DOI] [PubMed] [Google Scholar]
- 9.Carey, H. V., M. T. Andrews, and S. L. Martin. 2003. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol. Rev. 83:1153-1181. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33(Database iss.):D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collado, M. C., M. Derrien, E. Isolauri, W. M. de Vos, and S. Salminen. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73:7767-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deitch, E. A., J. Winterton, and R. Berg. 1987. Effect of starvation, malnutrition, and trauma on the gastrointestinal tract flora and bacterial translocation. Arch. Surg. 122:1019-1024. [DOI] [PubMed] [Google Scholar]
- 13.Deplancke, B., O. Vidal, D. Ganessunker, S. M. Donovan, R. I. Mackie, and H. R. Gaskins. 2002. Selective growth of mucolytic bacteria including Clostridium perfringens in a neonatal piglet model of total parenteral nutrition. Am. J. Clin. Nutr. 76:1117-1125. [DOI] [PubMed] [Google Scholar]
- 14.Derrien, M., E. E. Vaughan, C. M. Plugge, and W. M. de Vos. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54:1469-1476. [DOI] [PubMed] [Google Scholar]
- 15.Fujiwara, R., J. Watanabe, and K. Sonoyama. 2008. Assessing changes in composition of intestinal microbiota in neonatal BALB/c mice through cluster analysis of molecular markers. Br. J. Nutr. 99:1174-1177. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, G. R., J. H. Cummings, and G. T. Macfarlane. 1988. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl. Environ. Microbiol. 54:2750-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrica 40:237-264. [Google Scholar]
- 18.Gossling, J., W. J. Loesche, and G. W. Nace. 1982. Large intestine bacterial flora of nonhibernating and hibernating leopard frogs (Rana pipiens). Appl. Environ. Microbiol. 44:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossling, J., W. J. Loesche, and G. W. Nace. 1982. Response of intestinal flora of laboratory-reared leopard frogs (Rana pipiens) to cold and fasting. Appl. Environ. Microbiol. 44:67-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshi, S., T. Sakata, K. Mikuni, H. Hashimoto, and S. Kimura, S. 1994. Galactosylsucrose and xylosylfructoside alter digestive tract size and concentrations of cecal organic acids in rats fed diets containing cholesterol and cholic acid. J. Nutr. 124:52-60. [DOI] [PubMed] [Google Scholar]
- 21.Hume, I. D., C. Beiglboeck, T. Ruf, F. Frey-Roos, U. Bruns, and W. Arnold. 2002. Seasonal changes in morphology and function of the gastrointestinal tract of free-living alpine marmots (Marmota marmota). J. Comp. Physiol. B 172:197-207. [DOI] [PubMed] [Google Scholar]
- 22.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipman, T. O. 1995. Bacterial translocation and enteral nutrition in humans: an outsider looks in. J. Parenter. Enteral. Nutr. 19:156-165. [DOI] [PubMed] [Google Scholar]
- 24.Mainous, M., D. Z. Xu, Q. Lu, R. D. Berg, and E. A. Deitch. 1991. Oral-TPN-induced bacterial translocation and impaired immune defenses are reversed by refeeding. Surgery 110:277-283. [PubMed] [Google Scholar]
- 25.Miller, R. S., and L. C. Hoskins. 1981. Mucin degradation in human colon ecosystems. Fecal population densities of mucin-degrading bacteria estimated by a “most probable number” method. Gastroenterology 81:759-765. [PubMed] [Google Scholar]
- 26.Nettelbladt, C. G., M. Katouli, A. Volpe, T. Bark, V. Muratov, T. Svenberg, R. Möllby, and O. Ljungqvist. 1997. Starvation increases the number of coliform bacteria in the caecum and induces bacterial adherence to caecal epithelium in rats. Eur. J. Surg. 163:135-142. [PubMed] [Google Scholar]
- 27.Osborne, P. G., and M. Hashimoto. 2003. State-dependent regulation of cortical blood flow and respiration in hamsters: response to hypercapnia during arousal from hibernation. J. Physiol. 547:963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pamer, E. G. 2007. Immune responses to commensal and environmental microbes. Nat. Immunol. 8:1173-1178. [DOI] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto, K., H. Hirose, A. Onizuka, M. Hayashi, N. Futamura, Y. Kawamura, and T. Ezaki. 2000. Quantitative study of changes in intestinal morphology and mucus gel on total parenteral nutrition in rats. J. Surg. Res. 94:99-106. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, J. P. 1963. Microbiological aspects of hibernation in ground squirrels, p. 399. In E. G. Viereck (ed.), Influence of cold on host-parasite interactions. Arctic Aeromedical Laboratory, Fort Wainwright, AK.
- 32.Spaeth, G., R. D. Specian, R. D. Berg, and E. A. Deitch. 1990. Bulk prevents bacterial translocation induced by the oral administration of total parenteral nutrition solution. J. Parenter. Enteral Nutr. 14:442-447. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrand, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 34.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaughan, E. E., F. Schut, H. G. Heilig, E. G. Zoetendal, W. M. de Vos, and A. D. Akkermans. 2000. A molecular view of the intestinal ecosystem. Curr. Issues Intest. Microbiol. 1:1-12. [PubMed] [Google Scholar]
- 37.Zoetendal, E. G., A. D. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.