Abstract
Compost amendment and inoculations with specific microorganisms are fundamentally different soil treatment methods, commonly used in agriculture for the improvement of plant growth and health. Although distinct, both methods affect the rhizosphere and the plant roots. In the present study we used a 16S rRNA gene approach to achieve an overview of early consequences of these treatments on the assemblage of plant root bacterial communities. For this purpose, cucumber seedlings were grown, under controlled conditions, in perlite potting mix amended with biosolid compost or straw compost, or inoculated with Streptomyces spp. A uniform trend of response of root bacterial communities for all treatments was observed. Root bacterial density, measured as bacterial targets per plant tef gene by real-time PCR, was reduced in 31 to 67%. In addition, increased taxonomic diversity accompanied shifts in composition (α-diversity). The magnitude of change in these parameters relative to the perlite control varied between the different treatments but not in relation to the treatment method (compost amendments versus inoculations). Similarity between the compositions of root and of potting mix bacterial communities (β-diversity) was relatively unchanged. The abundance of Oxalobacteraceae was >50% of the total root bacterial community in the untreated perlite. Root domination by this group subsided >10-fold (straw compost) to >600-fold (Streptomyces sp. strain S1) after treatment. Thus, loss of dominance appears to be the major phenomenon underlining the response trend of the root bacterial communities.
Environmental concern over conventional agricultural fertilization and disease control measures has led to increased interest in finding environmentally friendly alternatives. The most explored ones include compost amendments (18, 36) and the application of different microbial preparations (11, 19, 37). These are widely distinct applications. The first approach adds to the amended medium not only a rich and diverse consortium of biological agents but also organic matter and nutrients. It was confirmed that the efficacy of such treatments involves the response of the soil, the plant, and the rhizosphere microbial communities (19, 56). The activities of rhizosphere microorganisms alter the rhizosphere and thus affect plant health and root growth and development (24). Therefore, one of the main objectives of compost amendment or of inoculation with specific microbial strains is manipulation of the plant rhizosphere conditions, particularly via manipulation of the microbial community composition (32).
The response of rhizosphere bacterial communities to different anthropogenic and other disturbances has been discussed in terms of resilience (3, 32). Generally, the introduction of new microorganisms produces only restricted spatial and temporal effects on the soil, rhizosphere, and root microbial communities (4, 29, 35). Thus, the plant growth-promoting effect of such treatments may be related to microbial events occurring during the early stages of plant development. Such early effects were pointed out for inoculants of different bacterial species (14, 42) and for compost amendment (15, 21, 50).
Consequences of compost amendment or of single species inoculation often include shifts in the plant roots hormonal balance or a plant systemic response, namely, induced systemic resistance (8, 38, 52). Thus, direct or indirect activities of the introduced microorganisms may result in similar modifications of the root habitat. If so, bacterial assemblages of treated roots may share qualitative and quantitative characteristics different from those exhibited by untreated roots.
The objective of the present study was therefore to describe and compare responses of bacterial communities of young plant roots to the application of compost or bacterial inoculants. This was performed in a simple model comprised of cucumber seedlings grown in potting mixes amended with compost or inoculated with Streptomyces spp. isolated from the two different composts.
MATERIALS AND METHODS
Streptomyces strains.
Two Streptomyces strains, designated S1 and S2, were isolated from biosolid (Cmp1) and straw (Cmp2) composts, respectively. Both strains were isolated based on their ability to adhere to cucumber seeds and were selected according to their ability to inhibit growth of Pythium aphanidermatum mycelia in vitro, as determined by dual-confrontation tests. According to sequence analysis of the partial 16S rRNA gene, the strain S1 16S rRNA gene sequence was 99% similar to that of Streptomyces thermocarboxydus (EU593721) and the strain S2 16S rRNA gene sequence was 100% similar to that of S. cyaneus (AY223254). 16S rRNA gene sequences of the two strains were 99% similar. Both strains were chromosomally transformed to retain resistance to the antibiotic apramycin. Transformation was achieved by conjugation with Escherichia coli ET12567/pIJ8641 as described by Kieser et al. (25). Plasmids and E. coli ET12567 strains were very kindly provided by Marvyn J. Bibb, John Innes Center, Norwich, England. The growth rate of the transformed strains in liquid yeast extract-malt extract (YEME) medium did not differ from that of the wild-type strains (data not shown). After transformation, both Streptomyces strains were routinely grown on mannitol-soy flour agar (MS) containing 50 mg of apramycin liter−1. Spores of S1 and of S2 were harvested from 14-day-old MS cultures grown at 30°C as described by Kieser et al. (26) and stored as glycerol stocks at −20°C. The stability of the transconjugants was tested in vitro. Spores of the transformed strains were plated on YEME agar and incubated at 30°C for 7 days. For each strain, 800 colonies were then picked at random and inoculated onto YEME agar containing 50 mg of apramycin liter−1. The in vitro transconjugant stabilities were >99.9 and 100% for strains S1 and S2, respectively. When used for inoculation of the potting mix, fresh spore suspensions of S1 and of S2 were prepared as described above. Spores were washed and resuspended in sterile saline solution before application in order to avoid carryover of growth medium components.
Potting mix preparation, plant material, and growth conditions.
Biosolid compost prepared from a mixture of sewage sludge and yard waste at 1:1 volumetric ratio was sampled from a commercial composting facility (Dlila Facility, Shacham, Givaat Ada, Israel). Biosolid compost characteristics were described by Termorshuizen et al. (49). Preparation and characteristics of straw compost, prepared from fresh separated cow manure and wheat straw at 2:1 volumetric ratio, were described by Yogev et al. (54). Both biosolid (Cmp1) and straw (Cmp2) composts were sieved through a 2-mm-pore-size sieve. In order to remove excess salts, composts were washed by the addition of 2 volumes of tap water. Composts were then drained and air dried. Perlite (4 mm) was likewise washed and air dried. Potting mixes containing either of the two composts were prepared by mixing perlite and compost at a 9:1 volume ratio. Where required, hydrated perlite was supplemented with ∼109 spores of S1 or S2 liter−1. Samples of the different potting mixes were taken for analysis of organic matter and the nitrogen and phosphorus content. The chemical characteristics of the final potting mixes are presented in Table 1. Potting mixes, including the perlite control, were then hydrated with half-strength Hoagland nutrient solution (17). The different mixes were covered and kept for 24 h before further use. Samples of each potting mix were taken 2 h after inoculation with the Streptomyces spores. The different potting mixes were distributed to plastic germination trays (15 ml, each pot). Cucumber (Cucumis sativus cv. ‘Kfir’, Zeraim Gedera, Israel) seeds were surface sterilized by soaking them in 3% sodium hypochlorite for 1.5 min and 70% ethanol for 1.5 min and then washed three times with sterile water. Seeds were germinated and grown for 7 days under controlled greenhouse conditions (30°C, continuous light). Plants were irrigated daily with half-strength Hoagland nutrient solution.
TABLE 1.
Chemical characteristics of the potting mixes used in this studya
| Mixb | Content
|
|||
|---|---|---|---|---|
| OM (%) | DOC (ppm) | N (%) | P (%) | |
| Perlite | 1.24 | 15.6 | NDc | ND |
| Cmp1 | 14.4 | 73.1 | 0.66 | 0.61 |
| Cmp2 | 9.32 | 50.0 | 0.42 | 0.42 |
| S1 | 1.31 | 25.5 | ND | ND |
| S2 | 1.27 | 23.3 | ND | ND |
Total organic matter (OM), dissolved organic carbon (DOC), total nitrogen (N), and phosphorus (P) were determined.
Cmp1, 10% (vol/vol) biosolid compost; Cmp2, 10% straw compost; S1 and S2; perlite inoculated with 106 spores of Streptomyces isolates S1 and S2 ml−1, respectively.
ND, not detected
Sampling procedure and enumeration of the inoculated Streptomyces.
Seven-day-old seedlings were harvested. For each treatment, three replicates were sampled, each composed of four individually grown plants randomly chosen and treated together. Plants were removed from the pots and shaken to remove loosely adhering particles. Roots were separated from shoots, placed in a sterile 50-ml plastic tube containing 30 ml of sterile saline (0.85% NaCl) and stirred by vortex mixing at maximum speed for 30 s to remove closely adhering particles (later described as the rhizosphere fraction). Roots were then removed from the tubes, dabbed dry on sterilized Whatman paper, sheared using sterilized scissors, and weighed. This was denoted as the root fraction (Rt). Half of the root material (designated for Streptomyces plate counts) was transferred into sterile 2-ml tubes containing 10 1-mm-diameter glass beads and 500 μl of sterile saline solution and then homogenized using a Bio 101 cell homogenizer for 25 s at 4.0 m s−1. The second half of the root material was transferred into DNA extraction tubes. The fraction containing closely adhering particle (above) was concentrated by centrifugation and was denoted as the rhizosphere potting mix (Rz) fraction. Next, 100-mg samples were weighed in DNA extraction tubes, and the remaining Rz fraction was suspended 1 to 10 (wt/vol) in sterile saline (designated for Streptomyces plate counts) and shaken for 30 min on a wrist action shaker at 300 rpm. Root-free potting mix (Pm) samples were also taken for DNA extraction (250 mg) and for Streptomyces plate counts and processed as described above for Rz samples. Samples used for Streptomyces enumeration were examined on MS agar containing 50 mg of apramycin liter−1. Plates were incubated at 30°C for 7 days. The cell density of S1 or S2 was expressed as CFU g−1 (fresh weight).
DNA extraction and PCR-DGGE of 16S rRNA gene fragments.
Total DNA was extracted from the samples in triplicate using an UltraClean soil DNA isolation kit (MoBio Laboratories, Inc.). The extracted DNA served as a template for PCR of bacterial 16S rRNA gene fragments using the primers 341FGC and 907R and analyzed by denaturing gradient gel electrophoresis (DGGE) as described by Green et al. (15). Streptomyces-specific PCR-DGGE was performed as described by Inbar et al. (22).
Cloning and sequencing of 16S rRNA gene fragments.
Clone libraries were constructed for root and for potting mix samples from each treatment (10 libraries). An equal-volume mixture of DNA from each of the replicates served as a template for PCR amplification of 16S rRNA gene fragment using primers the 341F (without GC clamp) and 907R. PCR products were cloned by using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Clones were examined by PCR and agarose (1.5%) gel electrophoresis and sequenced. Sequences recovered were submitted to the National Center for Biotechnology Information (NCBI) for BLAST analysis (2). Sequences were also examined by the CHECK_CHIMER program located at the Ribosomal Database Project (6), and suspected chimeric sequences were removed from further analyses.
Further analyses of the 16S rRNA gene sequences were performed using the program package ARB (31). A neighbor-joining tree, based on 493 alignment positions, was produced. Phylogenetic affiliation of the clone sequences was determined according to both ARB and NCBI-BLAST results. Sequences were considered as belonging to a single taxon if the dissimilarity between them was less than 1% (<5 mismatches). Sequences were further classified into distinct clades according to their inferred affiliation. Where possible, the family level of taxonomy was the basis for classification. Where family affiliation could not be determined, a 4% dissimilarity threshold was used. The sequences determined in the present study have been deposited in the GenBank database under accession numbers EU372270 to EU372626.
A phylogenetic tree of Oxalobacteraceae sequences detected in the clone libraries was generated by using the MEGA software (version 3.1). Closely related Oxalobacteraceae sequences, as well as other Oxalobacteraceae sequences, were imported from the NCBI and used to generate a neighbor-joining tree based on Kimura two-parameter model. Robustness of the proposed hierarchy was tested by bootstrap analysis with 1,000 resamplings.
Real-time PCR quantification of bacteria, Oxalobacteraceae, and the plant tef gene in root DNA samples.
Real-time PCR with SYBR green detection was used for the quantification of Oxalobacteraceae in root samples. The Oxaloabacteraceae-specific primers Oxal_225f (5′-GGAGCGGCCGATATCTGATTAG-3′ (16) and Oxalo_656r (5′-TTCTAGCCTTGCAGTCTCCATC-3′) (10), which amplify a 432-bp product, were used. In addition, the plant tef gene, coding for transcription elongation factor 1, was also quantified in root DNA samples and served as an internal normalizing gene (40, 53). The tef gene-specific primers tef_f (5′-ACTGTGCAGTAGTACTTGGTG-3′) and tef_r (5′-AAGCTAGGAGGTATTGACAAG-3′) (53), which amplify a specific 155-bp PCR product, were used. For quantification of bacteria, the primers 515_f (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (see above) were used to amplify a 411-bp PCR product. In order to correct bacterial target numbers for plant plastid-originating ones, a primer pair targeting the plant plastid was designed and applied. The primers Plast_f (5′-GAGGCAATAGCTTACCAAGGCG-3′) and Plast_r (5′-CTTGGTAGTTTCCACCGCCTG-3′) were used to amplify a 386-bp PCR product.
A plasmid standard containing the target region was generated for each primer set. For this purpose, PCR products for each primer pair were amplified from Rt-C DNA samples as a template. The PCR-amplified products were examined by gel electrophoresis to confirm the specificity of the amplification, and products were cloned by using a TOPO TA cloning kit. Plasmids were isolated by using a DNA-spin plasmid DNA extraction kit (iNtRON Biotechnology, Inc., Kyungki-Do, Korea). and 10 randomly selected cloned inserts were sequenced in order to assert their identity. Plasmid DNA concentrations were determined by NanoDrop ND1000 spectrophotometry. Copy numbers were calculated using the known DNA concentration and the specific plasmid plus the insert molecular weight, estimated from their lengths. Tenfold dilutions series within a range of 5 × 108 copies to 5 copies for Oxalobacteraceae standard and 109 to 10 copies for tef and bacterial standards were prepared.
Real-time PCR assays were conducted in polypropylene 96-well plates in a Mx3000P QPCR system (Stratagene, La Jolla, CA). Each 20-μl reaction contained 10 μl of Absolute Blue SYBR green ROX mix (Thermo Fisher Scientific, Surrey, United Kingdom), 1.25 μl of each primer (10 μM), 6.5 μl of H2O, and 1 μl of template DNA. The PCR conditions were 15 min at 95°C, followed by 40 cycles of 95°C for 30 s, 60°C (58°C for the bacterial primer pair) for 30 s, and 72°C for 30 s. Each plate contained triplicate reactions for each DNA sample, the appropriate set of standards, and no-template controls. Melting-curve analysis of the PCR products was conducted after each assay to confirm that the fluorescence signal originated from specific PCR products. PCR products were also examined by agarose gel electrophoresis to confirm the specificity of the amplification.
For all target genes, eight 10-fold dilutions of the calibration standards were measurable down to 100 copies of DNA for bacteria, plant plastid, and tef μl−1 and 50 copies of DNA μl−1 for Oxalobacteraceae. The standard curve slopes, correlation coefficients and amplification efficacies were calculated by using the MxPro QPCR Software analysis tools (Stratagene) and were, respectively, −3.194, 0.985, and 105.4% for bacteria; −3.326, 0. 991, and 99.8% for plant plastid; −3.327, 0.965, and 99.8% for tef; and −3.292, 0.969, and 101.3% for Oxalobacteraceae. Bacterial and Oxalobacteraceae densities are presented as the number of targets per tef targets and as the fold change from the untreated control by using the 2−ΔΔCT calculation method described by Livak and Schmittgen (28).
Data analysis.
PCR-DGGE patterns were aligned and analyzed by using Fingerprint II software (Bio-Rad). A neighbor-joining tree based on the Pearson r distance matrix was produced. Aligned densitometric curves were exported form the Fingerprint II software (Bio-Rad Laboratories), and multidimensional scaling analysis was done based on 1-Pearson r distance matrix using Statistica (version 7.1) software (Stat Soft, Inc., Tulsa, OK).
Clone libraries sequence data was analyzed by Unifrac metric analysis. Hierarchical clustering was performed in the UniFrac web interface (30), using an ARB-generated neighbor-joining tree containing the 358 clone sequences and file-mapping sequence labels to the specific sample type (Rt or Pm) and treatment. The unweighed pair group method with arithmetic means (UPGMA) was used as the hierarchical clustering algorithm in order to establish relationships between the composition of the different root and potting mix samples. The robustness of the clusters obtained was examined by the sequence jackknife method, wherein 100 replicate trees were examined; in each replicate tree the different samples were represented by 20 clone sequences each.
For each clone library, the coverage estimate was calculated by using Goods' equation (13): coverage = 1 − (n1/N), where n1 is the number of taxa that are represented only by a single clone sequence, and N is the total number of clone sequences in the specific library examined. Coverage estimates were lower when calculated at the taxon level than the respective estimates using the designated clade affiliation (see above) as a grouping factor (data not shown). Accordingly, further calculations of the different diversity and similarity indices were based on designated clade affiliation of the clone sequences.
The Shannon-Weaver index of diversity (H′) was calculated by using the equation: H′ = −Σpi·ln(pi), where pi is the proportion of the ith clade in the clone library. Approximated variance of H′ was calculated according to the method of Poole (39) and used for estimation of the 95% confidence intervals (95% CI). Dominance (D) was calculated by using the equation: D = Σ(pi)2. The Chao 1 richness estimate and its related 95% CI were calculated according to the method of Chao (5). The Chao-Jaccard abundance based similarity index (Jabd) was pairwise calculated by using EstimateS software (7) Jabd = UV/(U + V − UV), where U and V denote the total relative abundance of individuals belonging to the shared clades in the first and the second community libraries examined, respectively.
RESULTS
PCR-DGGE analysis of bacterial communities.
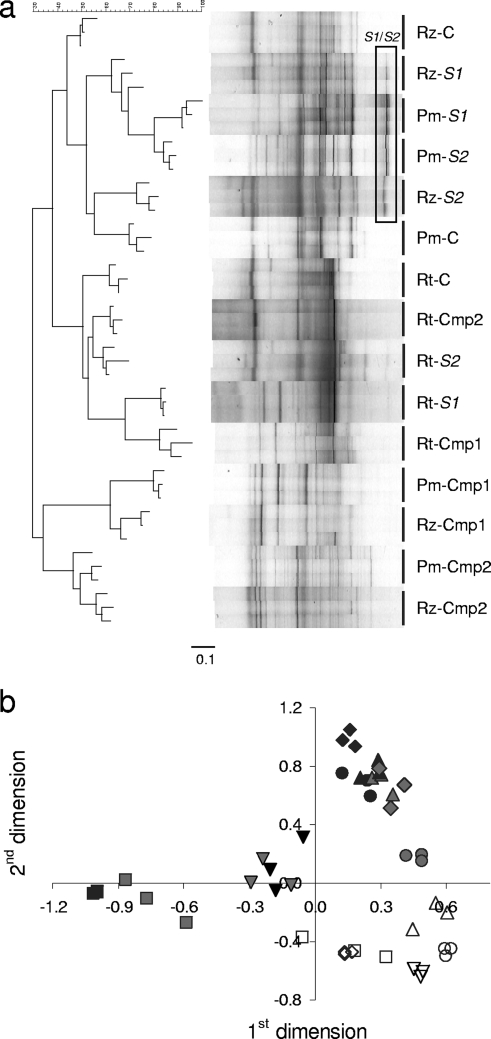
We compared the effects of biological interventions on bacterial community composition in potting mix, rhizosphere, and roots of 7-day-old cucumber seedlings. The treatments were potting mix amendment with one of two compost types (biosolid compost [Cmp1] or straw compost [Cmp2]) or inoculation with specific compost-derived Streptomyces spp. (S1 or S2, respectively). The control consisted of Hoagland solution hydrated perlite (unsterile). Shifts in bacterial community composition due to the different treatments were determined by PCR-DGGE analysis of 16S rRNA gene fragments using general bacterial PCR primers. Each sample combined four individual pots with one seedling in each. Three samples were analyzed for each treatment (making a total of 12 plants per treatment). High reproducibility was found for the triplicate PCR-DGGE patterns of the three independent samples obtained from each treatment (Fig. 1).
FIG. 1.
Effects of compost amendment and of Streptomyces species inoculation on bacterial community composition in potting mix (Pm), rhizosphere (Rz), and roots (Rt) of 7-day-old cucumber seedlings. Bacterial community compositions were compared based on PCR-DGGE patterns of 16S rRNA gene fragments. PCR-DGGE patterns were analyzed by using the Fingerprint II software. (a) A neighbor-joining tree was calculated from cosine correlation distance matrix. (b) MDSA of DGGE pattern densitometric curves. MDSA was performed based on 1-Pearson r distance matrices between the densitometric curves, using Statistica software. The first and the second dimensions are presented (stress = 0.014). The different treatments are denoted as follows: ○, C, untreated potting mix; □, Cmp1, biosolid compost (10% [vol/vol]); ⋄, S1, Streptomyces sp. isolated from Cmp1; ▿, Cmp2, straw compost (10% [vol/vol]); ▵ S2, Streptomyces sp. isolated from Cmp2. The box indicates bands corresponding to S1 or S2 populations (verified by sequences analysis of DNA from excised bands). Open, gray, and black symbols represent root, rhizosphere, and potting mix samples, respectively.
DGGE bands representing the Streptomyces strains were detected in the bacterial community DGGE patterns of the potting mix (Pm-S1 and Pm-S2) and rhizosphere (Rz-S1 and Rz-S2) samples where inoculated (Fig. 1). Neither of the Streptomyces strains appeared in bacterial community patterns of other samples (Fig. 1). However, both Streptomyces strains could be reisolated from root samples of the inoculated plants (Rt-S1 and Rt-S2) by plating on selective medium (see Fig. S1 in the supplemental material) and were also detected in these root samples in Streptomyces-specific PCR-DGGE patterns of the same samples (see Fig. S2 in the supplemental material). This implies that although the inoculated Streptomyces spp. colonized the cucumber roots, they were not among the dominant root populations.
Comparative analysis of community patterns was performed by using Fingerprint II image analysis software. A neighbor-joining tree, based on pairwise cosine correlation matrix was calculated (Fig. 1a). In order to further analyze associations between the bacterial communities patterns, multidimensional scaling analysis (MDSA) was performed by using pairwise Pearson correlations of densitometric curves obtained from the DGGE patterns (Fig. 1b). This analysis allows grouping of individual patterns avoiding the misrepresentation of distances inherent to hierarchical cluster analysis methodologies. The low-stress value given for the MDSA (0.014) indicated that the reproduced distances reliably represent the original pairwise distances. The clusters formed by both analysis methods were generally in agreement.
Root community patterns significantly differed from those of the potting mix and rhizosphere patterns (Fig. 1). Substantial shifts from the control consisting of perlite and Hoagland nutrient solution, similar in trend, were observed in root bacterial community patterns due to treatment with Cmp1 or S1. In contrast, relatively little change was observed in root bacterial community patterns as a result of Cmp2 amendment or S2 inoculation.
Both analysis methods discriminated between potting mix and rhizosphere samples of the control samples. No robust discrimination could be made between potting mix and rhizosphere samples in any of the treatments. Amendment with either of the compost types resulted in substantial shifts in potting mix community patterns with respect to the controls. The effect of inoculation with S1 or with S2 on both potting mix and rhizosphere DGGE patterns was less extensive than that induced by compost amendment (Fig. 1).
Community composition and structure based on partial 16S rRNA gene sequences.
A clone library of partial 16S rRNA gene sequences was constructed for bacterial communities of potting mix and of root samples of all treatments. This was done using PCR products directly amplified from total DNA obtained from each treatment. Of 431 sequences obtained, 48 originated from the plant plastid (11.13%), and 25 were suspected chimeras (5.8%). Omitting these, a total of 358 clone sequences were analyzed, of which 184 were obtained from roots and 174 were obtained from potting mixes. Sequences were analyzed by BLAST and by the ARB phylogenetic package and assigned an inferred taxonomic affiliation. Using a 1% sequence similarity threshold, sequences were grouped into 176 distinct taxa, affiliated with 56 designated clades. After the sequences were grouped according to clade affiliation, the coverage values obtained ranged between 0.61 (Pm-Cmp1) and 0.94 (Pm-S1) (Table 2). For potting mix libraries, the coverage levels were high in the absence and lower in the presence of compost. No such relationship between compost amendment and coverage levels was found for root libraries (Table 2). Table 3 details the relative abundances of the different taxonomic groups found in the clone libraries of potting mix and roots. The inoculated Streptomyces comprised 11.4 and 13.9% of the clone libraries of Pm-S1and Pm-S2, respectively. No Streptomyces sequences were detected in the other samples (Table 3). This result was consistent with the occurrence of S1 and S2 bands in DGGE patterns (Fig. 1a).
TABLE 2.
Diversity parameters of root and potting mix bacterial communities based on partial 16S rRNA gene sequences obtained from clone librariesa
| Communityb | Value (95% CI)
|
D | Coverage | No. of:
|
||
|---|---|---|---|---|---|---|
| H′ | Chao 1 | Clades | Clones | |||
| Rt-C | 1.41 (1.12-1.70) | 10.00 (9.3-10.9) | 0.332 | 0.93 | 7 | 42 |
| Pm-C | 2.05 (1.71-2.39) | 15.75 (14.1-18.8) | 0.18 | 0.82 | 12 | 34 |
| Rt-Cmp1 | 2.53 (2.30-2.75) | 17.50 (16.3-19.9) | 0.092 | 0.88 | 15 | 40 |
| Pm-Cmp1 | 2.77 (2.48-3.06) | 30.00 (27.6-33.1) | 0.076 | 0.61 | 19 | 31 |
| Rt-Cmp2 | 2.42 (2.17-2.67) | 18.75 (17.1-21.8) | 0.111 | 0.86 | 15 | 43 |
| Pm-Cmp2 | 2.75 (2.47-3.03) | 33.20 (29.5-38.4) | 0.082 | 0.68 | 20 | 38 |
| Rt-S1 | 2.46 (2.47-3.03) | 19.25 (17.1-22.9) | 0.099 | 0.75 | 14 | 28 |
| Pm-S1 | 1.99 (1.76-2.22) | 9.50 (9.1-11.8) | 0.161 | 0.94 | 9 | 35 |
| Rt-S2 | 2.06 (1.74-2.37) | 13.33 (11.5-17.2) | 0.17 | 0.84 | 10 | 31 |
| Pm-S2 | 1.72 (1.45-1.99) | 11.00 (10.3-11.9) | 0.227 | 0.92 | 8 | 36 |
Diversity indices were calculated after grouping the sequences according to the designated clade affiliation. Shannon-Weaver (H′), Chao 1 (5), dominance (D), and coverage (13) values were calculated. The 95% CI values for H′ were calculated from the approximated variance (39) and for Chao 1 according to the method of Chao (5).
C, untreated control; Cmp1, biosolid compost; Cmp2, straw compost; S1, Streptomyces sp. isolated from Cmp1; S2, Streptomyces sp. isolated from Cmp2.
TABLE 3.
Composition of root and potting mix bacterial communitiesa
| Taxon and organism (GenBank no.) | Relative abundance of partial 16S rRNA gene sequences
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All (%) | Root (%)
|
Potting mix (%)
|
|||||||||
| C | Cmp1 | Cmp2 | S1 | S2 | C | Cmp1 | Cmp2 | S1 | S2 | ||
| Actinobacteria | 4.2 | 2.3 | 6.5 | 5.3 | 11.4 | 13.9 | |||||
| Streptomyces | 2.5 | 11.4 | 13.9 | ||||||||
| Other Actinobacteria | 1.7 | 2.3 | 6.5 | 5.3 | |||||||
| Bacteroidetes | 19.8 | 11.9 | 27.5 | 16.3 | 25.0 | 16.1 | 5.9 | 45.2 | 44.7 | 2.9 | 5.6 |
| Flavobacteriaceae | 5.3 | 11.9 | 5.0 | 4.7 | 16.1 | 2.9 | 3.2 | 2.6 | 2.9 | 2.8 | |
| Flavobacteriumsp. (AJ786797) | 3.4 | 9.5 | 2.5 | 4.7 | 12.9 | 2.9 | |||||
| Cryomorphaceae | 1.9 | 7.5 | 10.7 | 3.2 | |||||||
| Crenotrichaceae | 0.3 | 2.6 | |||||||||
| Flexibacteraceae | 5.0 | 11.6 | 3.6 | 2.9 | 12.9 | 15.8 | 2.8 | ||||
| Sphingobacteriaceae | 1.1 | 6.5 | 5.3 | ||||||||
| Other Bacteroidetes | 6.1 | 15.0 | 10.7 | 19.4 | 18.4 | ||||||
| Alphaproteobacteria | 31.8 | 11.9 | 20.0 | 27.9 | 42.9 | 58.1 | 32.4 | 19.4 | 18.4 | 51.4 | 47.2 |
| Rhizobiaceae | 17.1 | 11.9 | 7.5 | 20.9 | 17.9 | 32.3 | 26.5 | 2.6 | 14.3 | 38.9 | |
| Rhizobium/Agrobacterium | 12.9 | 7.1 | 2.5 | 16.3 | 25.8 | 26.5 | 11.4 | 38.9 | |||
| Agrobacterium tumefaciens (AY972364) | 9.2 | 2.4 | 19.4 | 23.5 | 11.4 | 38.9 | |||||
| Caulobacteraceae | 1.7 | 4.7 | 3.6 | 3.2 | 5.7 | ||||||
| Phyllobacteriaceae | 0.3 | 2.6 | |||||||||
| Hyphomicrobiaceae | 1.4 | 3.6 | 6.5 | 5.3 | |||||||
| Rhodobacteraceae | 0.3 | 2.9 | |||||||||
| Rhodospirillaceae | 1.7 | 2.5 | 7.1 | 9.7 | |||||||
| Azospirillum lipoferumsp. (AY998242) | 1.4 | 2.5 | 7.1 | 9.7 | |||||||
| Sphingomonadaceae | 9.2 | 10.0 | 2.3 | 10.7 | 6.5 | 5.9 | 16.1 | 7.9 | 28.6 | 8.3 | |
| Unclassified | 0.3 | 3.2 | |||||||||
| Betaproteobacteria | 29.1 | 73.8 | 15.0 | 23.3 | 14.3 | 16.1 | 50.0 | 16.1 | 5.3 | 34.3 | 33.3 |
| Oxalobacteraceae | 7.0 | 52.4 | 7.0 | ||||||||
| Burkholderiaceae | 5.9 | 29.4 | 2.6 | 11.4 | 16.7 | ||||||
| Limnobacter thiooxidans (AJ289885) | 3.9 | 23.5 | 2.9 | 13.9 | |||||||
| Alcaligenaceae | 0.6 | 2.5 | 2.3 | ||||||||
| Comamonadaceae | 6.4 | 16.7 | 2.5 | 4.7 | 3.2 | 5.9 | 14.3 | 13.9 | |||
| Hydrogenophaga atypica(AJ585992) | 2.8 | 5.9 | 11.4 | 11.1 | |||||||
| Unclassified Burkholderiales | 1.9 | 4.8 | 2.3 | 2.9 | 8.6 | ||||||
| Methylophilaceae | 5.6 | 10.0 | 7.0 | 14.3 | 12.9 | 8.8 | 12.9 | ||||
| Methylophilussp. strain EHg7 (AY43679) | 2.5 | 4.6 | 12.9 | 8.8 | |||||||
| Methylovorus glucosetrophus(AY486133) | 1.9 | 10.0 | 7.1 | 3.2 | |||||||
| Nitrosomonadaceae | 0.3 | 2.6 | |||||||||
| Unclassified | 0.8 | 3.2 | 2.8 | ||||||||
| Gammaproteobacteria | 10.3 | 2.4 | 27.5 | 18.6 | 7.1 | 9.7 | 8.8 | 6.5 | 18.4 | ||
| Enterobacteriaceae | 1.1 | 10.0 | |||||||||
| Pseudomonadaceae | 3.4 | 2.4 | 18.6 | 3.2 | 5.9 | ||||||
| Xanthomonadaceae | 5.6 | 17.5 | 7.1 | 6.5 | 2.9 | 6.5 | 15.8 | ||||
| Unclassified | 0.3 | 2.6 | |||||||||
| Chloroflexi, unclassified | 0.8 | 3.2 | 5.3 | ||||||||
| Firmicutes | 3.9 | 10.0 | 11.6 | 10.7 | 2.9 | 3.2 | 2.6 | ||||
| Bacillaceae | 1.9 | 10.0 | 9.3 | ||||||||
| Clostridiaceae | 0.6 | 7.1 | |||||||||
| Thermoactinomycetaceae | 0.8 | 2.3 | 3.6 | 2.6 | |||||||
| Unclassified | 0.6 | 2.9 | 3.2 | ||||||||
The relative abundances of partial 16S rRNA gene sequences affiliated with the different taxonomic groups are presented. The different amendments are denoted as follows: C, untreated control; Cmp1, biosolid compost; Cmp2, straw compost; S1, Streptomyces sp. isolated from Cmp1; S2, Streptomyces sp. isolated from Cmp2. Relative abundance is expressed as the percentage of the total number of clone sequences from each library. “All” refers to the relative abundance in the complete set of sequences (358 clones). Specific subfamily taxa are denoted in boldface in column 1.
Composition of root and of potting mix bacterial communities was affected by all of the examined treatments (Table 3). While Betaproteobacteria dominated the root bacterial community in the untreated control (73.8%), in Rt-S1 and in Rt-S2 communities dominance shifted to Alphaproteobacteria (Table 3). Composition of Rt-Cmp1 and Rt-Cmp2 bacterial communities was more evenly distributed between the different bacterial groups. potting mix bacterial communities of Cmp1 and of Cmp2 were dominated by Bacteroidetes. Bacterial communities of Pm-C, Pm-S1, and of Pm-S2 were dominated by Alphaproteobacteria and Betaproteobacteria.
The extent of the change in the composition of root bacterial communities was more pronounced in Cmp1 and S1 treatments than in Cmp2 and S2. After Cmp1 amendment or S1 inoculation, <20% of the root sequences belonged to clades also detected in the Rt-C clone library. However, >50% of the root sequences of the Rt-Cmp2 and Rt-S2 communities belonged to clades detected in Rt-C. For all treatments, up to half of the “novel” root-associated clades were also detected in the control potting mix, in the treated potting mix, or in both (Table 3).
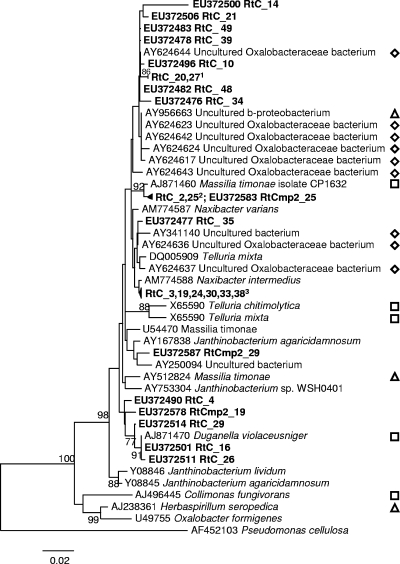
Oxalobacteraceae formed the dominant clade in Rt-C (52.4% of that clone library). Otherwise, Oxalobacteraceae were detected only in Rt-Cmp2 but at much lower relative abundance (7%). All Oxalobacteraceae sequences detected here were closely related to Oxalobacteraceae sequences previously retrieved from cucumber roots or rhizosphere (Fig. 3) (15, 16). Furthermore, apart from a single sequence (RtC_14 [EU372500]), all Oxalobacteraceae sequences shared above 96% similarity (see Fig. S3 in the supplemental material).
FIG. 3.
Composition of root-associated Oxalobacteraceae. The neighbor-joining phylogenetic tree was developed based on a Kimura two-parameter model of detected Oxalobacteraceae populations. The bootstrapped neighbor-joining tree was generated with 1,000 resamplings, and nodes with bootstrap values of >75% are indicated, as described in the text. RtC, roots of plants grown in untreated perlite; RtCmp2, roots of plants grown in Cmp2-amended perlite. Oxalobacteraceae sequences detected in the present study are indicated in boldface. Symbols: ⋄, cucumber root or rhizosphere-associated Oxalobacteraceae from other studies; ▵, root- or rhizosphere-associated Oxalobacteraceae originated from plants other than cucumber; □, soil isolates. Superscript numbers: 1, accession numbers EU372505 and EU372512, respectively; 2, accession numbers EU372488 and EU372510, respectively; 3, accession numbers EU372489, EU372504, EU372509, EU372515, EU372475, and EU372480, respectively.
Divergence between bacterial community clone libraries was assessed by pairwise calculation of the Chao-Jaccard abundance-based index (Jabd) (Table 4). This similarity index was calculated for potting mix and root samples using designated clade affiliation of the clone sequences. For potting mix samples, similarity to the unamended control was lower in the compost amendment treatments compared to the Streptomyces spp. inoculation treatments. Conversely, for roots, similarity levels were not related to the treatment type: Cmp1 or S1 treatment resulted in less similarity to the control than the Cmp2 or S2 treatments (Table 4). Within each treatment, the similarity between root and potting mix bacterial communities was generally low (control, Cmp1, Cmp2, and S1) but was higher for S2 (Table 4). After inoculation with S2, a single group of bacteria, related to the Rhizobiaceae, was found to dominate the root and potting mix clone libraries (Table 3). This shared dominance was the main contributor to the increased root-to-potting mix similarity in this particular treatment (Table 4).
TABLE 4.
Similarity between potting mix and root bacterial communities as affected by intervention
| Treatmentb | Treatment to control Jabda
|
Rt to Pm Jabd
|
||
|---|---|---|---|---|
| Pm | Rt | Jabdc | Difference (Δ) from controld | |
| Control | 1 | 1 | 0.266 | 0 |
| Cmp1 | 0.183 | 0.123 | 0.262 | −0.004 |
| Cmp2 | 0.396 | 0.521 | 0.232 | −0.034 |
| S1 | 0.55 | 0.077 | 0.24 | −0.026 |
| S2 | 0.681 | 0.317 | 0.437 | +0.171 |
Similarity was estimated by pairwise calculation of the Chao-Jaccard abundance-based similarity index (Jabd) using abundance data obtained from 16S rRNA gene sequences. Pm, potting mix; Rt, root.
Cmp1, biosolid compost; Cmp2, straw compost; S1, Streptomyces sp. isolated from Cmp1; S2, Streptomyces sp. isolated from Cmp2.
Jabd calculated using EstimateS software (7).
That is, the treatment Jabd (Rt to Pm) − the control Jabd (Rt to Pm).
In order to examine the effect of the different treatments on bacterial community structure, the Shannon diversity index (H′), Chao1 richness index (Chao1), and dominance (D) were calculated based on sequence data obtained from the clone libraries. These diversity parameters were calculated for all potting mix and root samples using designated clade affiliation of the clone sequences (Table 2). Root bacterial communities of all treatments had higher H′ and Chao1 values in parallel with lower D values compared to Rt-C. In contrast, the effect of the treatments on the potting mix bacterial community structure differed between compost amendment and Streptomyces spp. inoculation. Compost-amended potting mix bacterial communities had higher H′ and Chao1 values and lower D values compared to Pm-C. Streptomyces-inoculated potting mix bacterial communities had lower Chao1 values compared to Pm-C and had similar (S1) or higher (S2) D values (Table 2). In the control, Cmp1, and Cmp2 treatments, potting mix communities had higher Chao1 and H′ values than the respective root communities. An opposite trend was observed after inoculation with S1 or with S2 (Table 2).
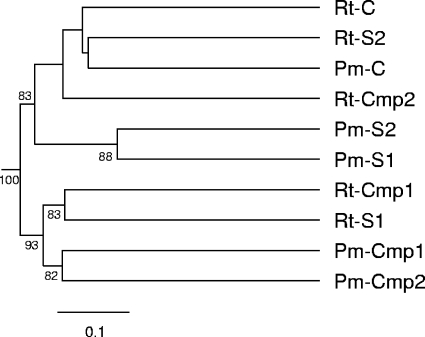
UniFrac metric analysis of partial 16S rRNA gene sequences.
A UniFrac metric algorithm was used for additional comparison of the root and potting mix clone libraries. UniFrac calculates relative divergence between the assigned communities according to shared or distinct branching within the phylogenetic tree. Figure 2 presents the UPGMA clustering of the different communities according to their pairwise UniFrac metrics, based on a neighbor-joining tree of the sequences. Divergence between the compositions of bacterial communities was considered robust if the calculated jackknife value was described above 75%. UniFrac related between the composition of Rt-C, Rt-Cmp2, Rt-S2, and Pm-C bacterial communities. Rt-Cmp1 and Rt-S1 were clustered together, separately from other root samples. Apart from Pm-C, all potting mix samples were separated from root samples (Fig. 2). Thus, UniFrac analysis results were generally in agreement with the results of the DGGE patterns analyses (Fig. 1) with some deviations.
FIG. 2.
UniFrac metric analysis of root (Rt) and potting mix (Pm) 16S rRNA gene sequences. A UPGMA tree, based on pairwise distances between the different bacterial communities (UniFrac metric, based on 358 partial 16S rRNA gene sequences), is shown. The different treatments are denoted as follows: C, untreated potting mix; Cmp1, biosolid compost (10% [vol/vol]); S1, Streptomyces sp. isolated from Cmp1; Cmp2, straw compost (10% [vol/vol]); S2, Streptomyces sp. isolated from Cmp2. Robustness of the proposed branching was examined by jackknife analysis (numbers at the nodes represent the percentage of occurrence of the node when each community was represented by 20 randomly chosen sequences in the distance matrix for n = 100 replicates).
Real-time PCR quantitative analysis of total bacteria in root communities.
We quantified bacterial targets in root samples using real-time PCR with SYBR green using a general bacterial primer pair targeted at the 16S rRNA gene (Eub515_f and 907_R). The plastid 16S rRNA gene was also amplified using general bacterial primer pairs. Therefore, we designed and applied an additional primer pair that specifically amplifies a 386-bp PCR product of the plant plastid gene. Plastid-originating targets accounted for up to 13.5% of the targets, and values were accordingly used for correction of the number of bacterial targets. The plant housekeeping gene tef was used as an internal reference to enable comparison between samples. All treatments reduced bacterial target numbers in root samples compared to the untreated control (Table 5). Reduction was highest for Rt-Cmp1 (66%) and was lowest for Cmp2 (31%). Bacterial target numbers per tef were significantly correlated with Jabd between the roots of treated samples and those of the untreated control (r2 = 0.86; P < 0.05).
TABLE 5.
Relative quantification of total bacteria and Oxalobacteraceae in root samplesa
| Sample | Total bacteriab
|
Oxalobacteraceae
|
|||
|---|---|---|---|---|---|
| Mean bacteria/tef ± SD | 2−ΔΔCT | Mean bacteria/tef ± SD | %Bc | 2−ΔΔCT | |
| Control | 373 ± 64 | 203 ± 58 | 55 | ||
| Cmp1 | 139 ± 25 | 0.33 (0.28-0.39) | 2.6 ± 0.65 | 0.61 | 0.005 (0.002-0.013) |
| Cmp2 | 264 ± 6 | 0.69 (0.67-0.71) | 15.4 ± 3.94 | 5.76 | 0.049 (0.033-0.073) |
| S1 | 169 ± 29 | 0.43 (0.36-0.52) | 0.32 ± 0.01 | 0.19 | 0.002 (0.0006-0.006) |
| S2 | 130 ± 20 | 0.34 (0.29-0.39) | 3.1 ± 0.82 | 1.93 | 0.011 (0.008-0.014) |
Total bacteria and oxalobacterial targets were normalized using the reference tef gene. The 2−ΔΔCT value is the fold change from the control and was calculated as described by Livak and Schmittgen (28). Values in parentheses represent the range of values within one standard deviation of the mean.
Eubacterial targets were determined after correction for plant plastid originating targets.
The %B value is the percentage of Oxalobacteraceae targets of the total bacteria.
Real-time PCR quantitative analysis of Oxalobacteraceae in root bacterial communities.
According to the clone library results, while Oxalobacteraceae made up more than 50% of root sequences in Rt-C, they accounted for only 7% of Rt-Cmp2 and were undetected in other root libraries (Table 3). We therefore examined the effect of the different treatments on Oxalobacteraceae population size. Using a specific primer pair, Oxalobacteraceae were detected in all root samples at various target densities per tef (Table 5). Due to the mixed nature of the amplified targets (Fig. 3) and the little knowledge available regarding rRNA operon copies in Oxalobacteraceae genomes (only two genomes are currently available), it is impossible to determine the actual cell numbers corresponding to the gene copy numbers obtained. It was, however, interesting that for both Rt-C and Rt-Cmp2 the relative abundance values obtained for Oxalobacteraceae by both clone libraries and real-time PCR were similar (Tables 2 and 4).
According to the 2−ΔΔCT method, real-time PCR results showed that relative to Rt-C, Oxalobacteraceae target numbers were significantly lower in all treated samples (Table 5). Furthermore, the extent of reduction was not equal for the different treatments, being highest for Rt-S1 (2.7 orders of magnitude) and lowest for Rt-Cmp2 (1.3 orders of magnitude). Oxalobacteraceae target numbers per tef were significantly correlated with those of bacteria (r2 = 0.78; P < 0.05).
DISCUSSION
Soil amendment with compost versus inoculations with a single species represents fundamentally distinct modifications. Compost improves soil structure, elevates soil content of organic matter, and supplies macro- and micronutrients (41, 55). Furthermore, compost introduces an abundant and highly diverse microbial input (49, 51). Thus, via a combination of physical, chemical and biological conditioners, compost may simultaneously influence the assemblage processes of root and rhizosphere bacterial communities. By comparison, soil inoculation with a single bacterial or fungal species may be viewed as a “simple” modification. Even so, the end effects of inoculation may extend beyond the target organisms—the plant or the phytopathogen (23, 29)—and may involve multiple mechanisms of action (8, 56). Here, we describe the consequences of both types of treatment on early assemblage of bacterial communities on plant roots. This was investigated in a simple model: cucumber seedlings grown under controlled conditions in perlite and Hoagland nutrient solution as a control and different potting mixes with compost or bacterial amendments.
The effects of compost amendment and of Streptomyces spp. inoculation on potting mix bacterial communities were explicit. The change in the composition of potting mix bacterial communities was specific to each treatment (Table 3). However, the extent of divergence from the composition of the untreated control potting mix was higher for the compost amendment treatments compared to Streptomyces spp. inoculation (Fig. 1 and Table 4). Furthermore, the consequences regarding community diversity parameters were opposite in trend between the two treatment types (Table 2). Such dichotomous responses to the different treatment types did not occur in the root bacterial communities.
Many studies have examined the effect of inoculation with a specific microbial species on root and rhizosphere bacterial communities. In some cases, a strong and persistent effect was observed (12, 39, 45, 46). However, in the majority of cases inoculation with plant growth-promoting bacterial strains resulted in undetectable or only minor and transient shifts in root and rhizosphere bacterial communities (4). These differences in effects may reflect the natural variance between the different experimental systems (including inoculant species, plant species, and soil type). In our experimental system, amendment with compost or inoculation with Streptomyces spp. had qualitative and quantitative effects on bacterial communities colonizing the roots of young cucumber plants. A significant shift in the composition of cucumber root bacterial communities after amendment with different composts was previously reported in greenhouse (15), as well as field experiments (50).
Although in the potting mix the responses of the bacterial community differed between compost amendment and Streptomyces inoculation, the consequences of the modifications applied to root bacterial communities were not related to the modification type. Surprisingly, the observed shifts in root bacterial community composition and diversity parameters were more related to the modification origin than to the modification type: Cmp1 and the derived Streptomyces strain S1 produced a related shift in the composition of root bacterial communities (Fig. 1 and 2 and Table 3). Detailed examination of the clone libraries revealed that the divergence of Rt-Cmp1 and Rt-S1 communities from Rt-C was due to displacement of the majority of Rt-C populations (Table 3). In contrast, Rt-C populations persisted in Rt-Cmp2 or Rt-S2 and supported linkage between the compositions of these root communities (Fig. 2).
An interesting phenomenon observed was the increase in root bacterial community diversity (Shannon H′ and Chao1 richness) after each of the treatments was applied (Table 2). The data obtained indicated that a loss-of-dominance situation may explain this phenomenon. In clone libraries the Oxalobacteraceae relative abundance was >50% in Rt-C. They were otherwise detected only in Rt-Cmp2 at a much lower relative abundance (Table 3). Quantitative PCR results confirmed the high abundance of Oxalobacteraceae in Rt-C, as well as the substantial reduction in the size of this population after any of the treatments (Table 5). Although we could not find parallel examples for bacteria in the literature, in plants and animals it has been demonstrated that the removal of a dominant species resulted in increased diversity (27, 44). In parallel, gain of dominance by invasive species (21, 47) or as a result of removal of a predator (1) was shown to decrease diversity. Hence, Oxalobacteraceae loss of dominance may, in part, explain the increase in the diversity of the root bacterial communities after treatment. It appears that although Oxalobacteraceae are successful root-colonizing bacteria, they are highly sensitive to environmental conditions (16).
Coinciding with the reduction in Oxalobacteraceae numbers, a 31 to 67% reduction in the total bacterial target numbers was observed in treated root samples (Table 5). Ruppel et al. (43) also reported a 33% decrease in Brassica oleracea root bacterial targets in response to inoculation with a specific bacterial strain. That decrease was dependent on the size of the inoculum and was found to be transient. Since Oxalobacteraceae and total bacterial numbers were significantly correlated, it could be assumed that the reduction in Oxalobacteraceae explains the reduction in root-colonizing bacteria. Due to the variance in the number of rRNA gene operons in the bacterial genome, in the composition of bacterial communities, even a 10-fold change in bacterial rRNA gene target numbers may theoretically be explained by a shift in composition. Indeed, the reduction in root bacterial target numbers was also correlated with the magnitude of the shift between the compositions of treated and untreated root bacterial communities (Tables 3 and 4).
Members of the Rhizobiaceae were the single most abundant group within the clone libraries (Table 3). Interactions between plant roots and Rhizobiaceae members (e.g., Rhizobium and Agrobacterium) are of particular importance and interest (24). Examination of the effect of the different treatments on relative Rhizobiaceae abundance emphasized the difference between the responses of root and of potting mix bacterial communities to modification. Although in the potting mix the response was directly related to the type of treatment, a complex response was observed in root communities ranging between a reduction in relative abundance and an apparent dominance gain (Table 3).
The taxonomic identity of different community members is to date the principle parameter used for description of soil, rhizosphere, and root microbial communities (33). However, patterns of global distribution and co-occurrence of different phylogenetic groups are only at the pioneering stage of research (20). Therefore, most of the knowledge available today is in essence situation-bound (40). Lynch et al. (33) considered the use of basic concepts of “above-ground” ecology in addressing soil microbial diversity. As pointed out by these authors, the soil habitat is composed of many essentially distinct niches. Indeed, in the present study, root bacterial communities significantly differed from those of potting mix in composition and diversity (α-diversity). Due to the nature of plant root activity, the root and the root-free soil habitats may be viewed as two separate niches dictated by physicochemical gradients. It was interesting to observe that the β-diversity, i.e., the extent of shift in microbial community composition between the root and the potting mix, was highly similar in four of the five potting mix formulas used here (Table 4). Consistent differences in α-diversity between the bulk soil and the rhizosphere bacterial communities were reported for Lolium perenne (48) and for oilseed rape and strawberry (9) when each of the plants was grown in different soil types. However, β-diversity was not considered in these studies, and its significance in the root-soil system is currently unknown.
Marilley et al. (34) found that root bacterial communities of L. perenne and Trifolium repens were characterized by lower diversity and higher dominance relative to the respective soils. In the present study, the same trend was observed in the untreated control as well as in both compost amendment treatments. In contrast, inoculation with Streptomyces spp. resulted in an opposite trend (Table 2). These results suggest that the complexity of the root bacterial community is not necessarily a consequence of the complexity of the respective soil bacterial community. Thus, the rhizosphere effect is demonstrated.
A combination of three approaches—PCR-DGGE, clone libraries, and quantitative PCR—was used here for 16S rRNA gene-based community analysis. The data obtained by each approach offered complementary insights into the response of the root bacterial communities to applied interventions. A simple perlite potting mix was used in the present study. This allowed clear-cut interpretation of the results, which are often too complicated when field soils are used. Although a simple model was investigated, highly complex root-soil-microbiota interactions were encountered. Fundamentally distinct treatments resulted in substantial shifts in cucumber root bacterial communities. These shifts were characterized by high divergence between the composition of treated and untreated roots, even at a relatively low resolution level of taxonomy. However, we demonstrate that examination of different diversity components revealed specific trends in the response of root bacterial communities to intervention.
Supplementary Material
Acknowledgments
Both Streptomyces strains were kindly provided by Ehud Inbar and were isolated as part of his M.Sc. project performed at the Hebrew University of Jerusalem, Jerusalem, Israel. We heartily thank Marvyn J. Bibb, John Innes Center, Norwich, England, for assistance and for supplying all of the means required for transformation of the Streptomyces strains.
This research was supported by research grant number US-3108-99 from BARD and by The Negev Foundation, Cleveland, OH.
Footnotes
Published ahead of print on 21 August 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Addicott, J. F. 1974. Predation and prey community structure: an experimental study of the effect of mosquito larvae on the protozoan communities of pitcher plants. Ecology 55:475-492. [Google Scholar]
- 2.Altschul, S.F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, and W. Miller. 1997. Gapped BLASR and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgarte, S., and C. C. Tebbe. 2005. Field studies on the environmental fate of the Cry1Ab Bt-toxin produced by transgenic maize MON810 and its effect on bacterial communities in the maize rhizosphere. Mol. Ecol. 14:2539-2551. [DOI] [PubMed] [Google Scholar]
- 4.Castro-Sowinski, S., Y. Herschkovitz, Y. Okon, and E. Jurkevitch. 2007. Effects of inoculation with plant growth-promoting rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiol. Lett. 276:1-11. [DOI] [PubMed] [Google Scholar]
- 5.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, et al. 2003. The Ribosomal Database Project RDP-II.: previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colwell, R. K. 2005. EstimateS: statistical estimation of species richness and shared species from samples, version 7.5. University of Connecticut, Storrs. http://viceroy.eeb.uconn.edu/estimates.
- 8.Compant, S., B. Duffy, J. Nowak, C. Clement, and E. Ait Barka. 2005. Use of plant growth promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71:4951-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa, R., M. Götz, N. Mrotzek, J. Lottmann, G. Berg, and K. Smalla. 2006. Effects of site and plant species on rhizosphere structure revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56:236-249. [DOI] [PubMed] [Google Scholar]
- 10.Dohrmann, A. B., and C. C. Tebbe. 2005. Effect of elevated tropospheric ozone on the structure of bacterial communities inhabiting the rhizosphere of herbaceous plants. Appl. Environ. Microbiol. 71:7750-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emmert, E. A. B., and J. Handelsman. 1999. Biocontrol of plant disease: a (gram-)positive perspective. FEMS Microbiol. Lett. 171:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Felici, C., L. Vettori, E. Giraldi, L. M. C. Forino, A. Toffanin, A. M. Tagliasacchi, and M. Nuti. 2008. Single and co-inoculation of Bacillus subtilis and Azospririllum brasilense on Lycopersicon esculentum: effects on plant growth and rhizosphere microbial community. Appl. Soil Ecol. 40:260-270. [Google Scholar]
- 13.Good, I. J. 1953. The population frequencies of species and the estimation of the population parameters. Biometrika 40:237-264. [Google Scholar]
- 14.Götz, M., N. C. Gomes, A. Dratwinski, R. Costa, G. Berg, R. Peixoto, et al. 2006. Survival of gfp-tagged antagonistic bacteria in the rhizosphere of tomato plants and their effects on the indigenous bacterial community. FEMS Microbiol. Ecol. 56:207-218. [DOI] [PubMed] [Google Scholar]
- 15.Green, S. J., F. C. Michel, Y. Hadar, and D. Minz. 2004. Similarity of bacterial communities in sawdust- and straw-amended cow manure composts. FEMS Microbiol. Lett. 233:115-123. [DOI] [PubMed] [Google Scholar]
- 16.Green, S. J., F. C. Michel, Y. Hadar, and D. Minz. 2007. Contrasting patterns of seed and root colonization by bacteria from the genus Chrysobacterium and from the family Oxalobacteraceae. ISME J. 1:291-299. [DOI] [PubMed] [Google Scholar]
- 17.Hoagland, D. R., and D. I. Arnon. 1950. The water-culture method for growing plants without soil. College of Agriculture, University of California, Berkeley.
- 18.Hoitink, H. A. J., and M. J. Boehm. 1999. Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37:427-446. [DOI] [PubMed] [Google Scholar]
- 19.Hoitink, H. A. J., A. G. Stone, and D. Y. Han. 1997. Suppression of plant diseases by composts. Hort. Sci. 32:184-187. [Google Scholar]
- 20.Horner-Devine, N., C. Silver, J. M. Leibold, et al. 2007. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88:1345-1353. [DOI] [PubMed] [Google Scholar]
- 21.Human, K. G., and D. M. Gordon. 1997. Effects of Argentine ants on invertebrate biodiversity in northern California. Conserv. Biol. 11:1242-1248. [Google Scholar]
- 22.Inbar, E., S. J. Green, Y. Hadar, and D. Minz. 2005. Competing factors of compost concentration and proximity to root affect the distribution of Streptomyces. Microb. Ecol. 50:73-81. [DOI] [PubMed] [Google Scholar]
- 23.Johansen, A., I. M. B. Knudsen, S. J. Binnerup, A. Winding, J. E. Johansen, L. E. Jensen, et al. 2005. Non-target effects of the microbial control agents Pseudomonas fluorescens DR54 and Clonostachys rosea IK726 in soils cropped with barley followed by sugar beet: a greenhouse assessment. Soil Biol. Biochem. 37:2225-2239. [Google Scholar]
- 24.Kapulnik, Y., and Y. Okon. 2002. Plant growth promotion by rhizospheric bacteria, p. 869-886. In Y. Waisel, A. Eshel, and U. Kafkafi (ed.), Plant roots: the hidden half, 3rd ed. Marcel Dekker, New York, NY.
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chatter, and D. A. Hopwood. 2000. Introduction of DNA into Streptomyces, p. 230-252. In Practical Streptomyces genetics. John Innes Centre, Norwich, United Kingdom.
- 26.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chatter, and D. A. Hopwood. 2000. Growth and preservation of Streptomyces, p. 43-61. In Practical Streptomyces genetics. John Innes Centre, Norwich, United Kingdom.
- 27.Kunte, K. 2008. Competition and species diversity: removal of dominant species increased diversity in Costa Rican butterfly communities. Oikos 117:69-76. [Google Scholar]
- 28.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 29.Lottmann, J., H. Heuer, J. de Vries, A. Mahn, K. Düring, W. Wackernagel, et al. 2000. Establishment of introduced antagonistic bacteria in the rhizosphere of transgenic potatoes and their effect on the bacterial community. FEMS Microbiol. Ecol. 33:41-49. [DOI] [PubMed] [Google Scholar]
- 30.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, H. Yadhukumar, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch, J. M. 2002. Resilience of the rhizosphere to anthropogenic disturbance. Biodegradation 13:21-27. [DOI] [PubMed] [Google Scholar]
- 33.Lynch, J. M., A. Benedetti, H. Insam, M. P. Nuti, K. Smalla, V. Torsvik, et al. 2004. Microbial diversity in soil: ecological theories, the contribution of molecular techniques, and the impact of transgenic plants and transgenic microorganisms. Biol. Fert. Soils 40:363-385. [Google Scholar]
- 34.Marilley, L., G. Vogt, M. Blanc, and M. Aragno. 1998. Bacterial diversity in bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as reveled by PCR restriction analysis of 16S rDNA. Plant Soil 198:219-224. [Google Scholar]
- 35.Nautiyal, C. S., J. K. Johri, and H. B. Singh. 2002. Survival of the rhizosphere-competent biocontrol strain Pseudomonas fluorescence NBRI2650 in the soil and phytosphere. Can. J. Microbiol. 48:588-601. [DOI] [PubMed] [Google Scholar]
- 36.Noble, R., and E. Coventry. 2005. Suppression of soil-borne plant diseases with composts: a review. Biocontrol Sci. Technol. 15:3-20. [Google Scholar]
- 37.Paulitz, T. C., and R. R. Belanger. 2001. Biological control in greenhouse systems. Annu. Rev. Phytopathol. 39:103-133. [DOI] [PubMed] [Google Scholar]
- 38.Persello-Cartieaux, F., L. Nussaume, and C. Robaglia. 2003. Tales from the underground: molecular plant-rhizobacteria interactions. Plant Cell Environ. 26:189-199. [Google Scholar]
- 39.Poole, R. W. 1974. An introduction to quantitative ecology. McGraw-Hill Book Co., New York, NY.
- 40.Prosser, J., I. Bohannan, B. J. Curtis, et al. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:382-384. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds, W. D., X. M. Yang, C. F. Drury, T. Q. Zhang, and C. S. Tan. 2003. Effects of selected conditioners and tillage on the physical quality of a clay loam soil. Can. J. Soil Sci. 83:381-393. [Google Scholar]
- 42.Roesti, D., R. Gaur, B. N. Johri, G. Imfeld, S. Sharma, K. Kawaljeet, and M. Aragno. 2006. Plant growth stage, fertilizer management and bio-inoculation of arbuscular mycorrhizal fungi and plant growth-promoting rhizobacteria affect the rhizobacterial community structure in rain-fed wheat fields. Soil Biol. Biochem. 38:1111-1120. [Google Scholar]
- 43.Ruppel, S., J. Rühlmann, and W. Merbach. 2006. Quantification and localization of bacteria in plant tissues using quantitative real-time PCR and online emission fingerprinting. Plant Soil 268:21-35. [Google Scholar]
- 44.Schutzenhofer, M. R., and T. J. Valone. 2006. Positive and negative effects of exotic Erodium cicutarium on an arid ecosystem. Biol. Conserv. 132:376-381. [Google Scholar]
- 45.Schwieger, F., and C. C. Tebbe. 2000. Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizosphere of a target plant Medicago sativa and a non-target plant (Chenopodium album)-linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl. Environ. Microbiol. 66:3556-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siciliano, S. D., and J. J. Garmida. 1998. BIOLOG analysis and fatty acid methyl ester profiles indicate that pseudomonad inoculants that promote phytoremediation alter the root-associated microbial community of Bromus biebersteinii. Soil Biol. Biochem. 30:1717-1723. [Google Scholar]
- 47.Silliman, B. R., and M. D. Bertness. 2004. Shoreline development drives invasion of Phragmites austrasil and the loss of plant diversity on New England salt marshes. Conserv. Biol. 18:1424-1434. [Google Scholar]
- 48.Singh, B. K., S. Munro, J. M. Potts, and P. Millard. 2007. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 36:147-155. [Google Scholar]
- 49.Termorshuizen, A. J., E. van Rijn, D. J. van der Gaag, C. Alabouvette, Y. Chen, J. Lagerlo, et al. 2006. Suppressiveness of 18 composts against 7 pathosystems: variability in pathogen response. Soil Biol. Biochem. 38:2461-2477. [Google Scholar]
- 50.Tiquia, S. M., J. Lloyd, D. A. Herms, H. A. J. Hoitink, and F. C. Michel. 2002. Effects of mulching and fertilization on soil nutrients, microbial activity and rhizosphere bacterial community structure determined by analysis of T-RFLPs of PCR-amplified 16S rRNA genes. Appl. Soil Ecol. 21:31-48. [Google Scholar]
- 51.Tiquia, S. M. 2005. Microbiological parameters as indicators of compost maturity. J. Appl. Microbiol. 99:815-828. [DOI] [PubMed] [Google Scholar]
- 52.Van Loon, L. C., P. A. H. M. Bakker, and C. M. J. Pieterse. 1998. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36:453-483. [DOI] [PubMed] [Google Scholar]
- 53.Wulf, A., K. Manthye, J. Doll, A. M. Perlick, B. Linke, T. Bekel, et al. 2003. Detection of highly specific transcriptional changes of the model plant Medicago truncatula in response to arbuscular mycorrhiza development. Mol. Plant-Microbe Interact. 16:306-314. [DOI] [PubMed] [Google Scholar]
- 54.Yogev, A., M. Raviv, Y. Hadar, R. Cohen, and J. Katan. 2006. Plant waste-based composts suppressive to diseases caused by pathogenic Fusarium oxysporum. Eur. J. Plant Pathol. 116:267-278. [Google Scholar]
- 55.Zebrath, B. J., G. H. Neilsen, E. Hogue, and D. Neilsen. 1999. Influence of organic waste amendments on selected soil physical and chemical properties. Can. J. Soil Sci. 79:501-504. [Google Scholar]
- 56.Zhang, W., D. Y. Han, W. A. Dick, K. R. Davis, and H. A. J. Hiotink. 1998. Compost and compost water extract-induced systemic acquired resistance in cucumber and Arabidopsis. Phytopathology 88:445-450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.