Abstract
During the last few years, genome-related information has become available for many microorganisms, including important food-related bacteria. Lactic acid bacteria (LAB) are important industrially in the production of fermented foods such as dairy products, sausages, sourdoughs, and vegetables. Despite their limited metabolic capacity, LAB contribute considerably to important characteristics of fermented foods, such as flavor and texture. In the present study, a species-independent functional gene microarray was developed that targets 406 genes that play key roles in the production of sugar catabolites, bacteriocins, exopolysaccharides, and aromas, in probiotic and biosafety characteristics, and in the stress response. Also, genes linked to negative traits, such as antibiotic resistance and virulence, are represented. As LAB ecosystems contain a variety of species, there was a more global focus on these specific functional properties. Thus, an algorithm was used to design gene-specific oligonucleotides that preferably hybridize with multiple LAB species, thereby allowing controlled cross-hybridization. For proof of concept, the microarray composed of 2,269 30-mer oligonucleotides focused on LAB species that are prevalent in sourdough ecosystems. Validation hybridizations using DNA and RNA from 18 LAB strains, covering 86% of all the oligonucleotides, showed that there were wide ranges in intensity and high reproducibility between microarrays.
The fast technological evolution of the last two decades in molecular biosciences, particularly in genome and transcriptome research, has resulted in increased use of molecular techniques in many research areas, including food microbiology and food biotechnology (24, 26). Thanks to this shift from conventional microbiology to molecular microbiology, genome-related information has become available for many microorganisms, including important food-related bacteria such as the lactic acid bacteria (LAB). LAB have great industrial importance in the production of fermented foods, such as dairy products, fermented sausages, and sourdoughs (29, 57). Also, in small-scale artisan fermented food products, natural LAB strains with interesting properties dominate the fermentation process (7, 15, 43, 46, 60). Despite their limited metabolic capacity, LAB contribute considerably to the microbial safety and organoleptic properties of fermented foods. They produce organic acids (mainly lactic acid) and bacteriocins (small antibacterial peptides) that contribute to the extended shelf-lives of fermented raw materials (29). Their production of exopolysaccharides influences the texture and/or mouthfeel of dairy products and sourdoughs (11, 13). Their ability to convert pyruvate and amino acids during various food fermentation processes results in flavor components and hence determines the sensory profiles of a variety of fermented food products (31). Besides these desired traits, LAB may have negative traits, such as the ability to produce toxic biogenic amines, possession of transmittable antibiotic resistance genes, and the potential for expression of putative virulence factors (29).
During the last few years, the full genome sequences of 25 LAB have been released into the public domain (6, 23, 35, 38, 40). This genomic information has provided better insight into the physiology and total metabolic capacity of LAB, particularly how specific LAB strains contribute to desired traits during food fermentations (31, 40). Furthermore, the genomic information has been used to develop several species-specific microarrays that allow workers to monitor the expression of all genes of a single LAB species and to examine interesting metabolic functions during monoculture experiments (2-4, 20, 26, 39, 41, 49). However, it is still a challenge to monitor gene expression in complex ecosystems, such as fermented foods. With a so-called functional gene microarray (63, 64), a microarray encompassing functional genes of different microbial strains belonging to different species, the expression of specific genes can be followed under certain environmental conditions, hence exceeding the strain level. Until now, only a few microarrays, all in the field of environmental research, were developed to monitor gene expression in complex ecosystems (for instance, soil) (18, 44, 58). One of these microarrays, GeoChip, encompassing functional genes of different soil microorganisms that play an important role in biogeochemical processes, is actually the first large-scale, comprehensive functional gene microarray that was described and utilized (18).
The present study was aimed at designing and validating a species-independent LAB functional gene microarray that allows examination of expression of functional genes in complex food ecosystems. The microarray developed was set up in such a way that the oligonucleotides in it allowed assessment of the expression of functional genes in an ecosystem which are involved in important enzymatic steps for desired traits as well as unwanted traits, regardless of the specific LAB contributing to the gene expression. For proof of concept, the sourdough ecosystem was used. Validation of this functional gene LAB microarray was performed using DNA and RNA from species represented on the microarray.
MATERIALS AND METHODS
Microarray production.
An oligonucleotide-based microarray covering 406 well-chosen genes that represent important pathways and phenotypic traits of fermented-food-related bacteria (47 LAB species and 4 non-LAB species) was constructed. All oligonucleotides were purchased from Isogen Life Science BV (Ijsselstein, The Netherlands). They were diluted to obtain a concentration of 25 μM in 125 mM sodium phosphate buffer containing 4.26 μM N-lauroylsarcosine (Sigma-Aldrich, St. Louis, MO). A MicroGrid II spotter (Genomic Solutions, Huntingdon, United Kingdom) was used to spot the oligonucleotides using 10-K pins on CodeLink activated slides (GE Healthcare, Bucks, United Kingdom). After spotting, the CodeLink activated slides were placed in a humid chamber containing saturated NaCl for at least 24 h. Unused active groups were blocked with a preheated blocking solution (50 mM ethanolamine, 0.1 M Tris; pH 9.0) at 50°C for 30 min. The slides were rinsed twice with ultrapure water and washed with preheated 4× sodium chloride-sodium citrate (SSC) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate (SDS) in a shaking water bath at 50°C for 30 min. The slides were rinsed with ultrapure water and dried by centrifugation at 115 × g for 3 min.
Bacterial strains and growth conditions.
The bacterial strains used for validation of the microarray are listed in Table 1. All strains were stored at −80°C and grown twice overnight in MRS-5 medium (de Man-Rogosa-Sharpe medium supplemented with a vitamin solution [34]) before use in experiments.
TABLE 1.
Strains used in the validation hybridizations
| Strain | Origin | Reference |
|---|---|---|
| Lactobacillus plantarum D06SS01T01-H18 | Belgian artisan bakery spelt sourdough | 47 |
| L. plantarum LMG 9211 | Human saliva | 25 |
| L. plantarum 80 | Ghanaian cocoa bean heap fermentation | 7 |
| L. plantarum ACA-DC 287 | Greek Xynotyri cheese | 14 |
| Lactobacillus fermentum LMG 8154 | Unknown | 36 |
| L. fermentum 222 | Ghanaian cocoa bean heap fermentation | 7 |
| L. fermentum IMDO 130101 | Rye laboratory sourdough fermentation | 54 |
| Lactobacillus johnsonii La1 | LC1 yoghurt (Nestlé, Switzerland) | 33 |
| Lactobacillus acidophilus IBB 801 | Romanian dairy product | 59 |
| Lactobacillus delbrueckii subsp. bulgaricus LMG 6901T | Bulgarian yoghurt | 53 |
| Lactobacillus sakei subsp. sakei CTC 494 | Spanish naturally fermented sausage | 28 |
| Lactobacillus curvatus D06SS01T01-H12 | Belgian artisan bakery spelt sourdough | 47 |
| Lactobacillus sanfranciscensis LMG 16002T | San Francisco sourdough | 10 |
| Lactococcus lactis subsp. lactis LDV 22186 | Dutch dairy starter | 12 |
| L. lactis subsp. cremoris MG 1363 | Plasmid-free NCDO 712 derivative (United Dairies, United Kingdom) | 16 |
| Enterococcus faecium RZS C5 | Belgian cheese | 27 |
| Enterococcus faecalis LMG 8222 | Urine | 37 |
| Leuconostoc mesenteroides subsp. mesenteroides LMG 6893 | Olive fermentation | 32 |
DNA and RNA extraction.
DNA extraction from the bacterial strains was performed as described previously (17). For RNA extraction, 10 ml of an overnight culture was collected in 40 ml RNAprotect (Qiagen, Hilden, Germany) that was diluted 2:1 with 1× phosphate-buffered saline (Invitrogen, Carlsbad, CA), mixed, and kept at room temperature for a minimum of 5 min. Subsequently, the sample was centrifuged at 5,000 × g for 15 min, and the supernatant was discarded. Each pellet was resuspended in 200 μl TE buffer (30 mM Tris-HCl, 1 mM EDTA; pH 8.0) containing 1.3 U μl−1 mutanolysin (Sigma-Aldrich) and 50 μg μl−1 lysozyme (Sigma-Aldrich). This mixture was incubated in a shaking water bath at 37°C for 1 h for enzymatic lysis of the cells. From this point on, an RNeasy mini kit (Qiagen) was used. Briefly, 700 μl of RLT buffer, provided with the kit, plus β-mercaptoethanol (10 μl ml−1) was added to the lysate, and the solution was mixed and transferred to a 2-ml tube containing approximately 50 mg acid-washed beads (Sigma-Aldrich). Vortexing for 5 min resulted in additional mechanical disruption of the cells. The tube was microcentrifuged at 13,000 rpm for 10 s, and 850 μl of the supernatant was added to 590 μl of 80% (vol/vol) ethanol. The manufacturer's standard instructions were followed from this point on. RNA quality was checked by determining the A260/A280 and A260/A230 ratios using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE) and by capillary electrophoresis using an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA).
Labeling and hybridization.
DNA samples from bacterial strains used for validation hybridization were labeled using an adapted protocol based on the BioPrime DNA labeling system (Invitrogen). Twenty microliters of a 2.5× mixture of a random primer and reaction buffer, which was provided with the kit, was added to 2 μg of DNA in a 21-μl mixture. The mixture was incubated at 90°C for 5 min and then immediately put on ice. Five microliters of a deoxynucleoside triphosphate mixture (1.2 mM dATP, 1.2 mM dGTP, 1.2 mM dTTP, and 0.6 mM dCTP; GE Healthcare), 3 μl of 1 mM Cy3-labeled dCTP (GE Healthcare), and 1 μl of Klenow fragments were added. The mixture was incubated in a thermal cycler at 37°C for 16 h. The reaction was stopped by adding 5 μl of stop buffer, which was part of the BioPrime DNA labeling system. The amplification product was purified using a MinElute PCR purification kit (Qiagen), and the concentration was measured using the NanoDrop spectrophotometer.
RNA from bacterial strains was linearly amplified using a Genisphere SensAmp kit (Genisphere, Hatfield, PA) with 200 ng of total RNA. The protocol described in the manufacturer's instructions was followed. The amplified RNA (aRNA) was purified using an RNeasy mini kit (Qiagen), and the purified aRNA was labeled with the Cy3 and Cy5 dyes in a reverse transcription reaction (42).
Hybridization mixtures were prepared using 40 pmol of labeled DNA or 50 pmol of labeled aRNA in 210 μl hybridization buffer (GE Healthcare) containing 50% (vol/vol) formamide (Sigma-Aldrich). The hybridization mixtures were denatured by heating them at 96°C for 3 min, put on ice for at least 5 min, held at 32°C for 5 min, and subsequently microcentrifuged at 12,000 rpm for 5 min. All samples were hybridized with an HS 4800 Pro hybridization station (Tecan Systems Inc., San Jose, CA) at 32°C for 16 h. Automated posthybridization washing was performed with 1× SSC, 0.2% SDS at 32°C, 27°C, and 23°C for 20 s, 20 s, and 30 s, respectively. This was followed by washing with 0.1× SSC, 0.2% SDS at 23°C for 1 min and with 0.1× SSC at 23°C for 30 s. Slides were dried using nitrogen gas at 30°C for 2 min and scanned using an Agilent scanner (Agilent Technologies) at 10 μm. Images were analyzed using ArrayVision v7 (GE Healthcare).
Experimental design and data analysis.
For validation hybridizations based on selected bacterial strains, DNA samples were hybridized twice using the same dye (Cy3), and aRNA samples were hybridized once with each dye (Cy3 and Cy5), using a loop design.
As each oligonucleotide was spotted four times on the array and each sample was hybridized twice (technical repeat), each oligonucleotide was measured eight times. The intensity of a spot was considered to be above the background level if the signal or foreground intensity (Fg) was greater than the background intensity (Bg) plus three times the standard deviation of the foreground intensity and background intensity, computed as the square root of the average of their variances [var(Fg) and var(Bg), respectively], as determined by the following equation:
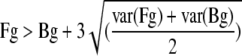 |
An oligonucleotide was considered present if the intensities for at least six of eight spots on the microarray were above the background level.
Microarray data accession numbers.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GPL5459 (microarray including detailed annotation), GSE9082 (DNA validation data), and GSE9140 (RNA validation data).
RESULTS
Microarray design.
A species-independent functional gene microarray that can be extended and upgraded on a regular basis was developed for food-related LAB, with a focus on sourdough LAB. Therefore, 406 genes encoding proteins and enzymes involved in desired and unwanted phenotypic traits of LAB used in the production of fermented foods were selected (Table 2).
TABLE 2.
Distribution of the key genes and corresponding oligonucleotides based on metabolic features
| Class | No. of genes | No. of oligonucleotides |
|---|---|---|
| Carbohydrate and pyruvate metabolism | 95 | 715 |
| Stress response | 57 | 534 |
| Amino acid metabolism | 59 | 282 |
| Proteolysis | 41 | 221 |
| Exopolysaccharide production | 44 | 215 |
| Glycosyltransferases | 49 | 64 |
| Lipoproteins | 4 | 52 |
| Antibiotic resistance | 17 | 41 |
| Virulence factors | 8 | 32 |
| Household genes | 4 | 30 |
| Phosphotransferase systems | 5 | 27 |
| Biogenic amines | 7 | 24 |
| Bacteriocin production | 7 | 11 |
| Others | 9 | 21 |
| Total | 406 | 2,269 |
As it was the purpose of the study to design a microarray containing oligonucleotides for conserved regions that could hybridize with several closely related LAB species and/or strains, a comparative analysis using BLAST (1) was performed for all coding sequences of 71 LAB strains that were retrieved from the GenBank database at the moment of oligonucleotide design, based on a search by the taxonomy identifier in the protein section (5; http://www.ncbi.nlm.nih.gov/GenBank/). These sequences, together with their annotations, were stored in an in-house relational database. The resulting BLAST hits were likewise stored in the database, referring to the BLAST query sequence, and selected for alignment length (>30 bp) and overall identity (>85%). To overcome inconsistencies in oligonucleotide annotation that might result from the sometimes incorrect and/or incomplete annotation of the coding sequences in public databases, the BRENDA (48; http://www.brenda-enzymes.info) and KEGGS (http://www.genome.ad.jp) databases were used as references for manual curation of the EC number and gene name information for the sequences of interest prior to oligonucleotide design.
An oligonucleotide design algorithm that used the results of the comparative sequence analysis was developed and was applied to the 406 key genes. For each gene, all related sequences were selected from the database, together with the corresponding BLAST hits obtained in the comparative sequence analysis mentioned above. For each of the gene-related sequences, homology clusters were designated based on the BLAST hits. For each cluster, a consensus sequence was obtained using ClustalW (8), allowing incorporation of nucleotide ambiguity codes. Depending on the length of the consensus sequence, 30-mer oligonucleotides were designed using Primer3 (45) by applying a 60-base sliding window in the original query sequence (i.e., a 60-base window in which an oligonucleotide was designed and which was moved 40 bases over the consensus sequence to design additional oligonucleotides). The 30-mer oligonucleotides obtained were ranked according to the number of species that could be hybridized and the number of possible mismatches. Altogether, a maximum of three possible mismatch positions were allowed, as long as a stretch of at least 15 bp was present (22). Query sequences for which no hits and/or oligonucleotides were found in the comparative sequence analysis were subjected to unique oligonucleotide design using OligoWiz (version 2.0.1) (56), which ensured that all gene-related sequences were represented by an oligonucleotide. The oligonucleotide design algorithm and adjacent scripts were written in Perl, making use of the Bioperl toolkit (51).
Applying the oligonucleotide design algorithm to the collected coding sequences resulted in 4,851 oligonucleotides. A subset of 2,269 oligonucleotides was selected for synthesis and hence production of the microarray based on the LAB microbiota prevailing during sourdough fermentations. This subset represented 406 key genes in a total of 46 LAB and 4 non-LAB species (Tables 2 and 3). Based on sequence annotation and oligonucleotide design information, 351 oligonucleotides could cross-hybridize with different LAB species and 649 oligonucleotides could cross-hybridize with different strains of the same species. The remaining 1,269 oligonucleotides were the result of unique oligonucleotide design, reflecting the rather low number of hits in comparative sequence analysis.
TABLE 3.
Number of oligonucleotides for each of the 46 represented LAB species, based on species annotation for the BLAST query sequences during oligonucleotide design, and number of target genes representeda
| Species | No. of oligonucleotides | No. of target genes |
|---|---|---|
| Lactococcus lactisb | 551 | 270 |
| Lactobacillus plantarumb | 302 | 220 |
| Lactobacillus acidophilusb | 207 | 147 |
| Enterococcus faecalisb | 206 | 152 |
| Lactobacillus sakeib | 187 | 145 |
| Leuconostoc mesenteroides | 120 | 79 |
| Lactobacillus delbrueckii | 117 | 75 |
| Enterococcus faecium | 113 | 76 |
| Lactobacillus johnsoniib | 96 | 79 |
| Pediococcus pentosaceus | 72 | 49 |
| Lactobacillus helveticus | 33 | 29 |
| Lactobacillus casei | 31 | 24 |
| Lactobacillus reuteri | 31 | 22 |
| Lactobacillus sanfranciscensis | 28 | 26 |
| Lactobacillus brevis | 17 | 13 |
| Lactobacillus crispatus | 14 | 9 |
| Lactobacillus fermentum | 14 | 13 |
| Enterococcus hirae | 11 | 11 |
| Lactobacillus hilgardii | 11 | 11 |
| Lactobacillus curvatus | 10 | 8 |
| Enterococcus casseliflavus | 8 | 7 |
| Enterococcus mundtii | 7 | 7 |
| Lactobacillus buchneri | 7 | 4 |
| Lactobacillus pentosus | 7 | 5 |
| Lactobacillus amylovorus | 6 | 4 |
| Pediococcus acidilactici | 6 | 3 |
| Lactobacillus gasseri | 5 | 5 |
| Streptococcus thermophilusb | 5 | 5 |
| Oenococcus oeni | 5 | 4 |
| Lactobacillus paracasei | 4 | 4 |
| Lactobacillus rhamnosus | 4 | 4 |
| Lactobacillus suntoryeus | 4 | 2 |
| Lactobacillus paraplantarum | 3 | 3 |
| Leuconostoc pseudomesenteroides | 3 | 3 |
| Weissella confusa | 3 | 3 |
| Lactobacillus alimentarius | 2 | 2 |
| Lactobacillus farciminis | 2 | 2 |
| Lactobacillus fructivorans | 2 | 2 |
| Lactobacillus parabuchneri | 2 | 2 |
| Leuconostoc citreum | 2 | 2 |
| Staphylococcus aureus | 2 | 2 |
| Bifidobacterium sp. strain ISO3519 (accession no. AAL30847) | 1 | 1 |
| Brevibacterium linens | 1 | 1 |
| Lactobacillus frumenti | 1 | 1 |
| Lactobacillus kimchii | 1 | 1 |
| Lactobacillus mindensis | 1 | 1 |
| Lactobacillus panis | 1 | 1 |
| Lactobacillus pontis | 1 | 1 |
| Lactobacillus rossiae | 1 | 1 |
| Staphylococcus lentus | 1 | 1 |
The oligonucleotides designed using sequences from four non-LAB species (Staphylococcus aureus, Bifidobacterium sp. strain ISO3519, Brevibacterium linens, and Staphylococcus lentus) represent genes involved in antibiotic resistance.
One or more genome sequences of this species were available when the oligonucleotides were designed.
Validation hybridizations.
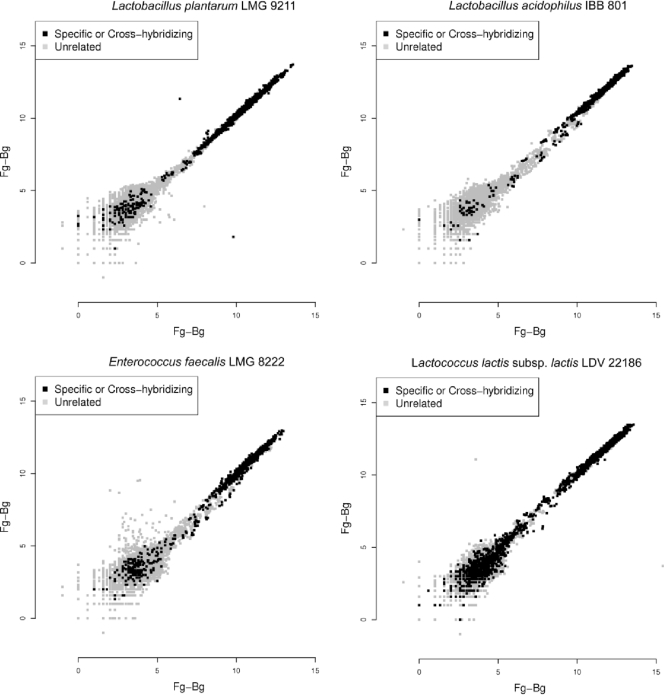
To validate hybridization reproducibility and oligonucleotide specificity, DNA and aRNA from 18 strains of 12 LAB species represented on the microarray were hybridized (Table 4). For each strain, hybridization reproducibility was monitored by plotting spot intensities for the duplicate hybridizations with DNA for all spots (Fig. 1). To determine the specificity of the microarray, hybridization intensities were analyzed for each of the 12 species to identify the oligonucleotides whose intensities were above the background level, and the results were compared with the annotations of the oligonucleotides (Table 4).
TABLE 4.
Results of the validation hybridizations using DNA and RNA from 18 LAB strains based on oligonucleotide typea
| Strain | No. of species-specific oligonucleotides
|
No. of cross-hybridizing oligonucleotides
|
No. of unrelated oligonucleotides
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Totalb | DNAc | RNAd | Totalb | DNAc | RNAd | Totalb | DNAc | RNAd | |
| Lactobacillus curvatus D06SS01T01-H12 | 10 | 4 | 4 | 14 | 8 | 5 | 2,245 | 250 | 38 |
| Lactobacillus fermentum 222 | 14 | 7 | 7 | 8 | 3 | 3 | 2,247 | 153 | 59 |
| L. fermentum LMG 8154 | 14 | 7 | 7 | 8 | 4 | 3 | 2,247 | 147 | 2 |
| L. fermentum IMDO 130101 | 14 | 7 | 7 | 8 | 2 | 2 | 2,247 | 218 | 51 |
| Lactobacillus plantarum D06SS01T01-H18 | 302 | 256 | 243 | 50 | 36 | 29 | 1,917 | 283 | 35 |
| L. plantarum 80 | 302 | 264 | 259 | 50 | 35 | 32 | 1,917 | 226 | 100 |
| L. plantarum LMG 9211 | 302 | 286 | 279 | 50 | 33 | 31 | 1,917 | 176 | 89 |
| L. plantarum ACA-DC 287 | 302 | 258 | 255 | 50 | 34 | 31 | 1,917 | 184 | 97 |
| Lactobacillus sanfranciscensis LMG 16002T | 28 | 25 | 25 | 7 | 6 | 6 | 2,234 | 155 | 12 |
| Lactobacillus johnsonii La1 | 96 | 94 | 89 | 46 | 34 | 31 | 2,127 | 215 | 117 |
| Lactobacillus acidophilus IBB 801 | 207 | 198 | 181 | 46 | 43 | 38 | 2,016 | 203 | 120 |
| Lactobacillus delbrueckii subsp. delbrueckii LMG 6901T | 117 | 81 | 75 | 26 | 15 | 11 | 2,126 | 213 | 66 |
| Lactobacillus sakei subsp. sakei CTC 494 | 187 | 165 | 160 | 48 | 33 | 27 | 2,034 | 176 | 73 |
| Lactococcus lactis subsp. lactis LDV 22186 | 551 | 386 | 344 | 30 | 18 | 14 | 1,688 | 137 | 59 |
| Lc. lactis subsp. cremoris MG1363 | 551 | 342 | 280 | 30 | 20 | 17 | 1,688 | 100 | 45 |
| Enterococcus faecium RSZ C5 | 113 | 85 | 70 | 27 | 7 | 7 | 2,129 | 157 | 77 |
| Enterococcus faecalis LMG 8222 | 206 | 181 | 171 | 37 | 19 | 14 | 2,026 | 154 | 65 |
| Leuconostoc mesenteroides subsp. mesenteroides LMG 6893 | 120 | 103 | 101 | 18 | 16 | 14 | 2,131 | 198 | 62 |
The oligonucleotide type was determined based on sequence annotation. Species-specific oligonucleotides were designed based on sequences of the corresponding LAB species; cross-hybridizing oligonucleotides were designed based on sequences from other LAB species and hybridized to the species indicated based on annotation; and the unrelated oligonucleotides did not show any homology to sequences from the species indicated.
Number of oligonucleotides that were available on the microarray.
Number of oligonucleotides whose intensities were above the background level for DNA hybridizations.
Number of oligonucleotides whose intensities were above the background level for RNA (a subset of the oligonucleotides whose intensities were above the background level for DNA).
FIG. 1.
Reproducibility of the background-corrected intensities (Fg − Bg, where Fg is the foreground intensity and Bg is the background intensity) for two hybridizations using DNA of each of the four LAB species with the highest number of oligonucleotides on the microarray: Lactobacillus plantarum LMG 9211, Lactobacillus acidophilus IBB 801, Enterococcus faecalis LMG 8222, and Lactococcus lactis subsp. lactis LDV 22186. The black symbols represent spots for species-specific and cross-hybridizing oligonucleotides. The gray symbols represent spots for unrelated oligonucleotides.
The validation hybridizations using DNA and aRNA from 18 strains of 12 LAB species, representing 86.0% of all oligonucleotides (1,951 of 2,269 oligonucleotides), showed wide ranges of intensity and high reproducibility for technical replicates. Altogether, the intensities of 79.0% (1,541 of 1,951) of all species-specific oligonucleotides for the 12 LAB species tested were above the background level when DNA was hybridized. For the LAB species for which more than one strain was hybridized, data for the strain with the lowest number of oligonucleotides whose intensities were above the background level were used. Of the 1,541 oligonucleotides whose intensities were above the background level, 1,406 (91.2%) produced spots whose intensities were above the background level for RNA as well, corresponding to a total of 72.1% of all oligonucleotides whose intensities were above the background level when aRNA was hybridized (Table 4). Of the 351 cross-hybridizing oligonucleotides on the microarray, 230 were covered in the validation hybridizations, resulting in 357 expected cross-hybridization occasions, as some oligonucleotides could cross-hybridize with several species tested. When DNA was hybridized, the intensities of 65.5% (234 of 357) of the oligonucleotides were above the background level, and the intensities of 85.5% of these oligonucleotides were also above the background level when they were hybridized with aRNA (Table 4). The percentage of unrelated oligonucleotides whose intensities were above the background level for DNA ranged from 5.9% (100 of 1,688 oligonucleotides for Lactococcus lactis subsp. cremoris MG 1363) to 14.8% (283 of 1,917 oligonucleotides for Lactobacillus plantarum D06SS01T01-H18). The percentage of unrelated oligonucleotides whose intensities were above the background level for both the DNA and aRNA hybridizations ranged from 0.1% to 6.0%.
As L. plantarum is an important species in food fermentations, the hybridization data obtained for the four strains of this species were used to look for strain-dependent hybridization behavior. Almost all of the 302 oligonucleotides for L. plantarum were designed based on sequences from L. plantarum WCFS1, whose genome sequence was available (25). L. plantarum strain LMG 9211 was in fact the same as the sequenced strain, although it came from a different culture collection. The percentage of oligonucleotides whose intensities were above the background level for this strain was 94.7%, which was much higher than the values obtained for the three other L. plantarum strains (84.8 to 87.4%) (Table 4). For all L. plantarum-specific oligonucleotides whose intensities were above the background level, the intensities of 94.9 to 98.8% of them were above the background level when they were hybridized with aRNA (Table 4).
During oligonucleotide design, a limited number of mismatches were tolerated in the search for cross-hybridizing oligonucleotides, as reflected by 357 cross-hybridizing occasions. For the species for which several strains were hybridized, an oligonucleotide was considered to have an intensity above the background level if the intensity for at least one of the strains was above the background level, in contrast to the rule mentioned above for the classification of oligonucleotides that were present or absent. Forty-two percent (39 of 93 oligonucleotides) of the perfectly matching cross-hybridizing oligonucleotides displayed a signal whose intensity was above the background level when they were hybridized with DNA, and the intensities of 87.2% of these oligonucleotides were also above the background level when the oligonucleotides were hybridized with aRNA. A higher percentage was obtained for oligonucleotides that had one mismatch when they were cross-hybridized; the intensities of the signals of 81.4% (57 of 70 oligonucleotides) were above the background level when DNA was hybridized, and the intensities of the signals of 96.5% of these were above the background level when aRNA was hybridized. For the oligonucleotides that had two mismatches when they were cross-hybridized, the intensities of the signals of 78.4% (76 of 97 oligonucleotides) were above the background level when DNA was hybridized, and 89.5% of these oligonucleotides were positive after hybridizations with aRNA. Of the 97 oligonucleotides that had three mismatches when they were cross-hybridized, 62 gave a positive signal. Of these 62 oligonucleotides, 75.8% (47 of 62 oligonucleotides) were positive when aRNA was hybridized.
DISCUSSION
In the present study, a species-independent functional gene LAB microarray was developed to study gene expression of LAB in fermented food ecosystems. This microarray covers 406 well-chosen genes that represent important pathways and phenotypic traits of food-related LAB. Genes involved in carbohydrate uptake and production of intermediates and end metabolites of sugar breakdown are represented, as are genes involved in the proteolysis and conversion of amino acids to favorable flavor components, in the production of exopolysaccharides that influence fermented food's texture, in the biosynthesis of bacteriocins as natural antibacterial peptides that are involved in mixed-strain competitiveness, and in stress responses, all including or not including regulatory genes. Also, genes to monitor unfavorable phenotypic traits, such as biogenic amines, antibiotic resistance, and potential virulence factors, are represented.
An algorithm able to design 30-mer oligonucleotides with limited and controlled cross-hybridization capabilities for species for a given gene was implemented by applying parameters for the amount and position of mismatches in hybridization processes (19, 21, 22, 30, 52, 55, 61, 62). Based on data obtained from a comparative analysis of all coding sequences for LAB strains that were present in the public domain (71 LAB species and 4 non-LAB species), a set of 4,851 oligonucleotides was designed, and 2,269 30-mer oligonucleotides for 46 LAB species and 4 non-LAB species were selected for synthesis. At the time of oligonucleotide design, eight full genome sequences were available for seven LAB species, including the genome sequences of two Streptococcus thermophilus strains. The length of the oligonucleotides was a well-balanced compromise between sensitivity and the aim to design oligonucleotides that could hybridize with several LAB species with a minimal number of mismatched positions and a maximal length of contiguous matching stretches.
Of the 2,269 oligonucleotides on the microarray, 15.5% met the initial criterion of the microarray design, namely, development of a species-independent microarray with gene-specific oligonucleotides that could cross-hybridize with multiple species. This could be ascribed to two factors. The first factor was the limited availability of gene sequence information for the 71 LAB strains selected, only 8 of which were fully sequenced and available in the public domain at the moment of oligonucleotide design. Indeed, six of the fully sequenced species accounted for 68.3% of all oligonucleotides on the microarray. Only 5 of the 2,269 oligonucleotides originated from S. thermophilus, the seventh fully sequenced species that was taken into account, but this was solely a matter of postdesign selection, as the microarray was to be used in the first instance for follow-up of sourdough fermentations. The second factor was the lower-than-expected homology at the gene sequence level. Indeed, from a taxonomic point of view, bacterial isolates with a global level of homology of at least 70%, as determined by DNA-DNA hybridization, are classified as members of the same species (50). Consequently, the amount of sequence variation at the gene level greatly influenced the search for proper species-independent oligonucleotides. Likewise, the fact that most of the 71 LAB strains selected belonged to the taxonomically heterogeneous genus Lactobacillus certainly contributed to this factor (9). The limited availability of gene sequence information for the 71 LAB strains selected also explains the number of unrelated oligonucleotides whose intensities were above the background level.
The validation hybridizations proved that 30-mer oligonucleotides hybridized well and showed good sensitivity and reproducibility. The validation hybridizations also provided insight into the specificity of the microarray, although it remains difficult to fully understand this specificity in terms of an ecosystem approach. The fact that 21.0% of the species-specific oligonucleotides tested had a hybridization intensity below the background intensity could be explained not only by experimental performance, such as random labeling and differences between hybridization conditions and thermodynamic parameters for the oligonucleotides (55), but also by sequence variations between the strains tested and the public sequences that were used for oligonucleotide design, which is well illustrated by the four L. plantarum strains that were hybridized. When the cross-hybridizing oligonucleotides were focused on, only 41.9% of the perfectly matching cross-hybridizing oligonucleotides resulted in a signal whose intensity was above the background level when DNA was hybridized. Cross-hybridizing oligonucleotides with incorporated mismatches seemed to result in better hybridization results than cross-hybridizing perfectly matching oligonucleotides. Probably, the incorporated mismatches, defined based on available sequence information, could be perfect matches and vice versa for the hybridized strains for which sequence information was missing.
In summary, to perform gene expression studies of fermented food ecosystems involving LAB, a species-independent functional gene microarray containing oligonucleotides that represent well-chosen key genes involved in favorable as well as unwanted phenotypic traits was designed and validated for the first time. The 30-mer oligonucleotides used did not compromise the sensitivity, and the oligonucleotides, including cross-hybridizing oligonucleotides with one mismatch, displayed good selectivity. Nevertheless, the results demonstrated that the aim to develop species-independent oligonucleotides was only met partially, due to highly variable genes involved in metabolic processes and hence low sequence similarity, as well as a lack of sufficient sequence information for the targeted species, explaining the incorporation of unique oligonucleotides besides species-independent oligonucleotides. As genome sequencing projects are ongoing and sequencing technology is becoming faster and cheaper, more sequence data will be available soon, resulting in a larger number of species-independent oligonucleotides in a future version. Consequently, the LAB functional gene microarray developed in the present study will be extended and upgraded regularly with additional species-independent oligonucleotides, so that the microarray will become a powerful tool to monitor gene expression in LAB communities during food fermentations.
Acknowledgments
This work was financed by SBO project IWT-030263 of IWT-Vlaanderen. L.D.V. and S.W. acknowledge financial support from the Research Council of the Vrije Universiteit Brussel (GOA and BOF projects). G.H. is a postdoctoral fellow of FWO-Vlaanderen.
We are grateful to Tom Bogaert, Kizi Coeck, Kirsten Deschouwer, Ruth Maes, Ilse Scheirlinck, and Ann Van Schoor for their contributions.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azcarate-Peril, M. A., R. Tallon, and T. R. Klaenhammer. 2009. Temporal gene expression and probiotic attributes of Lactobacillus acidophilus during growth in milk. J. Dairy Sci. 92:870-886. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA 103:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and E. W. Sayers. 2009. GenBank. Nucleic Acids Res. 37:D26-D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callanan, M., P. Kaleta, J. O'Callaghan, O. O'Sullivan, K. Jordan, O. McAuliffe, A. Sangrador-Vegas, L. Slattery, G. F. Fitzgerald, T. Beresford, and R. P. Ross. 2008. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 190:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camu, N., T. De Winter, K. Verbrugghe, I. Cleenwerck, J. S. Takrama, M. Vancanneyt, and L. De Vuyst. 2007. Dynamics and biodiversity of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentations of cocoa beans in Ghana. Appl. Environ. Microbiol. 73:1809-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claesson, M. J., D. van Sinderen, and P. W. O'Toole. 2008. Lactobacillus phylogenomics—towards a reclassification of the genus. Int. J. Syst. Evol. Microbiol. 58:2945-2954. [DOI] [PubMed] [Google Scholar]
- 10.Coton, M., F. Berthier, and E. Coton. 2008. Rapid identification of the three major species of dairy obligate heterofermenters Lactobacillus brevis, Lactobacillus fermentum and Lactobacillus parabuchneri by species-specific duplex PCR. FEMS Microbiol. Lett. 284:150-157. [DOI] [PubMed] [Google Scholar]
- 11.De Vuyst, L., and B. Degeest. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol. Rev. 23:153-177. [DOI] [PubMed] [Google Scholar]
- 12.De Vuyst, L., and E. J. Vandamme. 1992. Influence of the carbon source on nisin production in Lactococcus lactis subsp. lactis batch fermentations. J. Gen. Microbiol. 138:571-578. [DOI] [PubMed] [Google Scholar]
- 13.De Vuyst, L., and F. Vaningelgem. 2003. Developing new polysaccharides, p. 275-320. In B. M. McKenna (ed.), Texture in food, vol. 1. Semi-solid foods. Woodhead Publishing Ltd., Cambridge, United Kingdom. [Google Scholar]
- 14.De Vuyst, L., L. Makras, L. Avonts, J. Holo, Q. Yi, A. Servin, D. Fayol, C. Berger, G. Zoumpopoulou, E. Tsakalidou, D. Sgouras, B. Martinez-Gonzales, E. Panayotopoulou, A. Mentis, D. Smarandache, L. Savu, P. Thonart, and I. Nes. 2004. Antimicrobial potential of probiotic or potentially probiotic lactic acid bacteria, the first results of the international European research project PROPATH of the PROEUHEALTH cluster. Microb. Ecol. Health Dis. 16:125-130. [Google Scholar]
- 15.De Vuyst, L., V. Schrijvers, S. Paramithiotis, B. Hoste, M. Vancanneyt, J. Swings, G. Kalantzopoulos, E. Tsakalidou, and W. Messens. 2002. The biodiversity of lactic acid bacteria in Greek traditional wheat sourdoughs is reflected in both composition and metabolite formation. Appl. Environ. Microbiol. 68:6059-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gevers, D., G. Huys, and J. Swings. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31-36. [DOI] [PubMed] [Google Scholar]
- 18.He, Z., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, Z. Huang, W. Wu, B. Gu, P. Jardine, C. Criddle, and J. Zhou. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 19.He, Z., L. Wu, X. Li, M. W. Fields, and J. Zhou. 2005. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 71:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hüfner, E., R. A. Britton, S. Roos, H. Jonsson, and C. Hertel. 2008. Global transcriptional response of Lactobacillus reuteri to the sourdough environment. Syst. Appl. Microbiol. 31:323-338. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 22.Kane, M. D., T. A. Jatkoe, C. R. Stumpf, J. Lu, J. D. Thomas, and S. J. Madore. 2000. Assessment of the sensitivity and specificity of oligonucleotide (50-mer) microarrays. Nucleic Acids Res. 28:4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. F., H. Jeong, J.-S. Lee, S.-H. Choi, M. Ha, C.-G. Hur, J.-S. Kim, S. Lee, H.-S. Park, Y.-H. Park, and T. K. Oh. 2008. The complete genome sequence of Leuconostoc citreum KM20. J. Bacteriol. 190:3093-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaenhammer, T. R., R. Barrangou, B. L. Buck, M. A. Azcarate-Peril, and E. Altermann. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29:393-409. [DOI] [PubMed] [Google Scholar]
- 25.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuipers, O. P., A. de Jong, R. J. S. Baerends, S. van Hijum, A. L. Zomer, H. A. Karsens, C. D. den Hengst, N. E. Kramer, G. Buist, and J. Kok. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie van Leeuwenhoek 82:113-122. [PubMed] [Google Scholar]
- 27.Leroy, F., and L. De Vuyst. 2002. Bacteriocin production by Enterococcus faecium RZS C5 is cell density limited and occurs in the very early growth phase. Int. J. Food Microbiol. 72:155-164. [DOI] [PubMed] [Google Scholar]
- 28.Leroy, F., and L. De Vuyst. 2003. A combined model to predict the functionality of the bacteriocin-producing Lactobacillus sakei CTC 494 strain. Appl. Environ. Microbiol. 69:1093-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leroy, F., and L. De Vuyst. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15:67-78. [Google Scholar]
- 30.Liebich, J., C. W. Schadt, S. C. Chong, Z. He, S. K. Rhee, and J. Zhou. 2006. Improvement of oligonucleotide probe design criteria for functional gene microarrays in environmental applications. Appl. Environ. Microbiol. 72:1688-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, M., A. Nauta, C. Francke, and R. J. Siezen. 2008. Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria. Appl. Environ. Microbiol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, S. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makras, E., G. Van Acker, and L. De Vuyst. 2005. The probiotic strain Lactobacillus paracasei subsp. paracasei 8700:2 degrades inulin-type fructans of different degrees of polymerization. Appl. Environ. Microbiol. 71:6531-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meroth, C. B., J. Walter, C. Hertel, M. J. Brandt, and W. P. Hammes. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita, H., H. Toh, S. Fukuda, H. Horikawa, K. Oshima, T. Suzuki, M. Murakami, S. Hisamatsu, Y. Kato, T. Takizawa, H. Fukuoka, T. Yoshimura, K. Itoh, D. J. O'Sullivan, L. L. McKay, H. Ohno, J. Kikuchi, T. Masaoka, and M. Hattori. 2008. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveals a genomic island for reuterin and cobalamin production. DNA Res. 15:151-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naser, S., F. L. Thompson, B. Hoste, D. Gevers, K. Vandemeulebroecke, I. Cleenwerck, C. C. Thompson, M. Vancanneyt, and J. Swings. 2005. Phylogeny and identification of enterococci by atpA gene sequence analysis. J. Clin. Microbiol. 43:2224-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naser, S. M., P. Dawyndt, B. Hoste, D. Gevers, K. Vandemeulebroecke, I. Cleenwerck, M. Vancanneyt, and J. Swings. 2007. Identification of lactobacilli by pheS and rpoA gene sequence analysis. Int. J. Syst. Evol. Microbiol. 57:2777-2789. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 39.Pavlovic, M., S. Hormann, R. F. Vogel, and M. A. Ehrmann. 2005. Transcriptional response reveals translation machinery as target for high pressure in Lactobacillus sanfranciscensis. Arch. Microbiol. 184:11-17. [DOI] [PubMed] [Google Scholar]
- 40.Pfeiler, E. A., and T. R. Klaenhammer. 2007. The genomics of lactic acid bacteria. Trends Microbiol. 15:546-553. [DOI] [PubMed] [Google Scholar]
- 41.Pieterse, B., R. J. Leer, F. H. J. Schuren, and M. J. van der Werf. 2005. Unravelling the multiple effects of lactic acid stress on Lactobacillus plantarum by transcription profiling. Microbiology 151:3881-3894. [DOI] [PubMed] [Google Scholar]
- 42.Puskàs, L. G., A. Zvara, L. Hackler, and P. Van Hummelen. 2002. RNA amplification results in reproducible microarray data with slight ratio bias. BioTechniques 32:1330-1340. [DOI] [PubMed] [Google Scholar]
- 43.Ravyts, F., S. Barbuti, M. A. Frustoli, G. Parolari, G. Saccani, L. De Vuyst, and F. Leroy. 2008. Competitiveness and antibacterial potential of bacteriocin-producing starter cultures in different types of fermented sausages. J. Food Prot. 71:1817-1827. [DOI] [PubMed] [Google Scholar]
- 44.Rhee, S.-K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 46.Scheirlinck, I., R. Van der Meulen, A. Van Schoor, M. Vancanneyt, L. De Vuyst, P. Vandamme, and G. Huys. 2007. Influence of geographical origin and flour type on diversity of lactic acid bacteria in traditional Belgian sourdoughs. Appl. Environ. Microbiol. 73:6262-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheirlinck, I., R. Van der Meulen, A. Van Schoor, M. Vancanneyt, L. De Vuyst, P. Vandamme, and G. Huys. 2008. Taxonomic structure and stability of the bacterial community in Belgian sourdough ecosystems as assessed by culture and population fingerprinting. Appl. Environ. Microbiol. 74:2414-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schomburg, I., A. Chang, C. Ebeling, M. Gremse, C. Heldt, G. Huhn, and D. Schomburg. 2004. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 32:D431-D433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smeianov, V. V., P. Wechter, J. R. Broadbent, J. E. Hughes, B. T. Rodríguez, T. K. Christensen, Y. Ardö, and J. L. Steele. 2007. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 51.Stajich, J. E., D. Block, K. Boulez, S. E. Brenner, S. A. Chervitz, C. Dagdigian, G. Fuellen, J. G. R. Gilbert, I. Korf, H. Lapp, H. Lehvaslaiho, C. Matsalla, C. J. Mungall, B. I. Osborne, M. R. Pocock, P. Schattner, M. Senger, L. D. Stein, E. Stupka, M. D. Wilkinson, and E. Birney. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12:1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiquia, S. M., L. Wu, S. C. Chong, S. Passovets, D. Xu, Y. Xu, and J. Zhou. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664-675. [DOI] [PubMed] [Google Scholar]
- 53.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vrancken, G., T. Rimaux, L. De Vuyst, and F. Leroy. 2008. Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128:58-66. [DOI] [PubMed] [Google Scholar]
- 55.Weckx, S., E. Carlon, L. De Vuyst, and P. Van Hummelen. 2007. Thermodynamic behavior of short oligonucleotides in microarray hybridizations can be described using Gibbs free energy in a nearest-neighbor model. J. Phys. Chem. B 111:13583-13590. [DOI] [PubMed] [Google Scholar]
- 56.Wernersson, R., and H. B. Nielsen. 2005. OligoWiz 2.0—integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 33:W611-W615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood, B. J. B. 1998. Microbiology of fermented foods. Blackie Academic and Professional, London, United Kingdom.
- 58.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamfir, M., R. Callewaert, P. Calina Cornea, L. Savu, I. Vatafu, and L. De Vuyst. 1999. Purification and characterization of a bacteriocin produced by Lactobacillus acidophilus IBB 801. J. Appl. Microbiol. 87:923-931. [DOI] [PubMed] [Google Scholar]
- 60.Zamfir, M., M. Vancanneyt, L. Makras, F. Vaningelgem, K. Lefebvre, B. Pot, J. Swings, and L. De Vuyst. 2006. Biodiversity of lactic acid bacteria in Romanian dairy products. Syst. Appl. Microbiol. 29:487-495. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, L., M. F. Miles, and K. D. Aldape. 2003. A model of molecular interactions on short oligonucleotide microarrays. Nat. Biotechnol. 21:818-821. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, L., C. Wu, R. Carta, and H. Zhao. 2007. Free energy of DNA duplex formation on short oligonucleotide microarrays. Nucleic Acids Res. 35:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, J., and D. K. Thompson. 2002. Challenges in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 13:204-207. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]