Abstract
Cattle with high feed efficiencies (designated “efficient”) produce less methane gas than those with low feed efficiencies (designated “inefficient”); however, the role of the methane producers in such difference is unknown. This study investigated whether the structures and populations of methanogens in the rumen were associated with differences in cattle feed efficiencies by using culture-independent methods. Two 16S rRNA libraries were constructed using ∼800-bp amplicons generated from pooled total DNA isolated from efficient (n = 29) and inefficient (n = 29) animals. Sequence analysis of up to 490 randomly selected clones from each library showed that the methanogenic composition was variable: less species variation (22 operational taxonomic units [OTUs]) was detected in the rumens of efficient animals, compared to 27 OTUs in inefficient animals. The methanogenic communities in inefficient animals were more diverse than those in efficient ones, as revealed by the diversity indices of 0.84 and 0.42, respectively. Differences at the strain and genotype levels were also observed and found to be associated with feed efficiency in the host. No difference was detected in the total population of methanogens, but the prevalences of Methanosphaera stadtmanae and Methanobrevibacter sp. strain AbM4 were 1.92 (P < 0.05) and 2.26 (P < 0.05) times higher in inefficient animals, while Methanobrevibacter sp. strain AbM4 was reported for the first time to occur in the bovine rumen. Our data indicate that the methanogenic ecology at the species, strain, and/or genotype level in the rumen may play important roles in contributing to the difference in methane gas production between cattle with different feed efficiencies.
Microbial fermentation and ruminal nutrient absorption are key steps in the energy metabolism of cattle. The microbiota in the rumen is highly associated with the diet, age, antibiotic use, and health of host animals (32). Different types of symbiotic anaerobic microorganisms, including bacteria, archaea, ciliated protozoa, and fungi, inhabit the rumen (15), interact with each other, and play important roles in affecting the host's performance. The microbial-host relationships are highly complex and varied, ranging from mutually beneficial cooperation to competition (10). Among ruminal microbes, bacteria decompose the feed into short-chain (C1 to C5) fatty acids, amino acids, H2, and CO2, etc. (20). To maintain the low hydrogen level in this habitat, hydrogen-utilizing microbes, such as methanogens, utilize H2 and carbon substrates, mainly CO2, acetate, or methanol, to generate methane gas and hence to reduce hydrogen pressure in the rumen (8). However, this process causes a significant (6%) loss of dietary energy in the form of methane emission (14), which contributes to 13 to 19% of global greenhouse gas (16), and is one of the significant agricultural “causative sectors” contributing to global warming (13). Therefore, the energy loss and the consequent methane emission arouse both nutritional and environmental concerns in the livestock industry.
Archaeal methanogens are obligate anaerobes (38), and species of the order Methanobacteriales are the most common methanogens found in the rumen (11). Recent studies using culture-independent methods investigating the methanogenic communities in the rumens of sheep and cattle have identified 21 different strains belonging to 13 species in sheep (40, 41, 43, 44) and 13 different strains related to 8 species in cattle (23, 37, 42). In addition, the identification of novel uncultured methanogens in the rumen (23, 33, 40) suggests that the understanding of the methanogenic ecology is limited. Cattle with higher feed efficiencies are reported to produce 20 to 30% less methane (9, 24). However, the linkage between rumen methanogenic composition and the host's feed efficiency and methane production has not been studied and reported.
As one of the indicators of feed efficiency in cattle, residual feed intake (RFI) measures the difference between an animal's actual feed intake and the expected feed requirements for growth (1, 2). Cattle with low RFI (L-RFI) are designated “efficient,” while animals with high RFI (H-RFI) are designated “inefficient.” A recent study reporting a correlation between bacterial profiles and cattle RFI has suggested the probable linkage between rumen microbial ecology and feed efficiency in cattle (7). Therefore, we hypothesized that the structures and populations of methanogens may be also associated with RFI and methane gas production by the host. In this study, the compositions of methanogens in the rumens of cattle with different RFIs were compared by sequence analysis of the partial 16S rRNA genes (∼800 bp) generated from two constructed libraries, using pooled DNA from efficient (L-RFI) and inefficient (H-RFI) animals. The population of selected species in each steer was evaluated using quantitative real-time PCR (qRT-PCR) analysis, and the correlation between methanogenic structure/population and cattle RFI was investigated.
MATERIALS AND METHODS
Animal experiment and rumen sample collection.
Fifty-eight 10-month-old steers (Hereford crossed with Aberdeen Angus) were raised by following the guidelines of the Canadian Council on Animal care (4) under feedlot conditions at the Kinsella Research Station, University of Alberta, using a finishing diet described by Nkrumah et al. (24). The animal protocol was approved by the Animal Care and Use Committee (Moore-2006-55), University of Alberta. Feeding intake data were collected using the GrowSafe automated feeding system (GrowSafe Systems, Ltd., Airdrie, Alberta, Canada), a total mixed finishing composed of approximately 74% oats, 20% hay, and 6% feedlot supplement (32% crude protein beef supplement containing Rumensin [400 mg/kg of body weight] and 1.5% canola oil) (2). The feed efficiencies of steers were ranked as inefficient (H-RFI [RFI of >0.5]) or efficient (L-RFI [RFI of <−0.5]) on the basis of calculated RFI values as described by Nkrumah et al. (24). In this study, the RFI values for the examined steers (n = 58) were ranked as L-RFI (−0.68 ± 0.04 kg/day) and H-RFI (0.65 ± 0.05 kg/day) groups (P < 0.0001). Rumen sampling was performed within 1 week after RFI evaluation. Ruminal fluid was collected within 3 h after feeding by inducing flexible plastic tubing into the rumen and using the suction created with a 50-ml syringe to remove the fluid from the tubing. For each animal, 50 to 100 ml of rumen fluid was collected twice and transferred into a separate sterilized container, immediately frozen with liquid nitrogen, and stored at −80°C until processing.
DNA extraction.
Total DNA was extracted from 58 rumen fluid samples by using the methods outlined by Guan et al. (7). In brief, 0.5 ml of frozen rumen fluid was thawed on ice and washed with 4.5 ml of TN150 (10 mM Tris-HCl [pH 8.0], 150 mM NaCl) buffer, followed by 30 s of vortexing and 5 min of centrifugation at 200 × g at 4°C. Then, 1 ml of supernatant was transferred to a new microcentrifuge tube containing 0.3 g autoclaved zirconium-silica beads (0.1-mm diameter), and the cells were lysed by physical disruption in a model 8 BioSpec mini-bead beater at 4,800 rpm for 3 min. The supernatant of each sample was collected, DNA extraction was then performed with phenol-chloroform-isoamyl alcohol (25:24:1) extractions, and the DNA was precipitated with cold ethanol and dissolved in 20 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The concentration and quality of DNA were measured at A260 and A280 by using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Construction of 16S rRNA libraries.
Individual total DNA extracted from rumen fluid was diluted to a concentration of 50 ng μl−1 and was pooled by mixing 2 μl of each DNA sample from efficient animals (n = 29) (library 1) and inefficient animals (n = 29) (library 2) for library construction. The partial 16S rRNA gene (∼800 bp) was amplified with the universal primer pair Met 86f/Met 915r (Table 1), using the following program: an initial denaturation for 5 min at 94°C; 30 cycles at 94°C for 30 s, 57°C for 30 s, and 68°C for 1 min; and a final elongation for 7 min at 68°C. The PCR solution (50 μl) contained 1 μl of 20 pmol of each primer, 1 μl of 10 mM deoxynucleoside triphosphate, 2.5 U of Taq polymerase (Invitrogen, Carlsbad, CA), 1× PCR buffer, 1 μl of 50 mM MgCl2, and 1 μl of pooled DNA template. Amplified PCR products were then cloned into the TOP10 vector (TOPO TA cloning kit; Invitrogen) by using chemical transformation. Colonies with insertion were then selected on S-Gal (Sigma, St. Louis, MO) medium, and the plasmid DNA was extracted using a Millipore plasmid extraction kit (Millipore, Billerica, MA).
TABLE 1.
Primers used in this study to target methanogen 16S rRNA genes
| Primera | Sequence (5′ to 3′) | Reference |
|---|---|---|
| Met 86f | GCTCAGTAACACGTGG | 39 |
| Met 915r | GTGCTCCCCCGCCAATTCCT | 36 |
| 21F | TTCCGGTTGATCCYGCCGGA | 29 |
| 1389-1406R | ACGGGCGGTGTGTGCAAG | 17 |
| Met 1340r | CGGTGTGTGCAAGGAG | 38 |
“f” designates the forward primer and “r” the reverse primer.
Sequencing and phylogenetic analysis.
From libraries 1 and 2, 624 and 672 clones, respectively, were randomly selected and subjected to sequence analysis with an ABI 3730 sequencing system, using an ABI PRISM BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). The sequence reaction was performed with 10 μl of solution containing 0.5 μl of BigDye, 3.2 pmol of M13 Forward (CGCCAGGGTTTTCCCAGTCACGAC) or M13 Reverse (TTCACACAGGAAACAGCTATGAC) primer, 2.0 μl of 5× sequencing buffer, and 20 ng of plasmid DNA as the template. All sequences were subjected to BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches to determine the closest known taxon and were aligned using the ClustalW program (http://www.ebi.ac.uk/Tools/clustalw2/). The phylogenetic analysis was performed using the neighbor-joining method with the PHYLIP package (version 3.67) (http://evolution.genetics.washington.edu/phylip.html). Bootstrap numbers obtained from 1,000 replicates were assigned beside the nodes to verify the clustering of the sequences.
Clone library analysis.
The obtained libraries were then analyzed using the Mothur program (Mothur v1.3.0, http://www.mothur.org/wiki/Main_Page) by comparing operational taxonomic units (OTUs) on the basis of 97% similarity between sequences. Distance matrices were calculated by using the DNADIST program within the PHYLIP software package. Rarefaction analysis of library structure was conducted based on the principle in the DOTUR program (28). Diversity indices, such as the Shannon index, the Simpson index, and the Chao1 index, were used to measure the diversity of each library. Differences between the libraries were analyzed by comparing the levels of coverage of the samples, the similarities of community membership (Ochiai index), and the community structures (Bray-Curtis index) based on the principle in the ∫-LIBSHUFF program (27). Community diversity was compared in a phylogenetic context, using the UniFrac significance test and the P test within UniFrac (18).
qRT-PCR analysis.
The populations of selected species were determined by calculating the copy numbers of 16S rRNA genes. Three pairs of primers (Table 2) were used to detect Methanobrevibacter sp. strain AbM4, Methanosphaera stadtmanae, and total methanogens in each rumen sample. Species-specific and universal primers were designed based on the alignment of the identified targeted species sequences and all sequences, respectively, in two libraries, and the conserved region was targeted by using the software package Primer Express 3.0 (Applied Biosystems, Foster City, CA). qRT-PCR was performed with SYBR green chemistry (Fast SYBR green master mix; Applied Biosystems), using the StepOnePlus real-time PCR system (Applied Biosystems) with a fast cycle, a melting curve section, and the following program: 95°C for 10 min, followed by 40 cycles at 95°C for 3 s and 60°C for 30 s. For melting curve detection, the temperature was increased 0.3°C every 20 s from 60°C to 95°C. The standard curves were constructed by using species-specific primers based on a serial dilution of plasmid DNA from clones identified as Methanobrevibacter sp. strain AbM4 and Methanosphaera stadtmanae. The copy numbers of each standard curve were calculated based on the formula (NL × A × 10−9)/(660 × n), where NL is the Avogadro constant (6.02 × 1023 molecules per mol), A is the molecular weight of the molecule in standard, and n is the length of the amplicon (bp). The copy numbers of 16S rRNA genes of targeted methanogens per ml rumen fluid were calculated using the formula (MQ × C × VD)/(S × V), where MQ is the quantitative mean of the copy number, C is the DNA concentration of each sample, VD is the dilution volume of extracted DNA, S is the DNA amount (ng) subjected to analysis, and V is the rumen fluid volume subjected to DNA extraction. PCR efficiency (E) was calculated using the equation E = (10−1/slope −1) × 100, and the data generated from reactions with more than 90% efficiency were used for further analysis.
TABLE 2.
Primers used in this study for qRT-PCR analysis
| Organism(s) targeted | Primera | Sequence (5′ to 3′) | Product size (bp) |
|---|---|---|---|
| Methanobrevibacter sp. strain AbM4 | AbM4-F | TTTAATAAGTCTCTGGTGAAATC | ∼160 |
| AbM4-R | AGATTCGTTCTAGTTAGACGC | ||
| M. stadtmanae | Stad-F | CTTAACTATAAGAATTGCTGGAG | ∼150 |
| Stad-R | TTCGTTACTCACCGTCAAGATC | ||
| Total methanogens | uniMet1-F | CCGGAGATGGAACCTGAGAC | ∼160 |
| uniMet1-R | CGGTCTTGCCCAGCTCTTATTC |
“F” designates the forward primer and “R” the reverse primer. All sequences were determined in this study.
Statistical analysis.
Copy numbers and proportions of specific methanogen species were obtained from each individual, and the mean value was used for statistical analysis. Student's t test was used to verify the difference in each targeted species of methanogen between L-RFI and H-RFI animals. A simple covariance mixed model was used to correlate methanogen population with volatile fatty acid production and RFI by using the SAS system (version 9.1; SAS Institute, Cary, NC). Significance was defined at P values of <0.05.
Nucleotide sequence accession numbers.
The nucleotide sequences generated from this work have been deposited in GenBank under accession numbers FJ579097 to FJ580045.
RESULTS
Comparison of sequences generated from 16S clone libraries.
To identify methanogen profiles in the rumen, different combinations of reported universal methanogenic primers were used to amplify full or partial 16S rRNA gene products for library construction. But the attempt to generate a full 16S rRNA fragment with the combination of 21F (29) and 1389-1406R (17) as described by Ohene-Adjei et al. (25) was not successful, although these primers successfully targeted total methanogens in the ovine rumen. The usages of Met 86f/Met 1340r as outlined by Wright and Pimm (39) and the primer combination 21F/Met 1340r were not able to generate the PCR products from all animals. Only the primer pair Met 86f/Met 915r targeting a partial 16S rRNA gene product (∼800 bp) was found to generate amplicons from all 58 rumen samples. Therefore, this primer pair was used to amplify the pooled rumen DNA for library construction.
In total, 482 and 490 sequences were obtained from library 1 (pooled L-RFI animals) and library 2 (pooled H-RFI animals), respectively. From the rumens of L-RFI animals (library 1), 478 out of 482 sequences were identified to be methanogens, while 471 out of 490 sequences were identified to be methanogens from rumens of H-RFI animals (library 2). The sequences identified to be nonmethanogens, 4 sequences from library 1 and 19 sequences from library 2, were found to belong to 13 bacterial phylotypes. Since up to 16 bp of the primer sequences matched with the same region of bacteria, it is not surprising that the universal methanogen primers could also amplify some groups of bacteria. These sequences were not included for methanogenic community analysis.
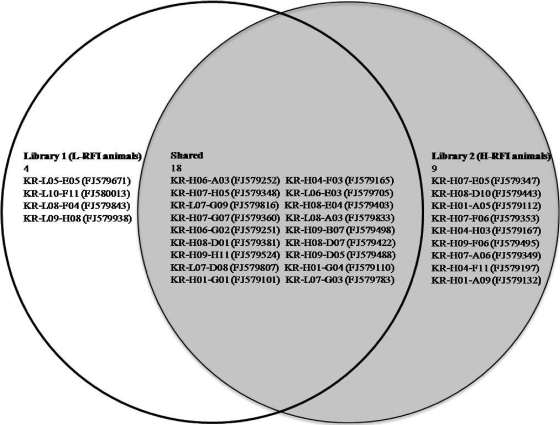
The taxonomy of each methanogen library was characterized first by determining the OTUs on the basis of 97% sequence similarity. In total, 31 unique OTUs were identified, with 22 OTUs from library 1 and 27 from library 2 (Table 3). Eighteen OTUs were found in both libraries (58.06% of total OTUs), while four and nine OTUs were found to be library 1- and library 2-specific, respectively (Fig. 1). When the structures and diversities of the two libraries were compared, higher values for Shannon index, diversity, and richness were observed in library 2, revealing that the methanogenic community of library 2, consisting of H-RFI animals, was more diverse than that of library 1, consisting of L-RFI animals. The differences in OTUs between the libraries at 100% similarity in a phylogenetic context were significant, with P values of <0.01 by both the P test (for transfer of lineages between libraries) and the UniFrac test (for evolutionary history shared between two libraries) in the UniFrac program (data not shown).
TABLE 3.
Comparison of structure diversities of sequenced clones in library 1 and library 2a
| Sample source | No. of sequences | No. of OTUs | Shannon index | Diversityb | Richnessc | Coverage (%)d |
|---|---|---|---|---|---|---|
| Library 1 | 478 | 22 | 0.9 (0.8-1.1) | 0.42 | 23 (20-41) | 100 |
| Library 2 | 471 | 27 | 1.5 (1.4-1.7) | 0.84 | 32 (25-68) | 96.2 |
Estimates of Shannon index, diversity, and richness are all based on 3% differences in nucleic acid sequence alignments. Values in parentheses are 95% confidence intervals as calculated by the Mothur program. The Ochiai index (for similarity between two communities) was 0.74. The Bray-Curtis index (for similarity between the structures of two communities) was 0.77.
Sample size-independent estimate of diversity based on negative natural log transformation of Simpson index values as calculated by the Mothur program.
Chao1 values, a nonparametric estimate of species richness.
Coverage values for a distance of 0.01, as calculated by the Mothur program.
FIG. 1.
Diagram of OTUs identified by the Mothur program at the 97% similarity level within and between libraries 1 (L-RFI animals) and 2 (H-RFI animals). Representative OTUs are presented by the clone identification numbers, with GenBank accession numbers in parentheses.
Taxonomy characterization of methanogenic ecology in the rumen.
To evaluate the identified difference in community structure between the two libraries, the taxonomies of all the OTUs were further investigated by a BLAST search based on an approach described by Ben-Dov et al. (3). The following criteria were used to determine the taxonomy of each OTU: a >97% match between the clone sequence and the GenBank data was considered to represent strains within the species level, and 93 to 96% identity represented different species at the genus level. All the OTUs obtained in this study resembled seven strains within five known species: Methanobrevibacter ruminantium, Methanobrevibacter thaueri, Methanobrevibacter smithii, Methanobrevibacter wolinii, and Methanosphaera stadtmanae.
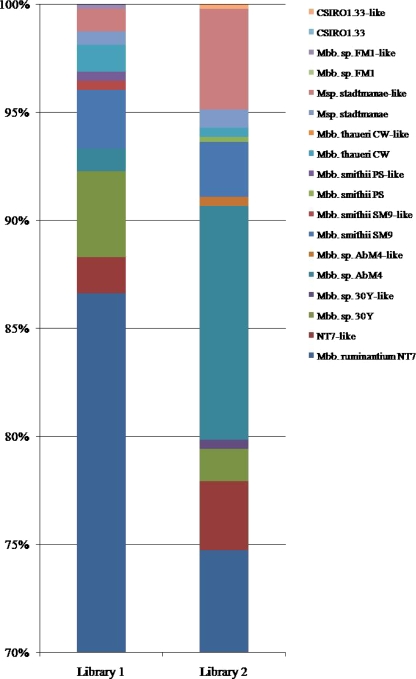
Four hundred twelve and 322 sequences in library 1 (L-RFI animals) and library 2 (H-RFI animals), respectively, were identical to M. ruminantium NT7 (AJ009959), which was predominant in both groups of animals, but with different distributions: 89.2% of the total clones from the library 1 and 73.0% of the total clones from library 2 (Fig. 2). The distributions of other species also varied between L-RFI and H-RFI animals. For example, for L-RFI animals, five sequences resembled Methanobrevibacter sp. strain AbM4 and eight sequences resembled M. stadtmanae, accounting for 1.0% and 1.7% of the total sequences, respectively (Fig. 2). For H-RFI cattle, 53 sequences resembled Methanobrevibacter sp. strain AbM4 and 27 sequences resembled M. stadtmanae, representing 10.8% and 5.7% of the total sequences, respectively (Fig. 2). M. wolinii-like Methanobrevibacter sp. strain AbM4 sequences have not previously been reported to occur in the bovine rumen. In addition, the distributions of different strains varied between the two groups of animals (data not shown).
FIG. 2.
Distribution of methanogenic species on the basis of their sequences, classified as methanogens from library 1 (L-RFI animals) and library 2 (H-RFI animals). NT7, M. ruminantium NT7; 30Y, Methanobrevibacter sp. strain 30Y; AbM4, Methanobrevibacter sp. strain AbM4; SM9, M. smithii SM9; PS, M. smithii PS; CW, M. thaueri CW; FM1, Methanobrevibacter sp. strain FM1; CSIRO1.33, Methanobacteriales archaeon CSIRO1.33 clone. The y axis shows that the percentages of >70% for more than 70% of the sequences were Methanobrevibacter ruminantium NT7 sequences in both libraries.
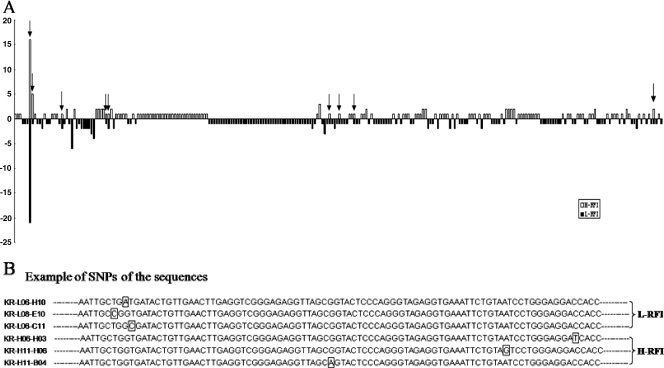
Furthermore, variation of methanogens at the genotype level in the two libraries was observed. For example, numerous genotypes in the sequences identified as M. ruminantium NT7 were observed to have high levels of diversity of single-nucleotide polymorphisms (SNPs). For instance, for the sequences with 99% identity with the M. ruminantium NT7 strain, 197 sequences in library 1 and 163 sequences in library 2 belonged to 264 genotypes. Figure 3 shows the alignment of six sequences with 99% identity with the M. ruminantium NT7 strain with SNPs observed in six representative locations (Fig. 3). When the association between the genotypes and cattle RFI was analyzed, some genotypes were detected only in L-RFI animals, while some were identified only in H-RFI animals. For example, clones KR-L06-H10, KR-L08-E10, and KR-L06-C11 were identified only in library 1 (L-RFI animals), and clones KR-H06-H03 and KR-H11-B04 were identified only in library 2 (H-RFI animals). Some genotypes, for example, clones KR-H11-H06 (FJ579567) and KR-H11-D04 (FJ579552), were identified in both groups of animals.
FIG. 3.
(A) Genotype analysis of all sequences with 99% identity with the Methanobrevibacter ruminantium NT7 strain. The bars indicate the number of sequences of each genotype in the 16S rRNA library generated from L-RFI and H-RFI animals. The arrows point out the genotypes that existed in both L-RFI and H-RFI animals. (B) Example of SNPs shown in the sequences belonging to this category. The position with an asterisk represents the nucleotide position with SNPs. The base with a square indicates the particular SNPs of each sequence.
Eight putative methanogens were identified at the genus level on the basis of sequences with 93 to 96% identity with the closest species. These OTUs may represent unidentified ruminal methanogens. Among them, the sequences similar to M. ruminantium NT7-like and M. stadtmanae-like OTUs were detected in both L-RFI and H-RFI animals, while M. smithii SM9-like, M. smithii PS-like, and Methanobrevibacter sp. strain FM1-like OTUs were detected only in L-RFI animals. Methanobrevibacter sp. strain 30Y-like, M. wolinii-like, and Methanobacteriales-like OTUs were detected only in H-RFI animals (Fig. 2).
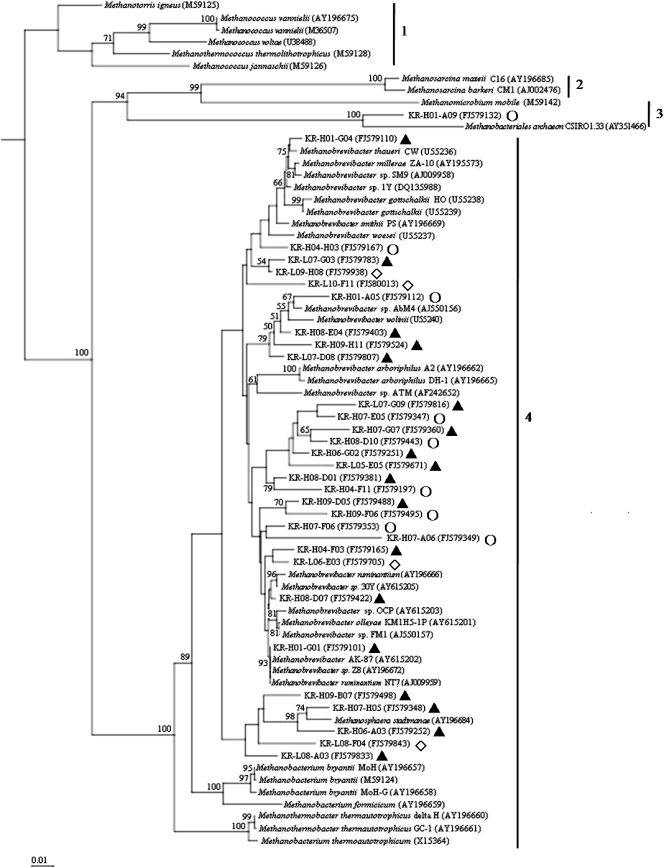
Phylogenetic analysis of the sequenced 16S rRNA libraries was performed based on the representative OTU sequences generated from the Mothur program and the typical methanogen species. As shown in Fig. 4, the major sequences clustered with their closest classification (the clone identification numbers are shown). Almost all major branches contained sequences from both L-RFI and H-RFI animals, with only one exception, KR-H01-A09. The methanogens detected in L-RFI animals and H-RFI animals did not differ greatly at the species level. However, some sequences with low levels of identity with the known species did not cluster with the closest species, as shown in the tree. For example, OTU KR-L10-F11, with M. ruminantium NT7 as the closest known species, was grouped with M. smithii instead of M. ruminantium. This confirmed our classification in which the genus-like sequences with <97% identities may represent new species within the genus Methanobrevibacter.
FIG. 4.
Phylogenetic analysis of methanogen partial 16S rRNA sequences obtained in this study. Representative sequences were generated by the Mothur program at a 3% difference level. GenBank sequences are identified by accession number. Bootstrap values (>50%) from 1,000 replications are indicated on the tree. 1, Methanococcales; 2, Methanosarcinales; 3, Methanomicrobiales; 4, Methanobacteriales; ▴, representative OTUs appearing in both libraries; ⋄, representative OTUs appearing only in library 1 (L-RFI animals); ○, representative OTUs appearing only in library 2 (H-RFI animals).
Comparison of methanogen populations between L-RFI and H-RFI animals.
The populations of total methanogens, Methanobrevibacter sp. strain AbM4, and M. stadtmanae were selected for qRT-PCR analysis to investigate these populations in 58 animals and the correlations with RFI. The mean total methanogen populations in L-RFI and H-RFI animals were 2.12 × 107 cells ml−1 and 2.52 × 107 cells ml−1, respectively (Table 4), confirming the similar quantities of the methanogens as previously reported (22, 26). The proportions and absolute copy numbers of 16S rRNA genes of M. stadtmanae and Methanobrevibacter sp. strain AbM4 were significantly lower (P < 0.05) in L-RFI animals than in H-RFI animals (Table 4). No significant difference between the two groups was observed for total methanogen population (P = 0.16).
TABLE 4.
Comparison of copy numbers of targeted methanogen 16S rRNA genes in L-RFI and H-RFI animalsa
| Organism group | No. of copies/ml
|
P | Proportion of total methanogens (%)
|
P | Amplification efficiency (%) | ||
|---|---|---|---|---|---|---|---|
| L-RFI | H-RFI | L-RFI | H-RFI | ||||
| Total methanogens | (2.12 ± 0.29) × 107 | (2.52 ± 0.29) × 107 | 0.16 | 100 | 100 | 93.55 | |
| M. stadtmanae | (1.33 ± 0.18) × 106 | (2.48 ± 0.59) × 106 | 0.03 | 6.02 ± 1.16 | 11.53 ± 2.55 | 0.02 | 94.46 |
| Methanobrevibacter sp. strain AbM4 | (5.69 ± 1.10) × 105 | (2.20 ± 0.59) × 106 | 0.02 | 3.73 ± 0.60 | 8.45 ± 2.16 | 0.02 | 94.30 |
Values shown are means ± standard errors. The L-RFI and H-RFI groups each contained 29 animals.
Statistical covariation analysis was performed for population of targeted species, total methanogen population, and RFI. The Methanobrevibacter sp. strain AbM4 and M. stadtmanae populations were positively correlated with total methanogen amount (P = 0.033 and 0.011, respectively). The total methanogen, Methanobrevibacter sp. strain AbM4, and M. stadtmanae populations were not linearly correlated with RFI ranking (P ranged from 0.17 to 0.69).
DISCUSSION
Previous studies have identified eight methanogenic species in the bovine rumen: M. ruminantium, M. thaueri, M. smithii, M. stadtmanae, Methanomicrobium mobile, Methanobacterium aarhusense, Methanobacterium formicicum, and Methanosarcina barkeri (11, 23, 37, 42). In this study, we identified M. ruminantium, M. thaueri, M. smithii, M. wolinii, and M. stadtmanae in the beef cattle that were examined. This concurs with previous studies showing that species belonging to Methanobrevibacter are the predominant methanogens in the rumens of ruminants (30, 31, 34, 40, 43). Contrasting with the results from previous studies, Methanobacterium aarhusense, Methanomicrobium mobile, Methanobacterium formicicum, and Methanosarcina barkeri were not detected in our study. This may be due to differences in many aspects, such as sampling procedures, types of rumen samples, DNA extraction methods, primers used, pooling approaches for construction of libraries, diets, animal hosts, and geographic regions. Previous studies have shown that the primers used for PCR amplification could affect the taxonomy identification of predominance of methanogens in the rumen. For example, Skillman and coworkers reported that using two different sets of primers revealed differences in methanogen predominance in rumen samples: 21f/958r amplified mainly M. stadtmanae-like sequences, whereas Arch f364/Arch r1386 generated mainly Methanobrevibacter sequences (31). Our results for sequencing and qRT-PCR analysis showed that the M. stadtmanae copy numbers accounted for <20% of the total methanogens (Fig. 2 and Table 4), confirming the observation that Methanobrevibacter is the dominant genus in the rumen. In addition, the characterization of unidentified methanogen groups (the genus-like sequences) (Fig. 2) supports the suggestion that a significant population of uncultured methanogens may inhabit the rumen (23). Since fewer then 700 clones were sequenced from each library, species with smaller populations might not be detected. Further experiments using more clones may improve the identification of sequences representing species with lower population densities. In addition, since we pooled DNA from each rumen sample for amplicons for library construction, this may reduce the amplification of the rare species. Pooling the amplicon from each rumen sample for library construction may also improve the identification of numbers of species.
Sequence analysis of methanogenic structures showed that the methanogen communities in the L-RFI and H-RFI animals differed at the species, strain, and genotype levels (Fig. 2 and 3). The identification of 148 and 125 genotypes with sequences 99% identical to Methanobrevibacter sp. strain NT7 in L-RFI and H-RFI animals, respectively, with only 9 genotypes conserved between the two groups of animals, revealed that very diverse genotypes of methanogens were represented in the rumen. It is not surprising that high numbers of genotypes of this particular strain were found, since the sequences were generated from the DNA pooled from samples derived from 29 individuals. Similarly, a study of identification of Escherichia coli in cows showed 240 different subtypes in 24 animals (12). Our data, in combination with those from the E. coli study, suggest that the variation of genotypes in methanogens may result from microbial mutation/adaptation to the specific host environment. Our discovery of the large portion of genotypes of methanogens indicates that the key members of the ruminal “methanogenbiome” are more complicated to define and are influenced by the host animal. Given that the genotypes were associated with RFI (Fig. 3), it may be suggested that the difference shown in genotypes of methanogens could also influence the metabolic energy traits of the host, leading to a variance in feed efficiency among the host animals. Future studies are needed to determine whether differences in genotype are associated with differences in methane production between L-RFI and H-RFI animals. Furthermore, the genotypes of a particular species may differ in each individual on account of many other factors, such as ruminal pH, the structures of other microbes (e.g., bacteria and protozoa), and the fermentation parameters in the rumen. Higher degrees of diversity at the species and genus levels have been reported for other microorganisms, such as bacteria (>40 species) and protozoa (15 different genera), compared to what was found for methanogens (8 species) (15), in the rumen. However, it is not clear whether the genotypes of bacteria and protozoa could be also be associated with the host animals and the methanogen structure in the rumen. Further studies correlating the methanogen diversity to that of other microbes, including bacteria and protozoa, may lead to the discovery of the roles of microbial-microbial interactions in feed efficiency in the host.
The unique combination of ruminal microbiota in each animal may have important roles in the host's nutrition uptake and energy metabolism, phenotypes that are usually regulated by the genetics, diet, and environment of the host. Host breed was found to have influence on ruminal bacterial structure and association between bacteria and cattle RFI within the breed (7), implying that methanogen structure may also be associated with host genetic variation. Diet is known to be another key factor that influences the microflora in the rumen. The impact of diet on methanogen profiles in the ovine rumen has been confirmed by identification of higher levels of methanogen diversity in pasture-grazing animals than in animals fed with oaten hay (40). Diet has also been reported to influence methane gas production and population changes for particular methanogens. Recent studies in which dietary fat supplementation was added to the feed showed reduced methane production in the rumen (19, 45). A preliminary study by Yu et al. revealed that dietary tallow might stimulate M. stadtmanae but inhibit Methanobrevibacter sp. strain AbM4 (44). Future studies of change in methanogenic structure in response to diet at the strain and/or genotype level for each animal will be essential. Furthermore, the environment may also contribute to differences in microbial diversity. It is not surprising that different species of methanogens were identified in the animals examined in our study, since different methanogenic components have been reported to occur in cattle raised in Canada (37, 42) and New Zealand (23).
Cattle with higher feed efficiencies have been reported to produce less methane (9). The total methanogen populations in L-RFI and H-RFI animals did not differ (Table 4), indicating that the quantity of total methanogens may not be vital for feed efficiency traits and may be associated with differences in methane yield. The methanogenic structures (species, strains, and genotypes) and populations of particular species/strains/genotypes may be associated with feed efficiency in cattle. The identification of higher populations of M. stadtmanae and Methanobrevibacter sp. strain AbM4 in H-RFI animals (Table 4) suggests a probable difference in methane production pathways in these inefficient animals. These two species were chosen because (i) their sequences were distributed at significantly different proportions between the two libraries (Fig. 2), (ii) Methanobrevibacter sp. strain AbM4 was identified for the first time in cattle in this study, and (iii) M. stadtmanae has been well studied for its methane production pathways. M. stadtmanae generates methane only by reduction of methanol with H2 (21). This species lacks the carbon monoxide dehydrogenase or acetyl-coenzyme A decarbonylase complex required for acetate substrate or acetyl-coenzyme A synthesis from substrates like CO2 and a methyl group (5). The population of Methanobrevibacter sp. strain AbM4 was negatively correlated with acetate concentration (P < 0.01) (unpublished data), indicating that acetate may be the substrate in the methanogenesis pathway of Methanobrevibacter sp. strain AbM4. However, more studies are required to verify such speculated mechanisms associated with methane yield and feed efficiency in the host. It has been shown that different strains of microorganisms of the same species could have distinct metabolic capacities and surface properties (6, 35); hence, the difference in methanogenesis substrates may be due to the results of the strain variation as we identified above. Thus, the strain level divergence of methanogens cannot be ignored, and the investigation of profiles of methanogen should include the strain variation.
In conclusion, this study demonstrated differences in methanogen ecology between rumens of beef cattle with different feed efficiencies with a low-energy diet. The methanogen communities were found to be different at the genus, species, strain, and genotype levels between efficient and inefficient animals. The cattle's feed efficiency was also correlated with the population of a particular species but not with the total quantity of methanogens. M. stadtmanae and Methanobrevibacter sp. strain AbM4 were found to have larger amounts and proportions of 16S rRNA genes in inefficient (H-RFI) animals, suggesting that organic-substrate-based methane biosynthesis pathways may be the cause of the low feed efficiency. Future studies for linking the methanogenic structure with the methane gas yield from cattle with different RFIs will be performed to verify and elucidate the different mechanisms of methanogenesis in the animals with higher feed efficiencies. This is the first study reporting the probable association between the “methanogenic biome” and feed efficiency in cattle. Our study of the linkage between the microbial ecology of methanogens and feed efficiency in cattle will allow better understanding of the gut microbiome and its impact on host physiology.
Acknowledgments
This study was supported by ALIDF/AARI (2007041R) and an NSERC discovery grant to L. L. Guan.
We thank S. Moore for supplying the animal information. We thank B. Irving, the manager of the Kinsella Ranch, and all the technical staff who assisted in the sampling. We also acknowledge U. Basu for real-time PCR analysis and Y. Meng for technical support.
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Archer, J. A., E. C. Richardson, R. M. Herd, and P. F. Arthur. 1999. Potential for selection to improve efficiency of feed use in beef cattle: a review. Aust. J. Agric. Res. 50:147-161. [Google Scholar]
- 2.Basarab, J. A., M. A. Price, J. L. Aalhus, E. K. Okine, W. M. Snelling, and K. L. Lyle. 2003. Residual feed intake and body composition in young growing cattle. Can. J. Anim. Sci. 83:189-204. [Google Scholar]
- 3.Ben-Dov, E., O. H. Shapiro, N. Siboni, and A. Kushmaro. 2006. Advantage of using inosine at the 3′ termini of 16S rRNA gene universal primers for the study of microbial diversity. Appl. Environ. Microbiol. 72:6902-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canadian Council on Animal Care. 1993. Guide to the care and use of experimental steers, vol. 1, 2nd ed. Canadian Council on Animal Care, Ottawa, Ontario, Canada. http://www.ccac.ca/en/CCAC_Programs/Guidelines_Policies/PDFs/ExperimentalAnimals_GDL.pdf.
- 5.Fricke, W. F., H. Seedorf, A. Henne, M. Kruer, H. Liesegang, R. Hedderich, G. Gottschalk, and R. K. Thauer. 2006. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 188:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan, L. L., J. D. Nkrumah, J. A. Basarab, and S. S. Moore. 2008. Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle's feed efficiency. FEMS Microbiol. Lett. 288:85-91. [DOI] [PubMed] [Google Scholar]
- 8.Hedderich, R., and W. Whitman. 2006. Physiology and biochemistry of the methane-producing Archaea, p. 1050-1079. In M. Dworkin (ed.), The prokaryotes, vol. 2, 3rd ed. Springer, New York, NY. [Google Scholar]
- 9.Hegarty, R. S., J. P. Goopy, R. M. Herd, and B. McCorkell. 2007. Cattle selected for lower residual feed intake have reduced daily methane production. J. Anim. Sci. 85:1479-1486. [DOI] [PubMed] [Google Scholar]
- 10.Hungate, R. E. 1984. Symposium on ‘Nutritional implications of microbial action in the non-ruminal alimentary tract’. Microbes of nutritional importance in the alimentary tract. Proc. Nutr. Soc. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis, G. N., C. Strompl, D. M. Burgess, L. C. Skillman, E. R. Moore, and K. N. Joblin. 2000. Isolation and identification of ruminal methanogens from grazing cattle. Curr. Microbiol. 40:327-332. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins, M. B., P. G. Hartel, T. J. Olexa, and J. A. Stuedemann. 2003. Putative temporal variability of Escherichia coli ribotypes from yearling steers. J. Environ. Qual. 32:305-309. [DOI] [PubMed] [Google Scholar]
- 13.Joblin, K. N. 1996. Potential options for reducing methane emissions from ruminants in New Zealand and Australia, p. 437-449. In W. J. Bouma, G. I. Pearman, and M. R. Manning (ed.), Greenhouse: coping with climate change. CSIRO Publishing, Canberra, New South Wales, Australia.
- 14.Johnson, K. A., and D. E. Johnson. 1995. Methane emissions from cattle. J. Anim. Sci. 73:2483-2492. [DOI] [PubMed] [Google Scholar]
- 15.Kamra, D. N. 2005. Rumen microbial ecosystem. Curr. Sci. 89:124-135. [Google Scholar]
- 16.Lassey, K. R., M. J. Ulyatt, R. J. Martin, C. F. Walker, and I. D. Shelton. 1997. Methane emissions measured directly from grazing livestock in New Zealand. Atmos. Environ. 31:2905-2914. [Google Scholar]
- 17.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machmuller, A., C. R. Soliva, and M. Kreuzer. 2003. Effect of coconut oil and defaunation treatment on methanogenesis in sheep. Reprod. Nutr. Dev. 43:41-55. [DOI] [PubMed] [Google Scholar]
- 20.Mackie, R. I., R. I. Aminov, B. A. White, and C. S. McSweeney. 2000. Molecular ecology and diversity in gut microbial ecosystems, p. 61-77. In P. B. Cronje, E. A. Boomker, P. H. Henning, W. Schultheiss, and J. G. van der Valt (ed.), Ruminant physiology: digestion, metabolism, growth and reproduction. CABI Publishing, Wallingford, Oxon, United Kingdom.
- 21.Miller, T. L., and M. J. Wolin. 1985. Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch. Microbiol. 141:116-122. [DOI] [PubMed] [Google Scholar]
- 22.Morvan, B., F. Bonnemoy, G. Fonty, and P. Gouet. 1996. Quantitative determination of H2-utilizing acetogenic and sulfate-reducing bacteria and methanogenic archaea from digestive tract of different mammals. Curr. Microbiol. 32:129-133. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson, M. J., P. N. Evans, and K. N. Joblin. 2007. Analysis of methanogen diversity in the rumen using temporal temperature gradient gel electrophoresis: identification of uncultured methanogens. Microb. Ecol. 54:141-150. [DOI] [PubMed] [Google Scholar]
- 24.Nkrumah, J. D., E. K. Okine, G. W. Mathison, K. Schmid, C. Li, J. A. Basarab, M. A. Price, Z. Wang, and S. S. Moore. 2006. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 84:145-153. [DOI] [PubMed] [Google Scholar]
- 25.Ohene-Adjei, S., R. M. Teather, M. Ivan, and R. J. Forster. 2007. Postinoculation protozoan establishment and association patterns of methanogenic archaea in the ovine rumen. Appl. Environ. Microbiol. 73:4609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saengkerdsub, S., R. C. Anderson, H. H. Wilkinson, W. K. Kim, D. J. Nisbet, and S. C. Ricke. 2007. Identification and quantification of methanogenic archaea in adult chicken ceca. Appl. Environ. Microbiol. 73:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin, E. C., B. R. Choi, W. J. Lim, S. Y. Hong, C. L. An, K. M. Cho, Y. K. Kim, J. M. An, J. M. Kang, S. S. Lee, H. Kim, and H. D. Yun. 2004. Phylogenetic analysis of archaea in three fractions of cow rumen based on the 16S rDNA sequence. Anaerobe 10:313-319. [DOI] [PubMed] [Google Scholar]
- 30.Skillman, L. C., P. N. Evans, G. E. Naylor, B. Morvan, G. N. Jarvis, and K. N. Joblin. 2004. 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 10:277-285. [DOI] [PubMed] [Google Scholar]
- 31.Skillman, L. C., P. N. Evans, C. Strompl, and K. N. Joblin. 2006. 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett. Appl. Microbiol. 42:222-228. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, C. S., H. J. Flint, and M. P. Bryant. 1997. The rumen bacteria, p. 10-72. In P. N. Hobson and C. S. Stewart (ed.), The rumen microbial system, 2nd ed. Blackie Academic and Professional, New York, NY.
- 33.Tajima, K., T. Nagamine, H. Matsui, M. Nakamura, and R. I. Aminov. 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200:67-72. [DOI] [PubMed] [Google Scholar]
- 34.Tokura, M., I. Chagan, K. Ushida, and Y. Kojima. 1999. Phylogenetic study of methanogens associated with rumen ciliates. Curr. Microbiol. 39:123-128. [DOI] [PubMed] [Google Scholar]
- 35.Walker, A. 2007. Say hello to our little friends. Nat. Rev. Microbiol. 5:572-573. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, T., S. Asakawa, A. Nakamura, K. Nagaoka, and M. Kimura. 2004. DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS Microbiol. Lett. 232:153-163. [DOI] [PubMed] [Google Scholar]
- 37.Whitford, M. F., R. M. Teather, and R. J. Forster. 2001. Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright, A. D., and C. Pimm. 2003. Improved strategy for presumptive identification of methanogens using 16S riboprinting. J. Microbiol. Methods 55:337-349. [DOI] [PubMed] [Google Scholar]
- 40.Wright, A. D., A. J. Williams, B. Winder, C. T. Christophersen, S. L. Rodgers, and K. D. Smith. 2004. Molecular diversity of rumen methanogens from sheep in Western Australia. Appl. Environ. Microbiol. 70:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright, A. D., A. F. Toovey, and C. L. Pimm. 2006. Molecular identification of methanogenic archaea from sheep in Queensland, Australia reveal more uncultured novel archaea. Anaerobe 12:134-139. [DOI] [PubMed] [Google Scholar]
- 42.Wright, A. D., C. H. Auckland, and D. H. Lynn. 2007. Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl. Environ. Microbiol. 73:4206-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright, A. D., X. Ma, and N. E. Obispo. 2008. Methanobrevibacter phylotypes are the dominant methanogens in sheep from Venezuela. Microb. Ecol. 56:390-394. [DOI] [PubMed] [Google Scholar]
- 44.Yu, Z., R. Garcia-Gonzalez, F. L. Schanbacher, and M. Morrison. 2008. Evaluations of different hypervariable regions of archaeal 16S rRNA genes in profiling of methanogens by Archaea-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 74:889-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zinn, R. A., and A. Plascencia. 1996. Effects of forage level on the comparative feeding value of supplemental fat in growing-finishing diets for feedlot cattle. J. Anim. Sci. 74:1194-1201. [DOI] [PubMed] [Google Scholar]