Abstract
Implementation of uranium bioremediation requires methods for monitoring the membership and activities of the subsurface microbial communities that are responsible for reduction of soluble U(VI) to insoluble U(IV). Here, we report a proteomics-based approach for simultaneously documenting the strain membership and microbial physiology of the dominant Geobacter community members during in situ acetate amendment of the U-contaminated Rifle, CO, aquifer. Three planktonic Geobacter-dominated samples were obtained from two wells down-gradient of acetate addition. Over 2,500 proteins from each of these samples were identified by matching liquid chromatography-tandem mass spectrometry spectra to peptides predicted from seven isolate Geobacter genomes. Genome-specific peptides indicate early proliferation of multiple M21 and Geobacter bemidjiensis-like strains and later possible emergence of M21 and G. bemidjiensis-like strains more closely related to Geobacter lovleyi. Throughout biostimulation, the proteome is dominated by enzymes that convert acetate to acetyl-coenzyme A and pyruvate for central metabolism, while abundant peptides matching tricarboxylic acid cycle proteins and ATP synthase subunits were also detected, indicating the importance of energy generation during the period of rapid growth following the start of biostimulation. Evolving Geobacter strain composition may be linked to changes in protein abundance over the course of biostimulation and may reflect changes in metabolic functioning. Thus, metagenomics-independent community proteogenomics can be used to diagnose the status of the subsurface consortia upon which remediation biotechnology relies.
Enzymatic reduction of U(VI) to insoluble U(IV) by dissimilatory Fe(III)-reducing bacteria (DIRB) can limit subsurface U(VI) migration (2, 19, 20). This observation provided the basis for an important new biotechnology that involves remediation of contaminated groundwater by organic amendment-based stimulation of the activities of DIRB. Despite the obvious high potential value, there are significant technological challenges that must be overcome before this approach can be deployed as a functional biotechnology. In situ U(VI) reduction rates are directly coupled with microbial physiology and community composition, both of which change as bioremediation progresses (22, 24, 28). Thus, a key need is the ability to monitor changes in microbial community membership and function as they occur, so that optimal management strategies can be implemented to achieve the desired geochemical outcomes. This challenge is significant because metal reduction occurs within pore spaces deep in the aquifer and the process is associated with vast numbers of microbial cells distributed in the subsurface.
Changes in the complements of DIRB proteins should reflect shifts in microbial physiology and could be used to monitor in situ bioremediation technologies if microbial proteomes could be tracked during organic amendment. Proteomic methods have previously been used to detect physiological responses of microorganisms growing in pure culture (8, 16, 17) and in genomically characterized natural microbial communities (7, 18, 26, 34). Strain-resolved proteomic approaches (18) have the potential to also track changes in microbial community composition. Previously, the use of proteomic analysis techniques to monitor subsurface bioremediation has been precluded by the lack of metagenomic data. Here, we used seven isolate Geobacter genomes to generate a reference database against which peptide data measured using two-dimensional (2D) liquid chromatography (LC)-based high-resolution tandem mass spectrometry (MS-MS) were compared for protein identification. Although differences between environmental and isolate peptide sequences may preclude peptide identification, peptides common to multiple Geobacter types and isolates enable protein identification, and peptides unique to one isolate constrain environmental genotypes. Simultaneous analysis of community structure and function in natural microbial communities that relies upon proteomics-derived genomic insights is referred to as proteogenomics (Fig. 1). In the current study, a proteogenomic approach was applied to monitor the progress of an in situ U bioremediation project carried out at the Department of Energy (DOE) Integrated Field Research Challenge site in Rifle, CO. Despite strain complexity in the recovered samples, proteomic data provided insights into the community structure and physiology of planktonic Geobacter isolates in aquifer solutions as groundwater U(VI) concentrations decreased.
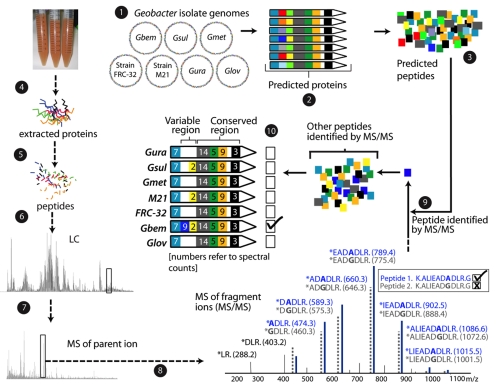
FIG. 1.
Strain-resolved proteogenomic techniques allow identification of unique peptides and constrain the genotypes present in environmental samples. (1) Proteins were predicted from the seven isolate Geobacter genomes. (2) Proteins were aligned. In the example shown, they share a conserved region (similar colors in all seven regions) and have a variable region (indicated by the variety of colors for each protein). (3) Tryptic digest patterns are used to predict peptide sequences. Proteins were extracted from the experimental sample (4) and digested into peptides by using trypsin (5). These peptides were separated using LC (6) before high-resolution mass spectrometry on a fraction of peptides from an elution peak (7) to determine the mass of the parent ions. (8) The peptides are fragmented and mass spectrometry measurements made on fragment ions. (9) Mass spectral data are matched to peptide sequences predicted in step 3 to allow peptide identification. Peptide sequences differing by only one amino acid can be identified, as shown in the spectral diagram. (10) Detected peptides are mapped onto aligned protein sequences to identify peptides shared or unique to each of the isolate species. Spectral counts can be mapped together with peptides. Where coexisting unique peptides are detected, ratios of unique spectral counts can be used to infer relative strain abundance. As shown, the identification of a unique peptide indicates that this protein is most closely related to that of G. bemidjiensis.
MATERIALS AND METHODS
Injection gallery design and operation.
The Winchester field experiment was carried out during August and September 2007 (8 August to 12 September) at the Rifle Integrated Field Research Challenge site in Western Colorado, where groundwater U concentrations are typically ∼1 μM. An injection gallery consisting of 10 injection wells, 12 down-gradient monitoring wells arranged in three rows, and three up-gradient monitoring wells was constructed using sonic rotary drilling (see Fig. S1 in the supplemental material). Acetate-bromide (50 mM:5 mM)-amended groundwater was injected into the subsurface to provide a target acetate concentration of ∼5 mM which acted as an electron donor over the course of the amendment experiment. Geochemical samples were taken from a 17-ft depth after 12 liters of groundwater was purged, and the samples were analyzed in the field and at Lawrence Berkeley National Laboratory. Ferrous iron and sulfide concentrations were analyzed immediately following sampling by using the HACH phenanthroline assay and a sulfide reagent kit, respectively (HACH, CO). Acetate was analyzed using a Dionex ICS1000 ion chromatograph equipped with a CD25 conductivity detector and a Dionex IonPac AS22 column (Dionex, CA). U(VI) values were determined using a kinetic phosphorescence analyzer (Chemchek, WA) at the DOE Grand Junction office.
Sample collection.
Three biomass samples were recovered from groundwater over the course of the in situ bioreduction experiment. Two samples were taken from well D07, and one was taken from well D05, both wells being located in the second row of the down-gradient monitoring wells. For each sample, 500 liters of groundwater pumped at approximately 2 liters min−1 from a well was filtered through a prefilter (1.4-μm-pore-size, 292-mm diameter Supor disc filter; Pall Corporation, NY), followed by a Pelicon tangential flow filtration system (0.2 μm) (Millipore, MA) to concentrate biomass. Groundwater was passed through a series of chilling baths containing an ice-rock salt mixture as soon as it was pumped to the surface to minimize changes to the proteome. Chilling baths ensured that the groundwater temperature was approximately 1°C as it passed through the filtration system which was located in an air-conditioned trailer. Biomass was concentrated to ∼200 ml in the retentate vessel and centrifuged at 4,000 rpm for 40 min at 4°C. The resulting pellet was resuspended in ∼5 ml of groundwater, immediately frozen in an ethanol-dry ice mix, and shipped overnight on dry ice to Oak Ridge National Laboratory (ORNL) and Pacific Northwest National Laboratory for proteomic analysis.
Proteomics.
In order to ensure that the proteomic analyses were as robust as possible, all experiments were conducted in two different laboratories, using parallel but distinct procedures and instrumentation. Results from both laboratories are provided in Table S2 in the supplemental material. Values with normalized spectral abundance frequency (NSAF) scores (see below for details) below 1 × 10−5 were removed, and values between this and 1 × 10−4 are shown in gray text. Despite many opportunities for discrepancies, the data indicate good replication and all conclusions are based on patterns evident in both datasets. Plots in the manuscript are generated from ORNL data.
ORNL shotgun proteomics via 2D LC-MS-MS.
Cells in ∼500 μl of a sample were lysed in a single tube and processed using a method optimized for small samples in which the proteins were denatured, reduced, and digested with sequencing grade trypsin (32). Approximately one-fifth of each digested sample was used for each of three technical replicates for the three samples. Samples were analyzed via 22-hour 2D nano-LC-MS-MS with a split-phase column (reversed phase-strong cation exchange-reversed phase) on a hybrid linear ion trap-Orbitrap mass spectrometer (Thermo Fisher Scientific, MA), as previously described (34). The linear ion trap (LTQ) and Orbitrap settings were as follows: 30K resolution for full scans using Orbitrap; all data-dependent MS-MS in LTQ (top five); two microscans for both full and MS-MS scans, with centroid data for all scans; and two microscans averaged for each spectra, with the dynamic exclusion set at 1.
All samples and technical replicates were searched against two databases; the first contained 862 isolate genomes with ∼3 million protein entries (http://img.jgi.doe.gov/) plus a Geobacter subdatabase that contained ∼26,000 Geobacter proteins (from seven species). Common contaminants, such as trypsin and keratin, were included in both databases. All MS-MS spectra were searched against both databases with the SEQUEST algorithm (10) and filtered with DTASelect/Contrast (30) at the peptide level with conservative filters [Xcorr values of at least 1.8 (+1), 2.5 (+2), and 3.5 (+3)]. Only proteins identified with two fully tryptic peptides at conservative filter levels were considered for further analysis.
We applied the accepted method of reverse database searching to determine false-positive levels on a subset of the data with the filters given above (9, 25). This reverse database searching method was designed and tested with single yeast genomes and proteomes. For the proteomics analysis presented here, the very large percentage of nonunique peptides (often the case in metaproteomics-type experiments) and the unknown degree of genomic sequence representation in the samples make the accuracy of false-positive-level estimation questionable. Nevertheless, we attempted this established method and calculated the false-positive levels twice, once with only unique peptides and once with all peptides. With unique peptides, the false-positive rate was between 5 and 8%, which is overestimated due to the massive reduction in real peptides that have nonunique status. With all the peptides considered, the rate falls to 0.6 to 0.9%, which is underestimated due to the addition of many nonunique peptides when truly only one peptide existed (up to seven maximum, the number of Geobacter species in the database). Since both datasets were then filtered again during the comparison stage to remove all proteins with very low NSAF values, it can be assumed that the final list used for biological analyses had a very low false discovery rate. All databases, peptide and protein results, MS-MS spectra, and supplementary tables for all database searches from the ORNL data set are archived and made available as open access publications via http://compbio.ornl.gov/ersp_rifle/ground_water_2007.
Pacific Northwest National Laboratory proteomics via LC-MS-MS and peptide putative mass and elution time tag assignments.
Global as well as soluble and insoluble protein fractions were extracted from cell pellets by using established protocols (1, 17). Briefly, frozen cells were thawed on ice, washed using 100 mM NH4HCO3, pH 8.4, buffer, and then suspended in a new aliquot of this buffer. Cells were lysed via pressure cycling technology using a barocycler (Pressure BioSciences, Inc., South Easton, MA). The suspended cells were subjected to 20 s of high pressure at 35 kilopounds per square inch, followed by 10 s of ambient pressure for 10 cycles. The protein concentration was determined by a Coomassie assay (Thermo Scientific, Rockford, IL). Protein extraction, digestion, and high-performance LC fractionation were performed as previously described (4). From each collected fraction, peptides were analyzed by reversed-phase high-performance LC separation coupled with the use of an LTQ ion trap mass spectrometer (ThermoFisher Scientific Corp., San Jose, CA) operated in a data-dependent MS-MS mode. Acquired spectra from approximately ∼400 MS-MS experiments were analyzed using the SEQUEST algorithm (10) in conjunction with predicted protein annotations concatenated from the seven Geobacter genomes. SEQUEST results were preliminarily filtered (Xcorr values of ≥1.9, ≥2.2, or ≥3.5 for 1+, 2+, or ≥3+ if seen once; Xcorr values of ≥1.9 if seen two or more times; no cleavage rules; minimum length, 6), extracted, and processed using the PRISM proteomics pipeline developed in-house (15).
Peptide mapping and strain variant analysis.
Probable orthologous proteins were identified by BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches between predicted proteins from the seven Geobacter isolates as proteins exhibiting 30% amino acid similarity over 70% of the alignment length (6, 27). Tables showing distribution across the seven Geobacter species were constructed as previously described (5). Spectral count data were normalized using the NSAF technique (36) and averaged over three technical replications per sample for the ORNL data set. While this technique was initially developed for analysis of isotope-labeled data, the statistical basis of the method allows for the normalization of data to account for biases arising from protein length in this instance. Peptides were mapped onto alignments of inferred orthologous proteins by using an in-house script which, at a protein-by-protein level, allows simultaneous visualization of unique (in single-isolate genomes) and nonunique (shared by one or more genomes) peptides as well as spectral count information. Within this program, identified peptides are rendered for individual samples, technical replicate runs, and multiple samples.
RESULTS
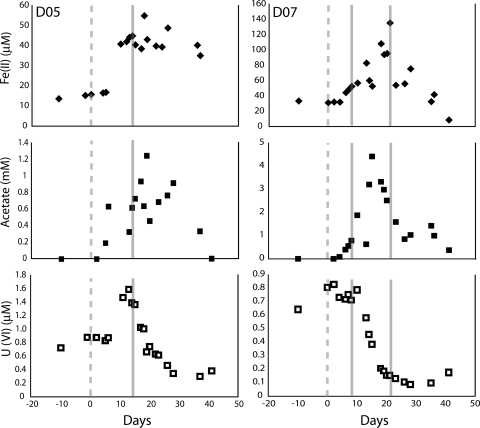
U(VI) groundwater concentrations were elevated at the time of collection of the D07(1) and D05 samples (∼0.7 and 1.4 μM, respectively) and lower (∼0.1 μM) when D07(2) was collected (Fig. 2). D07(1) and D05 were collected during the early phase of iron reduction, and D07(2) was collected later in iron reduction.
FIG. 2.
Acetate, Fe(II), and U(VI) geochemical data for the two proteomic sampling wells (D05 and D07) over the time course of the biostimulation experiment. The gray dashed line indicates the start of acetate amendment, whereas the full gray lines denote a proteomic sampling event.
Following protein extraction from the samples and 2D LC-MS-MS (see Materials and Methods), tandem mass spectra from all samples were searched against a database containing 862 isolate genomes (http://img.jgi.doe.gov/). Approximately 95% of all unique peptides identified matched Geobacter proteins, indicating that all samples were dominated by Geobacter species. The majority of the remaining unique peptides matched closely related microorganisms, such as Pelobacter species (12) (see the supplemental material). Subsequently, proteomic data were searched against a database (see the supplemental material) of predicted peptides from seven Geobacter isolate genomes: Geobacter bemidjiensis, G. metallireducens, G. sulfurreducens PCA, G. lovleyi SZ, G. uraniireducens Rf4, Geobacter strain FRC-32, and Geobacter strain M21. Between 2,000 and 2,500 proteins identified in each sample by using these subdatabase searches were then used in subsequent biological analyses (see Materials and Methods). While only three samples were collected, differences in proteomic patterns suggest some interesting changes in microbial community structure and activity.
Strain composition.
Mass spectral counts for spectra unique to each isolate Geobacter species in each sample were quantified (Fig. 3A) using a database with only the six Geobacter species that share between ∼60 and 70% amino acid similarity across the 1,480 to 2,291 homologous proteins (G. bemidjiensis, G. sulfurreducens, G. metallireducens, G. uraniireducens, G. lovleyi, and Geobacter strain FRC-32) (see Table S1 in the supplemental material). Unique spectral counts show that a strain or strains closely related to G. bemidjiensis dominated (60 to 70%) all three samples. Strain M21 was excluded from this initial analysis because the 3,098 proteins homologous with G. bemidjiensis share ∼95% average amino acid identity (see Table S1 in the supplemental material). Addition of the strain M21 genome would lead to classification of peptides shared with G. bemidjiensis as nonunique, prohibiting their inclusion in unique spectral counts for both strains. However, inclusion of strain M21 in the protein database for all subsequent analyses increased our ability to identify peptides and allowed higher-resolution analysis of strain makeup and activity.
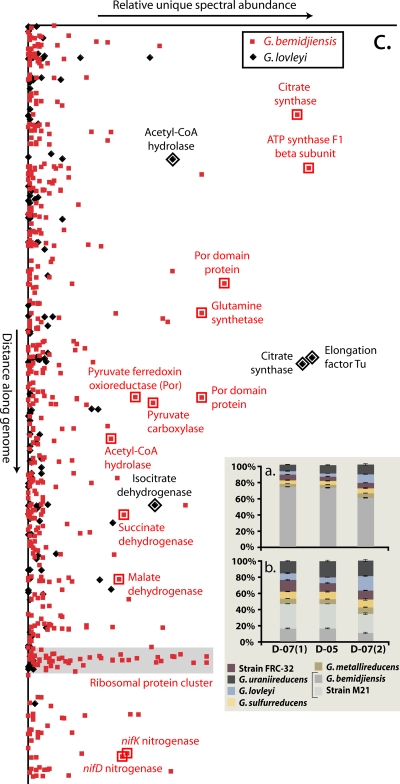
FIG. 3.
Counts of spectra unique to the Geobacter isolates as a percentage of the total number of unique spectra per sample. (a) Unique spectral count data observed when experimental results were searched using only the six Geobacter species with ∼60 to 70% amino acid identity (strain M21 excluded). (b) Spectral count patterns generated by a database search consisting of all seven Geobacter isolates (i.e., strain M21 is included). The percentage of spectra unique to G. bemidjiensis drops greatly when strain M21 is included due to the high amino acid sequence identity between G. bemidjiensis and strain M21. (c) Unique spectral counts for G. bemidjiensis and G. lovleyi in D07(2) plotted as a function of gene order. There are several loci where increased detection of unique spectra matching G. lovleyi indicates an environmental strain with similarity to both G. bemidjiensis and G. lovleyi.
While unique spectra for all Geobacter isolates were detected in all samples, each sample could, in principle, contain only one genotype. This “hypothetical” genotype would encode peptides classified as unique to each of all seven isolates. Thus, determination of strain makeup requires proteome-wide analysis of coexisting unique peptides of all seven Geobacter genotypes (Fig. 1). The identification of multiple homologous peptides that differ in their amino acid sequences indicates the presence of multiple genotypes. It is also possible to profile strain composition on the basis of the numbers of peptides characteristic of each isolate genotype.
In D07(2) compared to D07(1), the fraction of unique spectra matching G. lovleyi increased from ∼4% to ∼15% (Fig. 3B). Proteogenomic inference of the strain composition of the community in the D07(2) sample is indicated in Fig. 3C. The figure illustrates the relative even distribution along the genome of unique peptides matching G. bemidjiensis, but the presence of unique peptides characteristic of G. lovleyi at a few loci was observed, implying that the actual genotype has amino acid sequence similarity to G. lovleyi at these loci. Thus, late in iron reduction [D07(2)], we infer the proliferation of a strain(s) whose genotype remains most closely related to G. bemidjiensis and strain M21, but which has an increased fraction of loci shared with G. lovleyi or increased activity of such a strain(s).
Mapping of peptides onto alignments of homologous proteins predicted from the seven isolate genomes revealed that the best coverage of protein sequences by identified peptides was generally achieved by nonunique plus unique peptides of strains M21 and/or G. bemidjiensis. Over the entire proteomic data set, the combination of sequence coverage and spectral count data indicates dominance by a strain or strains slightly more closely related to M21 (isolated from the Rifle, CO, site) than to G. bemidjiensis. The coexistence of multiple strain variants in the Rifle subsurface under biostimulated conditions is indicated by the simultaneous identification of unique spectra from homologous peptides (Fig. 4; see also Fig. S3 in the supplemental material).
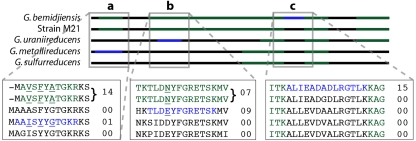
FIG. 4.
Identified peptide data from sample D07(1) mapped onto a schematic of aligned ribosomal S9 proteins. These data illustrate how inferences about strain makeup are achieved. Green regions indicate nonunique peptides, and blue regions indicate unique peptides. Expanded sections show the aligned amino acid sequence, with values displaying spectral counts for each peptide. Unique homologous G. metallireducens, G. uraniireducens, and G. bemidjiensis peptides and peptides unique to the G. bemidjiensis strain M21 group were present (regions a and b). The unique G. metallireducens peptide was identified much less frequently than the homologous G. bemidjiensis and strain M21 peptide (1 versus 14 spectral counts, respectively), suggesting the presence of a second protein variant at low abundance. Conversely, the unique G. bemidjiensis strain M21 and G. uraniireducens peptides have comparable spectral counts (seven versus nine spectral counts, respectively), consistent with two relatively equally abundant strain variants. The highest coverage is obtained using the G. bemidjiensis sequence due to the identification of a unique G. bemidjiensis peptide (section c). Given this and the high spectral count for this peptide, we infer that the dominant S9 ribosomal protein is from an organism more closely related to G. bemidjiensis than to other Geobacter isolates.
Microbial metabolism.
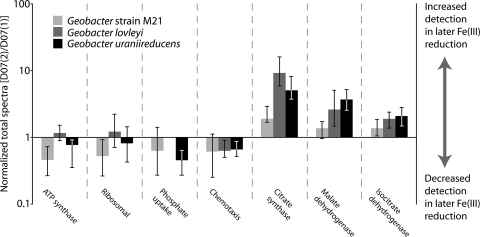
Relative protein abundances, inferred from numbers of spectral counts per protein (see Materials and Methods), indicate intriguing shifts in specific functional classes over the sampling period (Fig. 5). Rapid growth and protein production under biostimulated conditions were reflected in the abundance of detected peptides matching chemotaxis (see the supplemental material) and ribosomal proteins (35), while spectra matching ATP synthase F1 subunit proteins in all samples indicate the high energy demand of Geobacter species associated with this growth. However, these proteins, together with proteins associated with nutrient stress (metal efflux and P acquisition) were inferred to be more abundant in D07(1) than in D07(2), suggestive of a lowering growth rate later in Fe(III) reduction. Corresponding abundance increases from D07(1) to D07(2) for several tricarboxylic acid (TCA) cycle proteins (citrate synthase and isocitrate dehydrogenase) and the acetate-activating enzyme acetyl-coenzyme A (CoA) hydrolase (Fig. 5) suggest a shift in Geobacter energy metabolism, possibly tied to changes in strain makeup and lowering growth rates.
FIG. 5.
Changes in abundance of protein functional categories between early and late Fe(III) reduction samples, based on the ratio between NSAF values in D07(1) and D07(2). The patterns indicate a shift from a period of initial rapid growth to slower growth coupled with increasing energy demand later during biostimulation.
Increases in the abundance of citrate synthase, responsible for controlling flux into the TCA cycle by catalyzing the condensation of acetyl-CoA and oxaloacetate to citric acid (3), corresponded to the identification of more G. lovleyi and G. uraniireducens unique spectra (10- to 20-fold increases). However, spectral count data for conserved peptides confirm that the inferred increase was not simply due to a shift in strain type toward the genomically characterized isolates. There are more than twice as many spectral counts for a conserved peptide shared by all genotypes that contains a citrate binding site detected in D07(2) than in D07(1). The identification of coexisting homologous unique peptides associated with this protein indicates at least four Geobacter strain variants in D07(2) (see Fig. S3 in the supplemental material). Similar patterns were observed for unique peptides matching isocitrate dehydrogenase as the period of biostimulation increased. Increases in unique spectra in D07(2) relative to those in D07(1) (sevenfold for unique spectra matching G. lovleyi) were again suggestive of a shift toward strains related to G. lovleyi and G. uraniireducens, while there was a concurrent doubling of spectra matching a highly conserved peptide present in all isocitrate dehydrogenase variants.
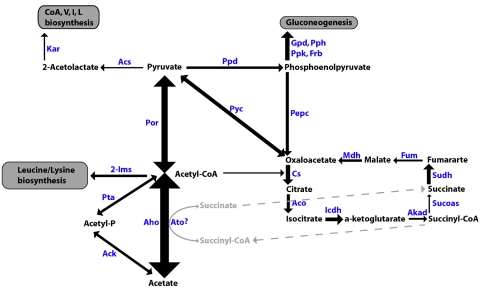
As expected during acetate biostimulation, peptides of acetyl-CoA hydrolase, which catalyzes the activation of acetate to acetyl-CoA for use in the TCA cycle, were highly detected in all three samples. An additional pathway for acetyl-CoA formation via acetate kinase and phosphotransacetylase was evident from the proteomic data, although lower spectral counts for this than for the acetyl-CoA hydrolase pathway may indicate that this was not the primary mechanism for acetyl-CoA formation. Peptides matching succinyl-CoA synthetase from M21 and G. bemidjiensis-like strains were identified, indicating the potential for operation of a complete TCA cycle in these organisms without acetyl-CoA transferase activity (29) (Fig. 6).
FIG. 6.
Geobacter central metabolism elucidated using identified peptides. Arrow thicknesses indicate relative abundances based on spectral count data for each protein. 2-Ims, 2-isopropylmalate synthase; Ack, acetate kinase; Acs, acetolactate synthase; Aco, aconitase; Aho, acetyl-CoA hydrolase; Akgd, alpha ketoglutarate dehydrogenase; Ato, acetyl-CoA transferase; Cs, citrate synthase; Frb, fructose bisphosphatase; Fum, fumarase; Gpd, glyceraldehyde-3-phosphate dehydrogenase; Icdh, isocitrate dehydrogenase; Kar, ketol-acid reductoisomerase; Mdh, malate dehydrogenase; Pepc, phosphoenolpyruvate carboxykinase; Por, pyruvate ferredoxin oxidoreductase; Ppd, pyruvate phosphate dikinase; Pph, phosphopyruvate hydratase; Ppk, phosphoglycerate kinase; Pta, phosphotransacetylase; Pyc, pyruvate carboxylase; Sucoas, succinyl-CoA synthetase; Sudh, succinyl-CoA dehydrogenase.
Detected at high abundance similar to that of acetyl-CoA hydrolase in all samples were peptides of pyruvate ferredoxin oxioreductase, which plays a key role in three-carbon synthesis from acetyl-CoA in Geobacter (21). Pyruvate metabolism opens an additional pathway for acetate utilization, with pyruvate carboxylase, pyruvate phosphate dikinase, and phosphoenolpyruvate carboxykinase all identified by proteomics from M21 and G. bemidjiensis-like strains. These enzymes convert pyruvate to oxaloacetate, which can enter the TCA cycle (Fig. 6) (33).
DISCUSSION
The proteomic analysis of three microbial planktonic samples here allows the identification of a range of important pathways utilized by Geobacter spp. in the subsurface during biostimulation and suggests possible ways that the community may evolve during the process. Intriguing shifts in the abundance of certain protein groups are also suggested by the data. Results indicate dominance of Geobacter strains from “subsurface clade I” (14) during the first 20 days of acetate amendment, the presence of multiple coexisting active strain variants, and high abundance of proteins involved in the efficient utilization of acetate (13). Acetyl-CoA hydrolase enables Geobacter to utilize acetate, and we infer that this is the primary route by which the subsurface community creates acetyl-CoA for energy (TCA cycle) and biomass production. In addition, acetate kinase and phosphotransacetylase offer another pathway for acetyl-CoA synthesis. All TCA cycle proteins were identified by proteomics, including succinyl-CoA synthetase from M21 and G. bemidjiensis-like strains. Given the absence of succinyl-CoA synthetase activity in G. sulfurreducens, the acetate kinase-phosphotransacetylase pathway was recently described as the mechanism by which this organism produces acetyl-CoA for biomass production (29). It has also been suggested that in G. metallireducens, acetyl-CoA accumulation forces the transformation of succinate to succinyl-CoA, limiting the rate of carbon metabolism through the TCA cycle (31). If correct, these observations may explain the slow growth of G. metallireducens- and possibly G. sulfurreducens-like strains in these acetate-amended communities and the dominance of M21 and G. bemidjiensis-like strains.
Another important observation is the high abundance of pyruvate ferredoxin oxidoreductase, which may enable Geobacter to “fix” some additional carbon by combining acetyl-CoA and carbon dioxide to form pyruvate (11). Modeling calculations suggest that this pathway allows G. sulfurreducens to synthesize amino acids more efficiently than Escherichia coli (21). While this enzyme also catalyzes the oxidative decarboxylation of pyruvate to acetate and CO2, given the nonlimiting carbon concentrations (H. Elifantz, L. A. N′Guessan, P. J. Mouser, K. H. Williams, M. J. Wilkins, J. E. Ward, P. E. Long, and D. R. Lovley, presented at the 108th General Meeting of the American Society for Microbiology, Boston, MA, 2008), the potential high activity of the pyruvate synthase pathway may be an important response of M21 and G. bemidjiensis-like strains to high acetyl-CoA availability during acetate amendment.
It is important to note that our perspective on changes in subsurface microbial activity is limited to planktonic cells. Geobacter isolates require direct access to ferric iron-bearing minerals (23), including ferric iron-bearing clays and oxyhydroxide minerals that are readily dispersed during drilling activities and well installation. As a result, Fe(III)-bearing phases of small particle size and hence large specific surface area are concentrated in the vicinity of well bores in advance of acetate amendment. The fraction of bioavailable minerals that are readily accessible to planktonic cells declines over time due to enzymatic reductive dissolution and settling. Thus, it is not surprising that the acetate-stimulated Geobacter bloom is associated with a higher initial investment in proteins required for active growth (ribosomal proteins) (35) and nutrient acquisition. While slowing cell growth [suggested by decreasing abundances of ribosomal proteins and ATP synthase subunits in D07(2)] might be expected later in biostimulation as concentrations of electron acceptors and nutrients drop, the corresponding increases in TCA cycle proteins may indicate a shift in energy investment. Many of these abundance increases were associated with increases in unique spectra matching G. lovleyi, suggesting a potential shift in strain composition toward Geobacter isolates that are still closely related to G. bemidjiensis and strain M21, but with some similarity to G. lovleyi. Understanding exactly how these changes in metabolic function and strain composition are linked to geochemical parameters will require the analysis of additional biomass samples to conclusively demonstrate these trends.
The current study is the first to use isolate genome sequences to enable extensive mass spectrometry-based proteomic analysis of natural microbial communities. There is no question that more peptides and proteins would have been identified had metagenomic data for these samples been available. Despite the limitations of the isolate sequence-based approach, over 13,000 peptides and 2,500 proteins were identified in each sample. Identification of each peptide confirmed the presence of that sequence in the sample, and each protein contributed to a rendering of the physiological state of Geobacter strains in the communities. As currently deployed, proteogenomics is a complicated approach that depends on sophisticated technological methods. However, by analogy with medical diagnostics, it is reasonable to anticipate that rapid, high-throughput methods can be developed to detect signals characteristic of the metabolic state of microbial communities in the subsurface and elsewhere.
Supplementary Material
Acknowledgments
We thank the city of Rifle, CO, the Colorado Department of Public Health and Environment, and the U.S. Environmental Protection Agency, Region 8, for their cooperation in this study. M. Lefsrud is thanked for his assistance with proteomic measurements at ORNL. ORNL is managed by the University of Tennessee—Battelle, LLC, for the DOE under contract DOE-AC05-00OR22725. Pacific Northwest National Laboratory is managed under contract DE-AC05-76RL01830 with Battelle Memorial Institute. Portions of this work were performed at the Environmental Molecular Sciences Laboratory, a DOE national scientific user facility located at the Pacific Northwest National Laboratory. R. Mahadevan (University of Toronto) is thanked for many helpful comments on the manuscript. The suggestions made by five anonymous reviewers are also appreciated.
This research was sponsored by the Environmental and Remediation Sciences Program, Biological and Environmental Research, Office of Science, U.S. DOE.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adkins, J. N., H. M. Mottaz, A. D. Norbeck, J. K. Gustin, J. Rue, T. R. W. Clauss, S. O. Purvine, K. D. Rodland, F. Heffron, and R. D. Smith. 2006. Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol. Cell. Proteomics 5:1450-1461. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond, D. R., T. Mester, C. L. Nesbo, A. V. Izquierdo-Lopez, F. L. Collart, and D. R. Lovley. 2005. Characterization of citrate synthase from Geobacter sulfurreducens and evidence for a family of citrate synthases similar to those of eukaryotes throughout the Geobacteraceae. Appl. Environ. Microbiol. 71:3858-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callister, S. J., M. A. Dominguez, C. D. Nicora, X. Zeng, C. L. Tavano, S. Kaplan, T. J. Donohue, R. D. Smith, and M. S. Lipton. 2006. Application of the accurate mass and time tag approach to the proteome analysis of sub-cellular fractions obtained from Rhodobacter sphaeroides 2.4.1. Aerobic and photosynthetic cell cultures. J. Proteome Res. 5:1940-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callister, S. J., L. A. McCue, J. E. Turse, M. E. Monroe, K. J. Auberry, R. D. Smith, J. N. Adkins, and M. S. Lipton. 2008. Comparative bacterial proteomics: analysis of the core genome concept. PLoS ONE 3:e1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chain, P. S. G., V. J. Denef, K. T. Konstantinidis, L. M. Vergez, L. Agullo, V. L. Reyes, L. Hauser, M. Cordova, L. Gomez, M. Gonzalez, M. Land, V. Lao, F. Larimer, J. J. Lipuma, E. Mahenthiralingam, S. A. Malfatti, C. J. Marx, J. J. Parnell, A. Ramette, P. Richardson, M. Seeger, D. Smith, T. Spilker, W. J. Sul, T. V. Tsoi, L. E. Ulrich, I. B. Zhulin, and J. M. Tiedje. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 103:15280-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denef, V. J., M. B. Shah, N. C. VerBerkmoes, R. L. Hettich, and J. F. Banfield. 2007. Implications of strain- and species-level sequence divergence for community and isolate shotgun proteomic analysis. J. Proteome Res. 6:3152-3161. [DOI] [PubMed] [Google Scholar]
- 8.Ding, Y. H. R., K. K. Hixson, C. S. Giometti, A. Stanley, A. Esteve-Nunez, T. Khare, S. L. Tollaksen, W. H. Zhu, J. N. Adkins, M. S. Lipton, R. D. Smith, T. Mester, and D. R. Lovley. 2006. The proteome of dissimilatory metal-reducing microorganism Geobacter sulfurreducens under various growth conditions. Biochim. Biophys. Acta 1764:1198-1206. [DOI] [PubMed] [Google Scholar]
- 9.Elias, J. E., and S. P. Gygi. 2007. Target-decoy search strategy for increased confidence in large-scale protein identification by mass spectrometry. Nat. Methods 4:207-214. [DOI] [PubMed] [Google Scholar]
- 10.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 11.Furdui, C., and S. W. Ragsdale. 2000. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J. Biol. Chem. 275:28494-28499. [DOI] [PubMed] [Google Scholar]
- 12.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. Comparison of 16S rRNA, nifD, recA, gyrB, rpoB and fusA genes within the family Geobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:1591-1599. [DOI] [PubMed] [Google Scholar]
- 13.Holmes, D. E., K. P. Nevin, R. A. O'Neil, J. E. Ward, L. A. Adams, T. L. Woodard, H. A. Vrionis, and D. R. Lovley. 2005. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl. Environ. Microbiol. 71:6870-6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes, D. E., R. A. O'Neil, H. A. Vrionis, L. A. N′Guessan, I. Ortiz-Bernad, M. J. Larrahondo, L. A. Adams, J. A. Ward, J. S. Nicoll, K. P. Nevin, M. A. Chavan, J. P. Johnson, P. E. Long, and D. R. Lovley. 2007. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J. 1:663-677. [DOI] [PubMed] [Google Scholar]
- 15.Kiebel, G. R., K. J. Auberry, N. Jaitly, D. A. Clark, M. E. Monroe, E. S. Peterson, N. Tolic, G. A. Anderson, and R. D. Smith. 2006. PRISM: a data management system for high-throughput proteomics. Proteomics 6:1783-1790. [DOI] [PubMed] [Google Scholar]
- 16.Kolker, E., A. F. Picone, M. Y. Galperin, M. F. Romine, R. Higdon, K. S. Makarova, N. Kolker, G. A. Anderson, X. Y. Qiu, K. J. Auberry, G. Babnigg, A. S. Beliaev, P. Edlefsen, D. A. Elias, Y. A. Gorby, T. Holzman, J. A. Klappenbach, K. T. Konstantinidis, M. L. Land, M. S. Lipton, L. A. McCue, M. Monroe, L. Pasa-Tolic, G. Pinchuk, S. Purvine, M. H. Serres, S. Tsapin, B. A. Zakrajsek, J. H. Zhou, F. W. Larimer, C. E. Lawrence, M. Riley, F. R. Collart, J. R. Yates, R. D. Smith, C. S. Giometti, K. H. Nealson, J. K. Fredrickson, and J. M. Tiedje. 2005. Global profiling of Shewanella oneidensis MR-1: expression of hypothetical genes and improved functional annotations. Proc. Natl. Acad. Sci. USA 102:2099-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipton, M. S., L. Pasa-Tolic, G. A. Anderson, D. J. Anderson, D. L. Auberry, K. R. Battista, M. J. Daly, J. Fredrickson, K. K. Hixson, H. Kostandarithes, C. Masselon, L. M. Markillie, R. J. Moore, M. F. Romine, Y. F. Shen, E. Stritmatter, N. Tolic, H. R. Udseth, A. Venkateswaran, L. K. Wong, R. Zhao, and R. D. Smith. 2002. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. USA 99:11049-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo, I., V. J. Denef, N. C. VerBerkmoes, M. B. Shah, D. Goltsman, G. DiBartolo, G. W. Tyson, E. E. Allen, R. J. Ram, J. C. Detter, P. Richardson, M. P. Thelen, R. L. Hettich, and J. F. Banfield. 2007. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446:537-541. [DOI] [PubMed] [Google Scholar]
- 19.Lovley, D. R., and E. J. P. Phillips. 1992. Bioremediation of uranium contamination with enzymatic uranium reduction. Environ. Sci. Technol. 26:2228-2234. [Google Scholar]
- 20.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 21.Mahadevan, R., D. R. Bond, J. E. Butler, A. Esteve-Nunez, M. V. Coppi, B. O. Palsson, C. H. Schilling, and D. R. Lovley. 2006. Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl. Environ. Microbiol. 72:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouser, P. J., L. A. N′Guessan, H. Elifantz, D. E. Holmes, K. H. Williams, M. J. Wilkins, P. E. Long, and D. R. Lovley. 2009. Impact of heterogeneous ammonium availability on community structure and the expression of nitrogen fixation and ammonium transporter genes during in situ bioremediation of uranium-contaminated groundwater. Environ. Sci. Technol. 43:4386-4392. [DOI] [PubMed] [Google Scholar]
- 23.Nevin, K. P., and D. R. Lovley. 2000. Lack of production of electron-shuttling compounds or solubilization of Fe(III) during reduction of insoluble Fe(III) oxide by Geobacter metallireducens. Appl. Environ. Microbiol. 66:2248-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng, J., J. E. Elias, C. C. Thoreen, L. J. Licklider, and S. P. Gygi. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43-50. [DOI] [PubMed] [Google Scholar]
- 26.Ram, R. J., N. C. VerBerkmoes, M. P. Thelen, G. W. Tyson, B. J. Baker, R. C. Blake, M. Shah, R. L. Hettich, and J. F. Banfield. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915-1920. [PubMed] [Google Scholar]
- 27.Rasko, D. A., G. S. A. Myers, and J. Ravel. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheibe, T. D., R. Mahadevan, Y. Fang, S. Garg, P. E. Long, and D. R. Lovley. 2009. Coupling a genome-scale metabolic model with a reactive transport model to describe in situ uranium bioremediation. Microb. Biotechnol. 2:274-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segura, D., R. Mahadevan, K. Juarez, and D. R. Lovley. 2008. Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens. PloS Comput. Biol. 4:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang, Y. J. J., R. Chakraborty, H. G. Martin, J. Chu, T. C. Hazen, and J. D. Keasling. 2007. Flux analysis of central metabolic pathways in Geobacter metallireducens during reduction of soluble Fe(III)-nitrilotriacetic acid. Appl. Environ. Microbiol. 73:3859-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, M. R., K. Chourey, J. M. Froelich, B. K. Erickson, N. C. VerBerkmoes, and R. L. Hettich. 2008. Experimental approach for deep proteome measurements from small-scale microbial biomass samples. Anal. Chem. 80:9517-9525. [DOI] [PubMed] [Google Scholar]
- 33.Utter, M. F., and D. B. Keech. 1960. Formation of oxaloacetate from pyruvate and carbon dioxide. J. Biol. Chem. 235:17-18. [PubMed] [Google Scholar]
- 34.VerBerkmoes, N. C., A. L. Russell, M. Shah, A. Godzik, M. Rosenquist, J. Halfvarson, M. G. Lefsrud, J. Apajalahti, C. Tysk, R. L. Hettich, and J. K. Jansson. 2009. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3:179-189. [DOI] [PubMed] [Google Scholar]
- 35.Xiao, L. J., and A. Grove. 2009. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr. Genomics 10:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zybailov, B., A. L. Mosley, M. E. Sardiu, M. K. Coleman, L. Florens, and M. P. Washburn. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5:2339-2347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.