Abstract
Pseudomonas sp. strain M18, an effective biological control agent isolated from the melon rhizosphere, has a genetic background similar to that of the opportunistic human pathogen Pseudomonas aeruginosa PAO1. However, the predominant phenazine produced by strain M18 is phenazine-1-carboxylic acid (PCA) rather than pyocyanin (PYO); the quantitative ratio of PCA to PYO is 105 to 1 at 28°C in strain M18, while the ratio is 1 to 2 at 37°C in strain PAO1. We first provided evidence that the differential production of the two phenazines in strains M18 and PAO1 is related to the temperature-dependent and strain-specific expression patterns of phzM, a gene involved in the conversion of PCA to PYO. Transcriptional levels of phzM were measured by quantitative real-time PCR, and the activities of both transcriptional and translational phzM′-′lacZ fusions were determined in strains M18 and PAO1, respectively. Using lasI::Gm and ptsP::Gm inactivation M18 mutants, we further show that expression of the phzM gene is positively regulated by the quorum-sensing protein LasI and negatively regulated by the phosphoenolpyruvate phosphotransferase protein PtsP. Surprisingly, the lasI and ptsP regulatory genes were also expressed in a temperature-dependent and strain-specific manner. The differential production of the phenazines PCA and PYO by strains M18 and PAO1 may be a consequence of selective pressure imposed on P. aeruginosa PAO1 and its relative M18 in the two different niches over a long evolutionary process.
Phenazines, known for over 150 years, are a group of the nitrogen-containing secondary metabolites synthesized mainly by Pseudomonas spp. and a few other bacterial strains. Advances within the past 2 decades have provided significant new insights into the genetics, biochemistry, and regulation of phenazine synthesis, as well as the mode of action and functional roles of these compounds in various environments (33, 44). Despite the fact that the phenazine biosynthesis locus is highly conserved among various Pseudomonas spp., individual strains differ in the range of phenazine compounds they produce and the relative amounts of the possible phenazines are strongly influenced by growth conditions (7, 30, 34, 61). The crucial roles of phenazine-1-carboxylic acid (PCA) in plant root disease suppression has been well documented in studies with several biological control strains, such as Pseudomonas fluorescens 2-79, P. chlororaphis 30-84, and P. chlororaphis PCL1391, where PCA can be converted into phenazine-1-carboxamide (PCN) (6, 33, 35, 37, 50). Nevertheless, PCA is considered a predominant phenazine (44) in these strains, and its secretion mainly contributes to the biocontrol activity against various fungal phytopathogens such as Gaeumannomyces graminis var. tritici (41, 49, 50). Chromosomal insertion of genes involved in the PCA biosynthetic pathway enhances the efficacy of damping-off disease control by P. fluorescens. The phenazine-deficient strains P. fluorescens 2-79 and P. chlororaphis 30-84 have reduced survival rates and a diminished ability to compete with the resident microflora (5, 51).
Pyocyanin (PYO), a derivative of PCA, is the other major phenazine compound produced by P. aeruginosa and functions as an important virulence factor, although PCA and PCN could also be produced by this species (26, 48). PYO imparts the characteristic blue or blue-green color to P. aeruginosa cultures and the fluids in sputum from cystic lung fibrosis patients (11). Recently, PYO was thought to be not required for fungal killing in P. aeruginosa (16). It has been postulated that the conversion of PCA to PYO is a two-step process that involves N-methylation by PhzM and hydroxylation by PhzS in pseudomonads. P. aeruginosa with phzM or phzS inactivated by gene-targeted insertion developed a PYO-deficient yellow-reddish phenotype (34).
The strain used in this study, Pseudomonas sp. strain M18, is an effective biocontrol agent against various soil-borne phytopathogens and was isolated from the sweet melon rhizosphere (20). Pseudomonas sp. strain M18 has a genetic background similar to that of P. aeruginosa PAO1; the nucleotide sequences of the 16S rRNA gene and several global regulatory genes such as gacA, rsmA, rpoS, rpoD, qscR, vqsR, lasI, and rhlI from strain M18 show high identities with those in P. aeruginosa PAO1 (4, 14, 15, 22, 54, 59, 62). In addition, there exist two sets of phz biosynthesis gene clusters (phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2), each encoding enzymes required to convert chorismic acid to PCA and two phenazine modification genes, phzM and phzS, flanking the phzA1B1C1D1E1F1G1 cluster in the strain M18 genome, which are also similar to those in P. aeruginosa PAO1 (unpublished data). Pseudomonas sp. strain M18 possesses las and rhl, two sets of acyl-homoserine lactone quorum-sensing (QS) systems that are the same as those in P. aeruginosa PAO1 as well (4, 59). Interestingly, strain M18 had developed several unusual features which were similar to those of biocontrol strains inhabiting soil and quite different from those of P. aeruginosa PAO1. First, the predominant phenazine produced in Pseudomonas sp. strain M18 is PCA rather than PYO, as shown in this report. Second, Pseudomonas sp. strain M18 is the first strain that was reported to produce two antibiotics, PCA and pyoluteorin (Plt). The Plt synthesis cluster of approximately 30 kb, including its flanking regulatory region, is highly homologous to that of biocontrol strain P. fluorescens pf-5. Third, several specific features of the regulatory mechanism of the two antibiotics of PCA and Plt production were described, such as that PCA is negatively regulated and Plt is positively regulated by a global regulator called GacA (14); however, the reverse relationship occurs in Pseudomonas sp. strain M18 through another global regulator, RsmA (62). In a more recent study, we demonstrated the negative effect of the regulator QscR on PCA production, but not on Plt production, in Pseudomonas sp. strain M18 (54). The differential regulation of PCA and Plt production by potential acyl-homoserine lactone QS signaling molecules secreted by this strain has also been studied (4, 59). Besides, the interrelationship between the las and rhl QS systems in Pseudomonas sp. strain M18 is also different from that in strain PAO1 (unpublished data).
As reported previously, PhzM is believed to be involved in the conversion of PCA into PYO in P. aeruginosa PAO1 (34, 40). PhzM is a predicted 36-kDa protein similar to enzymes involved in the methylation of polyketide antibiotics by Streptomyces spp. (60). Though the 1.8-Å crystal structure of PhzM from P. aeruginosa was determined by single-wavelength anomalous dispersion (40), little is known about the features of phzM gene expression and regulation in pseudomonads. All of these interesting phenomena found in strain M18, especially the different accumulation of PCA and PYO produced by strains M18 and PAO1, encouraged us to further investigate phzM gene expression and regulation and to clarify the differential production of the phenazines PCA and PYO. In this report, several characteristics of phzM gene expression with its regulation in strains M18 and PAO1 are newly described.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and chemicals.
Pseudomonas sp. strain M18 is a fluorescent pseudomonad strain isolated from the sweet-melon rhizosphere; it is an unusual strain that shares some distinct features with both P. aeruginosa and the other biocontrol pseudomonads (14, 20, 23). The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 deoR λ (φ80dlacZΔM15) | 46 |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr | 46 |
| Pseudomonas strains | ||
| P. aeruginosa PAO1 | Wild type | Dieter Haas |
| Pseudomonas sp. strain M18 | Wild type, PCA, Plt producer, Spr | Lab collection |
| M18pM | PYO defective; M18 phzM::Km Spr Kmr | This study |
| PAO1pM | PYO defective; PAO1 phzM::Km Spr Kmr | This study |
| M18PP | PYO overproducer; M18 ptsP::Gmr Spr Gmr | This study |
| PAO1PP | PYO overproducer; PAO1 ptsP::Gmr Spr Gmr | This study |
| PAOLSG | PAO1 lasI::Gmr Spr Gmr | This study |
| M18LSG | M18 lasI::Gmr Spr Gmr | 4 |
| Plasmids | ||
| pBluescript SK | ColE, cloning and sequencing vector, Ampr | Lab collection |
| pUCGm | Source of Gmr cassette, Gmr | Dieter Haas |
| pET28a | Expression vector, Kmr | Novagen |
| pUCKm | Source of Kmr cassette, Kmr | This study |
| pEX18Tc | Gene replacement vector with multiple cloning site from pUC18, oriT+sacB+ Tcr | 19 |
| pEXTphzM | pEX18Tc with EcoRI-HindIII insert of 1.06 kb, including phzM gene and partial flanking sequence, Tcr | This study |
| pEXTphzMKm | phzM:: Km in pEXTphzM, Tcr Kmr | This study |
| pEXTptsP | pEX18Tc with EcoRI-HindIII insert of 2.4 kb, including ptsP gene and partial flanking sequence, Tcr | This study |
| pEXTptsPGm | ptsP::Gm in pEXTptsP, Tcr Gmr | This study |
| pME6000 | Broad-host-range cloning vector, Tcr | 32 |
| pME6000ptsP | pME6000 with 2.4-kb EcoRI-HindIII insert, including ptsP gene with partial flanking sequence, Tcr | This study |
| pME6032 | pVS1-p15A E. coli-Pseudomonas shuttle vector, lacI′-ptac expression vector, Tcr | 18 |
| pME6032lasI | pME6032 with 1.2-kb BglI-EcoRI insert, including lasI gene with partial flanking sequence, Tcr | 4 |
| pME6015 | pVS1-p15A E. coli-Pseudomonas shuttle vector for translational lacZ fusions and promoter probing, Tcr | 18 |
| pME6522 | pVS1-p15A E. coli-Pseudomonas shuttle vector for transcriptional lacZ fusions and promoter probing, Tcr | 2 |
| pPML | phzM′-′lacZ translational fusion, 704-bp EcoRI-PstI PCR-amplified fragment containing 407 nucleotides upstream and 297 nucleotides downstream of transcriptional start site of phzM cloned into pME6015, Tcr | This study |
| pPMC | phzM′-′lacZ transcriptional fusion, 407-bp EcoRI-PstI PCR-amplified fragment from Pseudomonas sp. strain M18 cloned into pME6522, Tcr | This study |
| pPMCO | phzM′-′lacZ transcriptional fusion, 408-bp EcoRI-PstI PCR-amplified fragment from P. aeruginosa PAO1 cloned into pME6522, Tcr | This study |
| pPMCD | phzM′-′lacZ transcriptional fusion, 333-bp EcoRI-PstI PCR-amplified fragment lacking the lux box from Pseudomonas sp. strain M18 cloned into pME6522, Tcr | This study |
| pLIL | lasI′-′lacZ translational fusion plasmid, Tcr | 4 |
| pPPL | ptsP′-′lacZ translational fusion, 620-bp EcoRI-PstI PCR-amplified fragment containing 24-bp DNA sequence downstream of translational start site of ptsP cloned into pME6015, Tcr | This study |
Escherichia coli was routinely grown at 37°C in Luria-Bertani medium (46). The fungus Fusarium oxysporum var. cucumerinum was grown at 28°C on potato dextrose agar (PDA) plates (49). Pseudomonas sp. strain M18 and P. aeruginosa PAO1 were incubated in King's medium B (KMB) for growth (24). For secondary metabolite PCA and PYO production, pigment-producing medium (PPM) was used (28). Antibiotics were added at the following concentrations: gentamicin (Gm) at 40 μg ml−1, kanamycin (Km) at 50 μg ml−1, spectinomycin (Sp) at 100 μg ml−1, and tetracycline (Tc) at 125 μg ml−1 for pseudomonads and Km at 50 μg ml−1, Gm at 15 μg ml−1, Tc at 15 μg ml−1, and ampicillin (Ap) at 100 μg ml−1 for E. coli. Restriction endonucleases, DNA-modifying enzymes, Taq and Pfu DNA polymerases, DNA molecular mass markers, and other associated products were purchased and utilized as recommended by the manufacturers (TaKaRa, Dalian, China, and MBI Fermentas, Shenzhen, China). Small-scale preparations of plasmid DNA were performed with a MiniBEST plasmid purification kit, version 2.0 (Biodev-tech, Beijing, China). Genomic DNA was extracted and purified from Pseudomonas sp. strain M18 and P. aeruginosa PAO1 with an EZ-10 spin column genomic DNA isolation kit (Bio Basiv Inc., Shanghai, China). Restriction fragments were purified from agarose gels with a DNA gel extraction kit (Axygen Scientific, Inc., Hangzhou, China).
Sequencing of the phzM gene from Pseudomonas sp. strain M18 and mapping of its transcriptional start sites in strains M18 and PAO1.
Based on the P. aeruginosa PAO1 sequence, primers phzM1F and phzM1R were designed as listed in Table 2 (underlined letters are the EcoRI and HindIII restriction sites). The phzM gene coding region with its regulatory element from strain M18 was PCR amplified with Pfu DNA polymerase as recommended by the manufacturer. The PCR-amplified fragment was recovered and directly sequenced by Invitrogen (Shanghai, China).
TABLE 2.
Primers used in this study
| Primera | Nucleotide sequence | Restriction siteb |
|---|---|---|
| phzMsp1 | 5′-TGGAATGCCAGGTTGCTC-3′ | |
| phzMsp2 | 5′-CGCCGTAGAACAGCACCAT-3′ | |
| kan-F | 5′-CCGGGAGCTCGAATTCAGCAGAGCGAGGTATGTAGG-3′ | SacI |
| kan-R | 5′-CCGGGAGCTCGAATTCCAGGTGGCACTTTTCGGGGA-3′ | SacI |
| phzM1F | 5′-GGCCGAATTCTCTCCAGGTATGCCGGAGAA-3′ | EcoRI |
| phzM1R | 5′-GGCCAAGCTTGAAAGTTCCGATTCAGGCCC-3′ | HindIII |
| phzM2F | 5′-GGCCGAATTCGGTACTTCTCGGGTTACGGA-3′ | EcoRI |
| ptsP1 | 5′-CCGCGCGAATTCGACAGGTGGTGACCTTCAAG-3′ | EcoRI |
| ptsP2 | 5′-CCGCGC AAGCTT GAAATCAGGGCTGGACGGTA-3′ | HindIII |
| PM9F | 5′-GGCCGAATTCTCTCCAGGTATGCCGGAGAA-3′ | EcoRI |
| PM9R | 5′-CCGGCTGCAGCGCAGCAGCAAGATTCGAAT-3′ | PstI |
| DLasbF | 5′-GGCCGAATTC GCCAGACAGGGTATGCGGGA-3′ | EcoRI |
| TMPR | 5′-CCGGCTGCAG AGCATCAGCTTAGCAATCCC-3′ | PstI |
| TMPRO | 5′-CCGGCTGCAG AAGCATCAGCTTAGCAATCC-3′ | PstI |
| ptsPPF | 5′-GCGCGAATTCTGTATCCTCCCCGCACGAGT-3′ | EcoRI |
| ptsP620fR | 5′-CGGCCTGCAGGATCTTGCGCAGCGTGTTGA-3′ | PstI |
| RTphzM1 | 5′-GGATCGACAGCGACGAGACG-3′ | |
| RTphzM2 | 5′-CTCGCCGTAGAACAGCACCAT-3′ | |
| PSRMRPOD1 | 5′-GAGCGGGAGGAGCGTTTAC-3′ | |
| PSRMRPOD2 | 5′-CGGGCAAAAAATAAGCAGAGG-3′ | |
| RTlasI1 | 5′-AACTGCTGGGCGAGATGC-3′ | |
| RTlasI2 | 5′-CGCCAGCAACCGAAAACC-3′ | |
| RTptsP1 | 5′-AAGTGAACTCCGCCAAGG-3′ | |
| RTptsP2 | 5′-TGGAACGCTTGTTGAGGC-3′ |
All primers were obtained from Invitrogen (Shanghai, China).
The restriction sites used in the cloning procedure are underlined.
RNA was isolated at the early stationary growth phase (optical density at 600 nm [OD600], 1.0) from strain M18 and PAO1 cultures with RNAwiz and a DNA-Free kit (Invitrogen). Rapid amplification of the 5′ cDNA end was performed with the 5′ RACE (rapid amplification of cDNA ends) system (Invitrogen) to identify the transcriptional start site of phzM according to the manufacturer's instructions (29). Briefly, based on the phzM gene sequence, specific primer phzMsp1 was designed and reverse transcriptase was used for first-strand cDNA synthesis. A homopolymeric tail was then added to the 3′ end of the cDNA with TdT and dCTP/dATP. PCR amplification was accomplished with a nested gene-specific primer, phzMsp2, a novel deoxyinosine-containing anchor primer, and the poly(C) tail cDNA as the template. PCR products were directly sequenced, and transcriptional start sites were identified.
Construction of mutants with inactivated genes and fusion plasmids. (i) Construction of mutants.
All of the primers used for plasmid construction are listed in Table 2 with enzyme site sequences underlined. A 1,250-bp PCR fragment carrying the open reading frame of the Km resistance (Kmr)-encoding gene with its flanking regions was amplified with primers kan-F and kan-R from plasmid pET28a. The PCR product digested with SacI was ligated into plasmid pUCGm to generate a pUCKm vector. To inactivate the phzM gene, a 1,062-bp PCR fragment carrying the phzM open reading frame with flanking regions was amplified by the primer pair phzM2F-phzM1R from Pseudomonas sp. strain M18 chromosomal DNA, digested, and ligated into the vector pEX18Tc at the EcoRI and HindIII sites. The resulting plasmid, pEX18Tc-phzM, was digested with KpnI and SmaI to delete a 215-bp fragment in the phzM gene, blunted, and then ligated with a SmaI-digested Kmr cassette amplified from plasmid pUCKm to generate pEX18Tc-phzM-kan. Plasmid pEX18Tc-phzM-kan was transferred into Pseudomonas sp. strain M18 or P. aeruginosa PAO1 to generate the isogenic phzM inactivation mutant M18pM or PAO1pM with by allelic exchange and the Flp-FRT recombination protocol as described previously (19). The same method was used to construct chromosomal ptsP::Gm M18PP with primers ptsP1 and ptsP2. The successful inactivation of phzM::Km and ptsP::Gm in strains M18 and PAO1 was further verified by PCR. In addition, the lasI::Gm mutant M18LSG derived from wild-type strain M18 had been constructed previously (4). Plasmids pME6032lasI and pME6000ptsP (a 2.4-kb PCR product of the ptsP gene generated by primers ptsP1 and ptsP2 with EcoRI-HindIII and inserted into pME6000) were introduced into the lasI::Gm inactivation and ptsP::Gm mutants of M18, respectively, forming complemented mutants M18LSG and M18PP in trans.
(ii) Construction of translational and transcriptional phzM′-′lacZ fusion and ptsP′-′lacZ fusion plasmids.
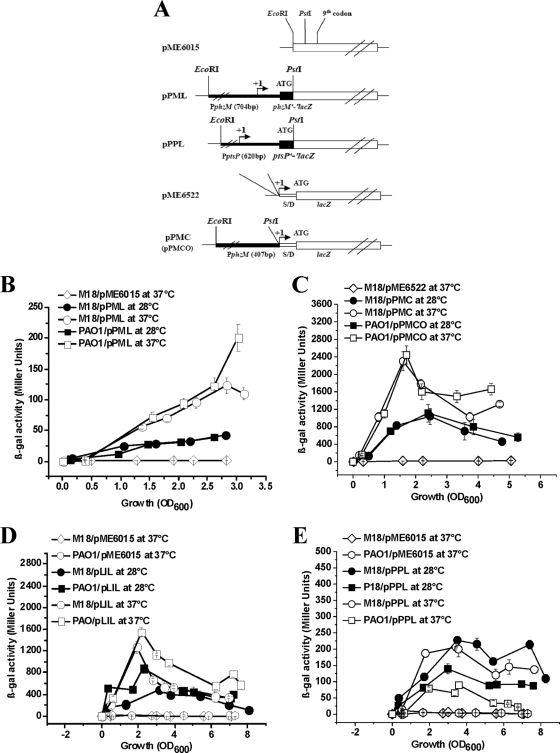
A 704-bp and a 620-bp PCR product, containing the promoter region with the first nine or eight codons of the phzM gene and the ptsP gene, respectively, from the M18 genome were generated with primer pairs PM9F-PM9R and ptsPPF-ptsP620fR and inserted into sites upstream of lacZ in pME6015, forming translational fusion plasmids pPML and pPPL, respectively. Meanwhile, to construct phzM′-′lacZ transcriptional fusion plasmids pPMC and pPMCO, the respective 407-bp or 408-bp PCR product containing the upstream region from the transcriptional start site was generated with strains M18 and PAO1 and primer pairs PM9F-TMPR and PM9F-TMPRO, respectively, and then inserted upstream of lacZ into pME6522 (see Fig. 4). The lasI′-′lacZ translational fusion plasmid pLIL was constructed in our previously published study (4).
FIG. 4.
Temperature-dependent and strain-specific expression of the phzM gene and its regulatory genes lasI and ptsP determined by assaying β-Gal activity in Pseudomonas sp. strain M18 and P. aeruginosa PAO1. (A) Physical maps of phzM′-′lacZ translational fusion plasmid pPML and transcriptional fusion plasmid pPMC in strain M18 and pPMCO and ptsP′-′lacZ translational fusion plasmid pPPL in strain PAO1. lasI′-′lacZ translational fusion plasmid pLIL was constructed by the same method as in our previous work (4). SD, putative Shine-Dalgarno sequence. phzM and ptsP sequences are shown by heavy black lines, and lacZ sequences are shown by white boxes. PphzM and PptsP are the phzM and ptsP gene promoters. (B) β-Gal activities of M18/pPML (circles) and PAO1/pPML (squares) at 28°C (solid symbols) and 37°C (empty symbols) and strain M18 (diamonds) containing control plasmid pME6015. (C) β-Gal activities of M18/pPMC (circles) and PAO1/pPMCO (squares) at 28°C (solid symbols) and 37°C (empty symbols) and strain M18 (diamonds) containing control plasmid pME6522. (D) β-Gal activities of M18/pLIL (circles) and PAO1/pLIL (squares) at 28°C (solid symbols) and 37°C (empty symbols) and strain M18 (diamonds) and PAO1 (hexagons) containing control plasmid pME6015. (E) β-Gal activities of M18/pPPL (circles) and PAO1/pPPL (squares) at 28°C (solid symbols) and 37°C (empty symbols) and M18 (diamonds) and PAO1 (hexagons) containing control plasmid pME6015. Growth was in PPM broth. The values are means ± standard deviations of triplicate cultures.
Quantitative real-time PCR (qRT-PCR).
Total RNA was extracted from the various target strains, and the amounts of target cDNAs obtained by reverse transcription were quantified with the MiniOpticon RT-PCR system (Bio-Rad) with SYBR green I stain. Primer pairs RTphzM1-RTphzM2, RTlasI1-RTlasI2, RTptsP1-RTptsP2, and PSRMRPOD1-PSRMRPOD2 were designed based on the phzM (193 bp), lasI (169 bp), ptsP (156 bp), and rpoD (173 bp) sequences, respectively, in Pseudomonas sp. strain M18 and P. aeruginosa PAO1. The housekeeping gene rpoD was used as a reference. Twenty-five microliters of each reaction mixture was prepared according to standard PCR protocols. No-template controls containing distilled water and no-amplification controls containing total RNA instead of the cDNA as the template were included in each RT-PCR run to check for DNA contamination. The target cDNA (phzM, lasI, or ptsP) and reference cDNA (rpoD) were amplified in separate wells. PCRs were run in an MJ Mini personal thermal cycler (Bio-Rad) with the following program: one step of 10 min at 95°C and 40 cycles of 95°C for 15 s, 60°C for 10 s, and 72°C for 20 s. The evolution of fluorescence intensity in each reaction mixture was recorded continuously with a MiniOpticon RT-PCR detector (Bio-Rad). The PCR products amplified from cDNA were further confirmed by sequencing. PCR analyses of each target strain in different growth phases were repeated three times. The data analysis method used was described previously (59). The amount of phzM, lasI, or ptsP cDNA, normalized to the level of the reference rpoD and calibrated relative to strain M18, was expressed as a 2−ΔΔCT value, which represents the change in target gene expression in a given strain relative to that in strain M18. Statistical significance was computed by an unpaired Student t test; a P value of <0.05 was considered statistically significant.
Assays of β-Gal activity and PCA and PYO production.
phzM′-′lacZ, lasI′-′lacZ, and ptsP′-′lacZ translational fusion plasmids pPML, pLIL, and pPPL, respectively, and phzM′-′lacZ transcriptional fusion plasmid pPMC were transformed into the various target strains, while only plasmid pPMCO was introduced into strain PAO1. Subsequently, these transformed strains harboring the various fusion plasmids were cultivated at 28°C or 37°C with shaking at 200 rpm in 150-ml conical flasks containing 50 ml PPM broth supplemented with 0.05% Triton X-100. Samples were collected after a certain time of growth, and β-galactosidase (β-Gal) specific activity was assayed according to the method of Miller (38).
PPM was used for PCA and PYO production. PCA was extracted at pH 4 and quantified in chloroform as described by Ge et al. (14). Quantification of PYO production was done at neutral pH in chloroform as described by Essar et al. (12, 45, 58).
Fungal inhibition assay.
Fungal inhibition by strains M18 and PAO1 was determined by measuring their ability to inhibit mycelium plug growth of F. oxysporum var. cucumerinum on a PDA agar plate as follows. Briefly, a 5-mm plug from the leading edge of a culture of F. oxysporum var. cucumerinum grown for 5 days at 28°C on a PDA plate was placed in the center of a fresh PDA plate. Bacterial cultures grown overnight on a KMB plate were placed 25 mm from the PDA plate center. The coculture plates were incubated at 28°C and scored after 4 or 5 days by measuring the distance in millimeters between the edges of the bacterial colony and the fungal mycelium plug as described by Thomashow and Weller (16, 49). Each experiment was repeated twice with three replicates.
Sequences analysis.
The primers used in this study are listed in Table 2. PCR products were recovered and sequenced directly by a commercial service (Invitrogen).
Nucleotide sequence accession numbers.
The nucleotide sequences of the phzM and ptsP genes from the Pseudomonas sp. strain M18 chromosome were deposited in GenBank with accession no. FJ535573 and FJ790321, respectively.
RESULTS
The PCA and PYO pigments are produced by strains M18 and PAO1 in a temperature-dependent and strain-specific manner.
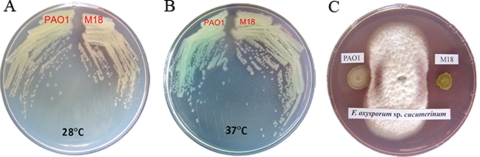
In our previously published works, we demonstrated that strain M18 isolated from rhizosphere niches showed a genetic background similar to that of strain PAO1 and was a strong biocontrol strain (15, 54, 59). When Pseudomonas sp. strain M18 and P. aeruginosa PAO1 were inoculated onto KMB plates as described in Materials and Methods and incubated at 28°C or 37°C, respectively, for 1 to 2 days, the two strains produced distinct pigments, as shown in Fig. 1A and B. Strain M18 produced a yellow-reddish pigment, while strain PAO1 produced a yellow pigment with a little blue color at 28°C. However, when the plates were incubated at 37°C, strain M18 still produced the yellow-reddish pigment, while strain PAO1 produced a blue-green pigment. To assay fungal inhibition by the two strains, the fungus F. oxysporum var. cucumerinum was cocultured with strains M18 and PAO1 on a fungal growth PDA plate at 28°C. Mycelium growth of the fungus F. oxysporum var. cucumerinum was totally inhibited by strain M18, but strain PAO1 showed almost no fungal killing activity (Fig. 1C). These results indicated that strains M18 and PAO1 showed different biological control abilities. It is well known that phenazine compounds are the major contributors to the colony color of various pseudomonads (17, 33, 44). PYO produces a characteristic blue-green color, while PCA generates a yellow-reddish color, where the final colony color appearance on the plates suggests that different phenazines are produced by the two strains in a temperature-dependent and strain-specific manner.
FIG. 1.
Pseudomonad growth and plate cocultures of F. oxysporum var. cucumerinum with Pseudomonas sp. strain M18 and P. aeruginosa PAO1. Shown are (A) pigments produced by strains M18 and PAO1 grown for 16 to 24 h at 28°C and (B) pigments produced by strains M18 and PAO1 grown for 16 to 24 h at 37°C on KMB plates. (C) A 5-mm plug of the fungus F. oxysporum var. cucumerinum was placed in the center of a PDA plate, and cultures of Pseudomonas strains M18 and PAO1 were inoculated 25 mm from the center on each side of the plate.
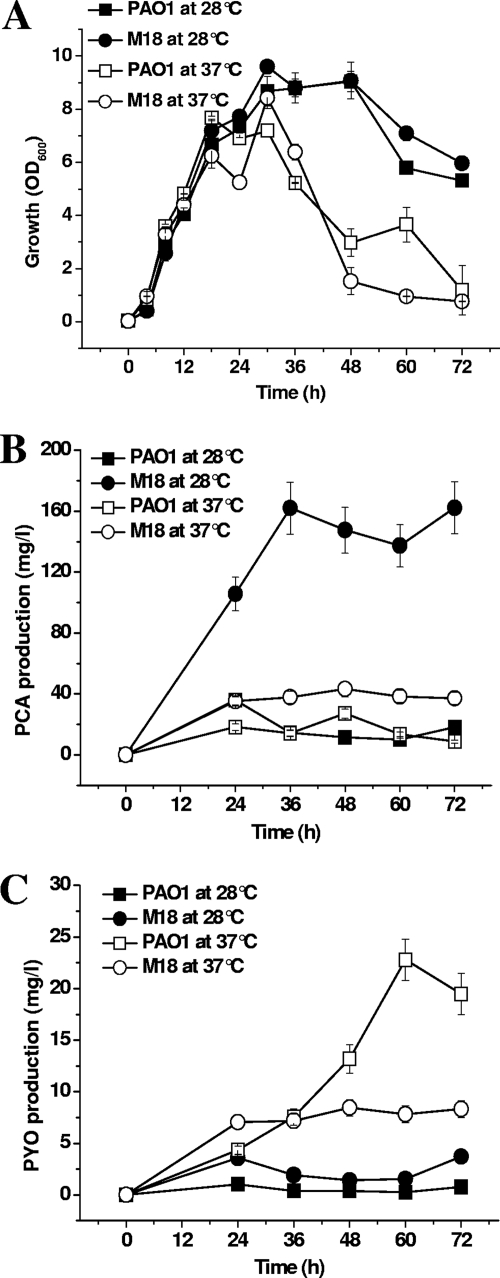
The PCA and PYO production of the two strains was further measured at 28°C and 37°C, respectively, and the results are shown in Fig. 2. The growth curves of strains M18 and PAO1 were similar at both 28°C and 37°C (Fig. 2A). However, 28°C was more suitable for cell growth of the two strains, and cell density dropped rapidly at 37°C during the late logarithmic phase. It was evident that the predominant phenazine produced by strain M18 and strain PAO1 was different at the different temperatures, as shown in Fig. 2B and C. The suitable temperature for PCA production was apparently 28°C, where the major phenazine produced by strain M18 was PCA, reaching up to 147 mg/liter or ninefold over that of P. aeruginosa PAO1 at that temperature. The quantitative ratio of PCA to PYO was very high, reaching up to 105:1 at 28°C and was 5:1 even at 37°C in strain M18. PYO was largely produced by strain PAO1 at 37°C, and the quantitative ratio of PYO to PCA was 2:1 at this temperature. The PYO production of strain PAO1 reached 24 mg/liter and was 3.5-fold greater than that in strain M18 at 37°C. In addition, the production of PCA by strain M18 was 4-fold lower at 37°C than at 28°C, while PYO production by strain PAO1 at 37°C was 20-fold greater than that at 28°C. There was no PYO detected in phzM::Km inactivation mutants M18pM and PAO1pM, but the total PCA produced by the two strains accumulated differently (data not shown).
FIG. 2.
Dynamic growth curves and different accumulations of PCA and PYO in strains M18 and PAO1. Shown are (A) the dynamic growth curves and (B) PCA and (C) PYO production of Pseudomonas sp. strain M18 (circles) and P. aeruginosa PAO1 (squares) grown in PPM broth at 28°C (solid symbols) and 37°C (open symbols). Values are the means ± standard deviations of triplicate cultures.
Mapping of the phzM gene transcriptional start sites in strains M18 and PAO1.
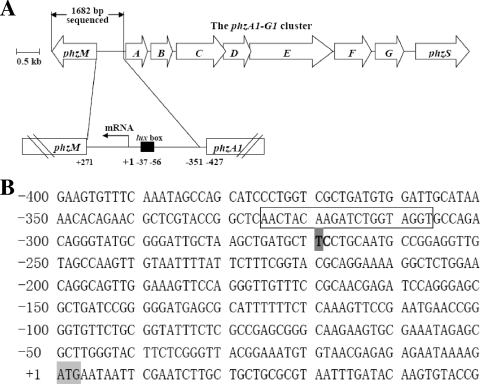
To clarify whether the temperature-dependent and strain-specific phzM gene expression in the two strains comes from differences in their gene coding regions, we first compared the nucleotide sequences of the phzM gene coding region with its flanking sequences in strains M18 and PAO1. A 1,694-bp fragment of a PCR product including the 1,005-bp gene coding region from the chromosomal DNA of strain M18 was sequenced directly by Invitrogen and then complemented with that from strain PAO1 by using NCBI BLASTX. We found that the full nucleotide sequence of the PCR fragment from strain M18 covering the whole gene structure and its flanking region showed 99% similarity to the phzM gene-containing region in P. aeruginosa PAO1. The phzM genes of both strains were closely located upstream of the phzA1B1C1D1E1F1G1 operon and transcribed divergently from it (Fig. 3A). The functional roles of the phzM gene were confirmed in the two strains by insertional inactivation of the gene. The results indicated that the production of PYO was abolished completely and the quantity of the PCA product increased in both mutant strains M18pM and PAO1pM (data not shown). We then investigated the transcriptional start sites of the phzM genes of Pseudomonas sp. strain M18 and P. aeruginosa PAO1 by 5′ RACE as described in Materials and Methods. With the poly(C)-tailed cDNA as a template, PCR amplification with the nested gene-specific primer and the novel deoxyinosine-containing anchor primer resulted in a single-band PCR product. This PCR product was analyzed by DNA sequencing, and the result revealed that there was only a single nucleotide difference between the two transcriptional start points in strains M18 and PAO1. The phzM transcriptional start site was at 269 bp upstream (C nucleotide) of the translation start site in strain PAO1 but was 270 bp upstream (T nucleotide) of the translation start site in strain M18, as shown in Fig. 3B. A putative lux box was found between bp 326 and 307 upstream of the translational start site in both genes (Fig. 3B) and exhibited substantial homology to other reported lux box sequences (29, 39, 45, 55, 59).
FIG. 3.
Location of the phzM gene with its transcriptional start site and the nucleotide sequence of the phzM promoter region in Pseudomonas sp. strain M18. (A) Location of phzM with its transcriptional start site. Numbering is relative to the transcriptional start site. A putative lux-like box (black box) is located upstream of the transcription start site. The length of the space between phzM and phzA1 is 696 bp. (B) Nucleotide sequence of the phzM promoter region. In strain M18, the mRNA start site of phzM, mapped by 5′ RACE, is at position −270, which is occupied by the T nucleotide indicated by a dark gray box in bold, while in strain PAO1, it is at position −269, which is occupied by the C nucleotide in bold. The potential lux-like box is enclosed in a rectangle. The ATG translational start codon is shaded light gray.
The temperature-dependent and strain-specific expression of the phzM gene occurred at the transcriptional level.
Since the phzM product is involved in the conversion of PCA to PYO in P. aeruginosa PAO1 (34, 40) and production of PCA and PYO by the two strains was determined to be temperature dependent and strain specific (Fig. 2), we investigated further to see if the phzM gene is expressed in a the temperature-dependent manner in the two strains and to determine if the phzM gene expressional regulation occurs at the translational, transcriptional, or both levels. Based on the phzM transcriptional start sites determined as described above, phzM′-′lacZ translational and transcriptional fusions were constructed to identify the phzM regulatory expression levels in strains M18 and PAO1. The same phzM′-′lacZ translational fusion plasmid, pPML, was introduced into strains M18 and PAO1, and β-Gal activity was measured at both 28°C and 37°C. Compared with the β-Gal activity in strain M18/pPML at 28°C, threefold and fivefold increased activities were detected in strains M18/pPML and PAO1/pPML, respectively, at 37°C, and the β-Gal activity increased sharply in PAO1/pPML when the OD600 reached 3.0 (Fig. 4B). The same trends were also found by analysis of the activities of the two transcriptional fusion products in strains M18/pPMC and PAO1/pPMCO; the β-Gal activities in both strains were about three to four times higher at 37°C than that at 28°C (Fig. 4C). The β-Gal activity in PAO1/pPMCO was threefold higher than that in M18/pPMC at the late logarithmic phase. Thus, we can conclude that the temperature-dependent expression of the phzM gene in strains M18 and PAO1 occurred mostly at the transcriptional level, since we obtained nearly the same increased expression ratio at both the translational and transcriptional levels at the two temperatures. It was found that the strain-specific expression of the phzM gene occurred mainly at the late logarithmic growth phase, when the OD600 of the two strains reached 3.0.
The expression of the phzM gene was positively regulated by LasI at the transcriptional level and negatively regulated by PtsP at both the transcriptional and translational levels in strain M18.
We further investigated the effects of six global regulatory genes, gacA, rsmA, rhlI, qscR, lasI, and ptsP, on phzM gene expression. It was found that the regulators GacA and RsmA had no obvious effect on phzM gene expression, while LasI and RhlI had a positive effect but QscR and PtsP had a negative effect on phzM gene expression (unpublished data). Interestingly, the lasI and ptsP regulatory genes, which exerted a strong regulatory effect on phzM, were also expressed in a temperature-dependent and strain-specific manner. The evidence showing the regulatory effect of lasI and ptsP on phzM expression is presented in the following sections.
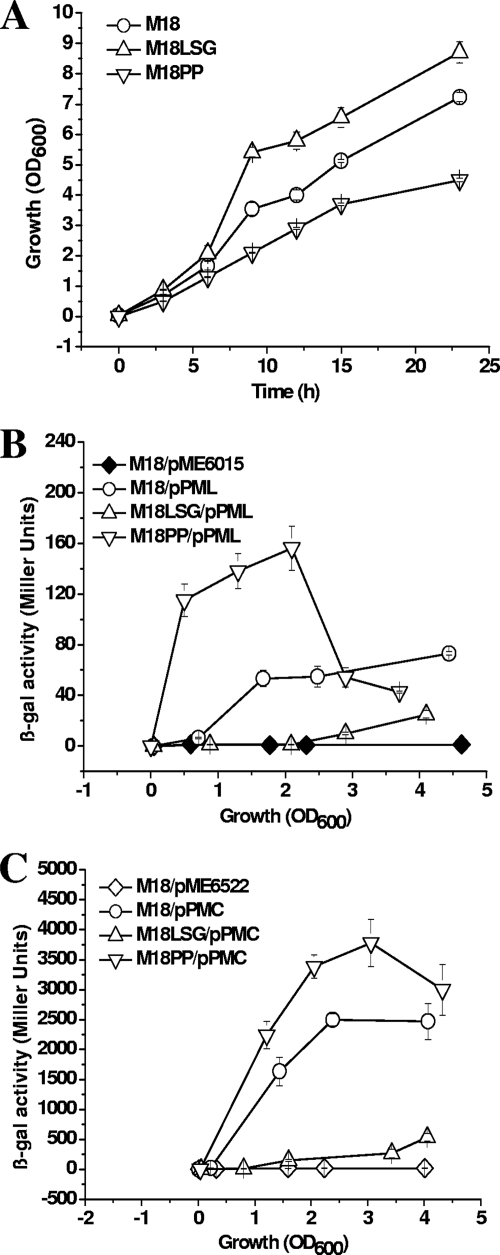
The construction of mutant M18PP with ptsP::Gm inactivated is described in Materials and Methods. The full nucleotide sequence of the whole ptsP gene coding region with its flanking regions and the encoded PtsP protein sequence of strain M18 show 97% and 100% identities, respectively, with those of P. aeruginosa PAO1. The mutant M18LSG with lasI::Gm inactivated was constructed in our previously published work (4). The cell growth rate was promoted by lasI inactivation and inhibited by ptsP inactivation, compared with that of wild-type strain M18 (Fig. 5A).
FIG. 5.
Effects of LasI and PtsP on phzM gene expression in Pseudomonas sp. strain M18. Shown are the growth curves of wild-type M18 (circles), corresponding lasI::Gm mutant M18LSG (triangles), and ptsP::Gm mutant M18PP (inverted triangles) (A) and the β-Gal activities of translational fusion plasmid pPML (B) and transcriptional fusion plasmid pPMC (C) in wild-type strain M18 (circles), lasI::Gm mutant M18LSG (triangles), and ptsP::Gm mutant M18PP (inverted triangles). pME6015 and pME6522 are the control plasmids transformed into strain M18 (diamonds). Growth was in PPM broth. The values are means ± standard deviations of triplicate cultures.
To determine whether LasI and PtsP regulate phzM expression, we tested the translational and transcriptional phzM′-′lacZ fusions in wild-type M18, lasI::Gm mutant M18LSG, and ptsP::Gm mutant M18PP (Fig. 5). In lasI::Gm mutant M18LSG, phzM gene expression was strongly inhibited and delayed at both the translational and transcriptional levels. β-Gal activity was nearly undetectable at the early logarithmic phase. Only 20 to 30% activity was found at the late logarithmic phase, when the OD600 reached 3.0 to 4.0, compared with that in wild-type M18. In addition, the β-Gal activity from putative lux box deletion-containing fusion plasmid pPMCD was undetectable in strains M18 and M18LSG (data not shown). These data suggested that phzM expression was positively controlled by the las system at the transcriptional level mostly via the lux box upstream of the transcriptional start site, in agreement with other reports on the regulation of LasI on vqsR (29). Inactivation of ptsP induced large amounts of PYO production and promoted cell lysis (data not shown); therefore, cell growth was inhibited and the OD600 of M18PP could not exceed 5.0 (Fig. 5A). The β-Gal activity from translational phzM′-′lacZ fusion plasmid pPML increased by 3-fold in ptsP::Gm mutant M18PP when the OD600 reached 2.0, compared with that in wild-type M18 (Fig. 5B), while at the transcriptional level in M18PP it was only 1.5-fold greater than that of wild-type M18 (Fig. 5C). This result suggested that PtsP inhibited phzM expression mainly at the translational level and not at the transcriptional level. This negative regulation of phzM by PtsP disappeared at later growth phases at an OD600 of about 4.0, perhaps due to the impaired cell growth ability of strain M18PP. The induction of phzM expression corresponded to the increase in PYO production by the ptsP::Gm mutant.
To confirm the proposed regulatory roles of LasI and PtsP in phzM expression, the phzM transcripts were assessed in wild-type M18, the lasI::Gm mutant, the ptsP::Gm mutant, and the complemented strains in trans by qRT-PCR at both the early exponential phase and the late exponential phase, as summarized in Table 3. When cells reached an OD600 of 2.0 to 2.4 in the early exponential phase, the normalized phzM levels in strains M18LSG and M18PP were 0.28 and 4.26, respectively, relative to that in strain M18. At the late exponential phase, the normalized phzM levels in M18LSG and M18PP reached 0.37 and 4.66, respectively. The results indicated that phzM expression was reduced in the lasI mutant and increased in the ptsP mutant. The transcriptional level of phzM was recovered in the complemented mutants in both the early exponential phase (P < 0.05) and the late exponential phase (P < 0.05).
TABLE 3.
Relative phzM transcription levels in wild-type strain M18, lasI::Gm mutant M18LSG, complemented M18LSG, ptsP::Gm mutant M18PP, and complemented M18PP determined by qRT-PCR during early and late exponential phase
| Strain | Early log phase
|
Late log phase
|
||||||
|---|---|---|---|---|---|---|---|---|
| CT,rpoDa | CT,phzMa | ΔCTb | Mc | CT,rpoDa | CT,phzMa | ΔCTb | Mc | |
| M18 | 20.05 ± 0.5 | 23.94 ± 0.1 | 3.89 ± 0.1 | 1.00 | 20.14 ± 0.01 | 22.09 ± 0.2 | 1.71 ± 0.05 | 1.00 |
| M18LSG | 19.78 ± 0.09 | 25.48 ± 0.12 | 5.70 ± 0.1 | 0.28 | 20.28 ± 0.03 | 23.65 ± 0.05 | 3.37 ± 0.04 | 0.37 |
| Complemented M18LSG | 20.07 ± 0.02 | 23.96 ± 0.07 | 3.89 ± 0.02 | 1.01 | 20.10 ± 0.02 | 21.81 ± 0.03 | 1.702 ± 0.03 | 1.19 |
| M18PP | 20.06 ± 0.27 | 21.05 ± 0.89 | 0.99 ± 0.1 | 4.26 | 19.76 ± 0.08 | 19.20 ± 0.66 | -0.56 ± 0.04 | 4.66 |
| Complemented M18PP | 19.60 ± 0.03 | 21.84 ± 0.26 | 2.24 ± 0.1 | 1.79 | 19.68 ± 0.09 | 20.67 ± 0.70 | 0.99 ± 0.07 | 1.59 |
Values were measured during early exponential phase at an OD600 of 2.0 to 2.4 and during late exponential phase at an OD600 of 5.0 to 6.0. All values are means ± standard deviations of three independent experiments.
ΔCT = mean CT, phzM − mean CT, rpoD.
M = 2−ΔΔCTphzM normalized amount of cDNA from the phzM gene in different strains relative to that in wild-type Pseudomonas sp. strain M18; ΔΔCT = mean ΔCT − mean ΔCT, M18.
These data, together with the results of analyses by qRT-PCR and phzM′-′lacZ fusional expression, clearly confirmed that the expression of phzM is positively regulated mainly at the transcriptional level by the QS protein LasI and negatively regulated at both the transcriptional and translational levels by the PtsP protein.
Quantitative analysis of temperature-regulated phzM, lasI, and ptsP transcripts by qRT-PCR in strains M18 and PAO1.
According to the previous results of phzM′-′lacZ fusional expression shown in Fig. 4, the phzM gene was expressed in a temperature-dependent and strain-specific manner. We further investigated the expression of phzM with its regulatory genes lasI and ptsP by measuring the transcripts by qRT-PCR at both 28°C and 37°C in strains M18 and PAO1, with the housekeeping gene rpoD as a reference (Table 4). In early and late exponential phase, compared with the transcripts in strain M18, the normalized phzM levels in strain PAO1 at 37°C were high, at six- and sevenfold over that at 28°C and increased to over twofold in strain M18 at 37°C or in strain PAO1 at 28°C. The results indicated that temperature-dependent phzM expression at the transcriptional level occurs in a strain-specific manner since phzM transcriptional activity was much higher in strain PAO1 than in strain M18, especially at 37°C. The lower expression of the phzM gene in strain M18, especially at 28°C, corresponded to the higher quantity of PCA produced by Pseudomonas sp. strain M18 at 28°C. Therefore, we can conclude that the different amounts of phenazine produced by strains M18 and PAO1 are related to phzM gene expression in a temperature-dependent and strain-specific manner.
TABLE 4.
Relative transcriptional levels of phzM and its regulatory genes lasI and ptsP in strains M18 and PAO1 determined by qRT-PCR during early and late exponential phase
| Strain and temp (°C) | Early log phase
|
Late log phase
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CT, rpoDa | CT, phzMa | CT, lasIa | CT, ptsPa | Mb | Lb | Pb | CT, rpoDa | CT, phzMa | CT, lasIa | CT, ptsPa | Mb | Lb | Pb | |
| M18 | ||||||||||||||
| 28 | 19.75 ± 0.2 | 24.76 ± 0.1 | 18.91 ± 0.4 | 20.65 ± 0.15 | 1.00 | 1.00 | 1.00 | 20.06 ± 0.1 | 23.08 ± 0.2 | 19.14 ± 0.4 | 27.10 ± 0.5 | 1.00 | 1.00 | 1.00 |
| 37 | 20.05 ± 0.13 | 23.94 ± 0.1 | 18.70 ± 0.1 | 23.01 ± 0.52 | 2.17 | 1.42 | 0.24 | 20.14 ± 0.3 | 22.09 ± 0.05 | 18.06 ± 0.3 | 28.52 ± 0.6 | 2.10 | 2.23 | 0.40 |
| PAO1 | ||||||||||||||
| 28 | 19.61 ± 0.32 | 23.44 ± 0.1 | 18.05 ± 0.2 | 20.73 ± 0.40 | 2.27 | 1.65 | 0.86 | 20.17 ± 0.2 | 21.91 ± 0.13 | 18.84 ± 0.5 | 27.27 ± 0.8 | 2.43 | 1.33 | 0.96 |
| 37 | 19.36 ± 0.54 | 21.77 ± 0.2 | 16.28 ± 0.2 | 23.35 ± 0.95 | 6.06 | 4.72 | 0.12 | 20.03 ± 0.3 | 20.20 ± 0.15 | 17.48 ± 0.3 | 28.94 ± 0.9 | 7.21 | 3.10 | 0.27 |
Values were measured during early exponential phase at an OD600 of 2.0 to 2.4 and during late exponential phase at an OD600 of 5.0 to 6.0. All values are means ± standard deviations of three independent experiments.
M = 2−ΔΔCTphzM, L = 2−ΔΔCTlasI, and P = 2−ΔΔCTptsP (normalized amounts of cDNA from the phzM, lasI, and ptsP genes in different strains relative to that in wild-type Pseudomonas sp. strain M18 at 28°C; ΔΔCT = mean ΔCT − mean ΔCT, M18 at 28°C).
In addition, the transcriptional expression of phzM, as well as its positive regulatory gene lasI and negative regulatory gene ptsP, was also assayed by qRT-PCR. As expected, the levels of the lasI transcripts at 37°C were twofold greater than those at 28°C and were much higher in strain PAO1 than in strain M18 at both temperatures. The results revealed that lasI had a similar temperature-dependent and strain-specific expression pattern, with the highest level in strain PAO1 at 37°C and the lowest level in strain M18 at 28°C. However, ptsP expression had an adverse temperature-dependent and strain-specific pattern; the highest level of ptsP transcripts was observed in strain M18 at 28°C, while the lowest level of ptsP transcripts was observed in strain PAO1 at 37°C. The normalized ptsP levels decreased to 0.24, 0.86, and 0.12, respectively, in strain M18 at 37°C and strain PAO1 at 28°C and 37°C, relative to that in strain M18 at 28°C, when the OD600 reached 2.0 to 2.4.
Temperature-dependent and strain-specific expression of lasI and ptsP in strains M18 and PAO1 as determined by assaying fusion β-Gal activities.
The temperature-regulated transcripts of lasI and ptsP were first identified in strains M18 and PAO1 by qRT-PCR as shown in Table 4; further, lasI′-′lacZ and ptsP′-′lacZ translational fusion plasmids pLIL and pPPL were constructed and then transformed into strains M18 and PAO1, respectively. Their β-Gal activities were assayed at 28°C and 37°C, as shown in Fig. 4D and E. The β-Gal activity detected in PAO1/pLIL was 2.5-fold higher than that in M18/pLIL, and the β-Gal activities were 2.5 to 3 times higher at 37°C than at 28°C in both strains M18/pLIL and PAO1/pLIL. The highest β-Gal activity was detected in PAO1/pLIL at 37°C (Fig. 4D). Meanwhile, the β-Gal activities produced by the ptsP′-′lacZ translational fusion in strains M18 and PAO1 were assayed. The results (Fig. 4E) showed that the β-Gal activities were threefold higher at 28°C than at 37°C in both strains M18/pPPL and PAO1/pPPL. At the same temperature, the β-Gal activity was two to three times higher in M18/pPPL than in PAO1/pPPL. The highest β-Gal activity was detected in M18/pPPL at 28°C. These measurements of β-Gal activity (Fig. 4D and E), together with the transcriptional analysis by qRT-PCR (Table 4), show clearly that the expression of lasI and ptsP is also regulated in a temperature-dependent and strain-specific manner.
DISCUSSION
An unexpected finding of this study was that phzM and its regulatory genes lasI and ptsP were expressed in a temperature-dependent and strain-specific manner in P. aeruginosa PAO1 and the related strain Pseudomonas sp. strain M18. P. aeruginosa is the most-studied phenazine-producing microorganism, and the characteristic blue-green pigment PYO is the most well-described phenazine and is associated with 90 to 95% of P. aeruginosa isolates (33, 48). P. aeruginosa is a ubiquitous environmental bacterium that is one of the top three causes of opportunistic human infections, while the major PYO produced by P. aeruginosa functions as an important virulence factor (26, 27, 43). Several strains of P. aeruginosa have also been found to be good biocontrol agents by being well adapted to the rhizosphere (1), where they can display strong activities against fungal phytopathogens (52). However, the antifungal metabolites produced by P. aeruginosa have been identified as PCN, PCA, or 5′-methyl-PCA, rather than PYO (5, 16, 20, 25). In our previously published work (20, 21, 59), we reported that Pseudomonas sp. strain M18 is another effective biocontrol agent and that the major virulence factor against soil-borne phytopathogens is PCA. Pseudomonas sp. strain M18 has a genetic background similar to that of P. aeruginosa PAO1, but the different phenotypic characteristics and phenazines produced from strain M18 have not been studied in detail. In the present study, we used P. aeruginosa PAO1, a representative strain isolated from a human patient, and Pseudomonas sp. strain M18, a strain related to P. aeruginosa and isolated from the rhizosphere, as two models from different ecological niches to characterize their phenazine production phenotypes and their biosynthetic regulatory genes.
We first described the differences in phenotype and fungal inhibition between these two strains (Fig. 1). Much stronger activity against fungal growth was detected in strain M18 than in strain PAO1 under suitable fungal growth conditions. It is known that PCA and PYO are the two important varieties of phenazines produced in various pseudomonads, although the relative quantities and functional roles of PCA and PYO produced in pseudomonads were not strictly distinguished and carefully studied in many publications. PCA and PYO were considered to have similar physiological and biochemical functions in cells (13); however, the different functional roles of PCA and PYO have been described in a few publications recently (10, 16, 28, 57). Based on the methods used in this study, it would be quite difficult to compare our PCA and PYO measurements with those in other studies, where PCA was mainly assayed at pH 4.0 by UV spectroscopy of high-performance liquid chromatography at 248 nm, while PYO was assayed at pH 7.0 by measuring OD520 (9, 14, 45, 58, 59). We first quantified the relative amounts of PCA and PYO produced by strains M18 and PAO1 and, interestingly, found that the main phenazine accumulated in strain M18 was PCA, especially at 28°C, while it was PYO in strain PAO1, especially at 37°C. PCA is considered to play critical roles in the protection of plants by biocontrol pseudomonad strains in the rhizosphere against infection and phytopathogenic disease (37, 44), while PYO is not essential for fungal killing (16). The PDA plate assay confirmed the different fungal killing abilities of PCA and PYO, and the purified PCA and PYO products need to be tested in fungal inhibition experiments to further validate the results. The differential production of the phenazines PCA and PYO by strains M18 and PAO1 might be a consequence of selective pressures imposed on P. aeruginosa PAO1 and its relative strain M18 in different ecological niches over a long evolutionary process.
PCA is believed to be converted to PYO by the sequential actions of the putative S-adenosylmethionine-independent N-methyltransferase PhzM and the putative flavin-containing monooxygenase PhzS in P. aeruginosa PAO1 during in vitro experiments (34, 40). The characteristics of phzM expression and regulation were first detailed in this report, although the expression and regulation of the phzS gene remain to be characterized. Nucleotide sequence determination indicated that the coding region and the upstream regulatory sequence of phzM show 99% similarity between strains M18 and PAO1, with only a nucleotide difference between the two transcriptional start sites of phzM in strains M18 and PAO1. The one nucleotide difference between the two transcriptional start sites had no effect on the transcriptional expression of phzM in the two strains (data not shown). We found that phzM expression is regulated in a temperature-dependent and strain-specific manner by quantitative analysis of phzM transcript levels by qRT-PCR and by analyzing the β-Gal activities from phzM′-′lacZ translational and transcriptional fusion constructs in these two strains. In contrast to M18, strain PAO1 displayed substantially higher levels of phzM expression at 37°C, especially in the late logarithmic phase. These results were consistent with data showing the differential accumulation of different phenazines and suggested that the regulation of phzM in P. aeruginosa PAO1 and Pseudomonas sp. strain M18 evolved to meet the host-driven survival and fitness requirements dictated by different environmental conditions, although further evidence is required to determine the activities of the PhzM protein at 28°C and 37°C. However, the relationship and interaction between the precise sequences in cis in the phzM gene regulatory region and the concrete factor or factors in trans which are involved in the temperature-dependent and strain-specific expression of phzM at both the transcriptional and translational levels in the two strains await further investigation. Other temperature-related regulators will be investigated by performing genome-wide transcriptome profiling in our laboratory as described by Chan et al. (3).
Besides the temperature-dependent and strain-specific expression of phzM, the results provided in this study suggest that the differential accumulation of PCA in the two strains might result from multiple mechanisms involved in the expression of the phenazine biosynthetic genes developed during evolution in different niches. We demonstrated that significantly greater amounts of PCA were produced in both wild-type M18 and the phzM::Km inactivation mutant M18pM than in both wild-type PAO1 and the phzM::Km inactivation mutant PAO1pM, although PYO was completely abolished in both phzM inactivation mutant strains M18pM and PAO1pM (data not shown). The amount of PCA produced by both strains M18 and M18pM at 28°C was almost 10-fold higher than that produced by both strains PAO1 and PAO1pM. Meanwhile, PYO was predominant at 37°C only in wild-type strain PAO1. These results suggest that another factor or factors besides phzM gene expression may be involved in the differential accumulation of phenazine production by the two strains. Regardless, expression of the phz phenazine biosynthetic gene cluster is strongly influenced by environmental conditions, as well as the other global regulators and QS systems (17, 42). Two phz biosynthetic clusters were found in both strains M18 and PAO1. Recently, 32 genes were found to be involved in phzA1B1C1D1E1F1G1 gene expression, which functions mainly for PYO synthesis in PAO1 (8, 31), but the phzA2B2C2D2E2F2G2 cluster functions mainly for PCA synthesis in Pseudomonas sp. strain M18 (unpublished data). The environmental conditions and other regulators involved in phz gene cluster expression and the different amounts of PCA accumulation should also be further investigated.
The functions of the global regulatory genes gacA, rsmA, rhlI, lasI, ptsP, and qscR in phzM gene expression were also investigated. Only lasI and rhlI showed strong positive effects, while ptsP and qscR showed a negative effect on phzM expression in both strains M18 and PAO1 (Table 3 and Fig. 5). Interestingly, we found that the expression of lasI and ptsP also occurs in a temperature-dependent and strain-specific manner, as shown in Table 4 and Fig. 4D and E. These results indicated that the two strains from different niches had developed their delicate regulatory systems to adapt to their environments. This is the first report of the temperature-dependent and strain-specific expression of lasI and ptsP and their regulatory effects on phzM. From previous reports (4, 41, 56), LasI produces the diffusible signal molecule 3-oxo-C12-HSL, which binds to the transcriptional regulator protein LasR to form a complex, which can then activate the transcription of the lux operon by binding to the lux box in the promoter region. A conserved dyad symmetry DNA sequence, termed the putative lux box, is present upstream of phzM (Fig. 3), suggesting that the las system directly activates phzM by recognizing the putative lux box. In previous studies (36, 58), inactivation of ptsP resulted in PYO overproduction by P. aeruginosa PAO1, leading to a significant decrease in the expression of qscR and twofold and threefold increases in the expression of lasI and rhlI, respectively, and suggested that PtsP inhibited PYO production by activating qscR and repressing the las and rhl system in P. aeruginosa. Furthermore, PtsP may regulate phzM indirectly by other regulators. As far as we know, the exact regulation mechanism of PtsP has not yet been clarified completely and may involve the utilization of carbon and nitrogen, rather than a promoter-specific regulatory factor (36, 53, 58). In this report, we provide evidence that the conversion of PCA to PYO by PhzM activity, especially at 37°C, is activated by LasI and inhibited by PtsP, although the precise regulatory mechanism remains to be further investigated in detail.
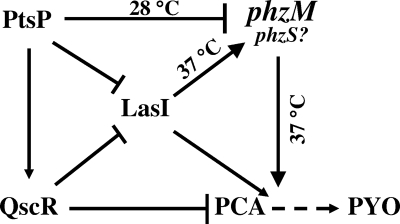
In conclusion, we demonstrated several characteristics of phzM gene expression and regulation. It was revealed that phzM gene expression occurs in a temperature-dependent and strain-specific manner and the phzM gene was positively regulated by the las system and negatively regulated by ptsP, while both regulators were also expressed in a temperature-dependent manner. The network related to phzM gene expression and its regulators involved in the conversion of PCA into PYO are summarized in Fig. 6. The results of this study provide preliminary information for future studies to more fully understand phzM expression and global regulatory networks involved in the virulence factors of and antibiotic production by strain P. aeruginosa and its relative Pseudomonas sp. strain M18 (47). Although strain M18 has a genetic background similar to that of strain PAO1, it has distinct unusual phenotypic and biochemical features to meet the requirements of its ecological fitness and competitiveness in its living niche. We cannot make the final determination of whether strain M18 belongs to a new subspecies of P. aeruginosa or a new pseudomonad species at present. The final decision about the classification of strain M18 is dependent on the pending results of whole-genome nucleotide sequencing, which is currently being carried out in our laboratory. Only when that has been completed can we determine the differences between strains M18 and PAO1, as well as other pseudomonad strains, on the whole-genome level and determine the taxonomic position of strain M18 based on phylogenetic analysis.
FIG. 6.
Network of the regulation on phzM expression and phenazine PCA and PYO production by pseudomonads. Arrows indicate positive regulation, lines with flat ends indicate negative regulation, and the broken arrow indicates a putative pathway.
Acknowledgments
This work was supported in part by grants from the National Key Basic Research Program (973 Program) (2009CB118906), the National High Technology Research and Development Program of China (863 Program) (2006AA10A209, 2006AA02Z228, and 2007AA02Z215), and the Shanghai Science and Technology Program (08391911900).
We thank Dieter Haas for his comments on the manuscript.
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Berg, G., L. Eberl, and A. Hartmann. 2005. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7:1673-1685. [DOI] [PubMed] [Google Scholar]
- 2.Blumer, C., S. Heeb, G. Pessi, and D. Haas. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 96:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, Y. C., S. Raengpradub, K. J. Boor, and M. Wiedmann. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73:6484-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y., X. Wang, X. Huang, X. Zhang, and Y. Xu. 2008. Las-like quorum-sensing system negatively regulates both pyoluteorin and phenazine-1-carboxylic acid production in Pseudomonas sp. M18. Sci. China C Life Sci. 51:174-181. [DOI] [PubMed] [Google Scholar]
- 5.Chin-A-Woeng, T. F., G. V. Bloemberg, I. H. Mulders, L. C. Dekkers, and B. J. Lugtenberg. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 6.Chin-A-Woeng, T. F., J. E. Thomas-Oates, B. J. Lugtenberg, and G. V. Bloemberg. 2001. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant-Microbe Interact. 14:1006-1015. [DOI] [PubMed] [Google Scholar]
- 7.Chin-A-Woeng, T. F., D. van den Broek, G. de Voer, K. M. van der Drift, S. Tuinman, J. E. Thomas-Oates, B. J. Lugtenberg, and G. V. Bloemberg. 2001. Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factors secreted into the growth medium. Mol. Plant-Microbe Interact. 14:969-979. [DOI] [PubMed] [Google Scholar]
- 8.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cugini, C., M. W. Calfee, J. M. Farrow III, D. K. Morales, E. C. Pesci, and D. A. Hogan. 2007. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 65:896-906. [DOI] [PubMed] [Google Scholar]
- 10.Debode, J., K. De Maeyer, M. Perneel, J. Pannecoucque, G. De Backer, and M. Höfte. 2007. Biosurfactants are involved in the biological control of Verticillium microsclerotia by Pseudomonas spp. J. Appl. Microbiol. 103:1184-1196. [DOI] [PubMed] [Google Scholar]
- 11.Denning, G. M., L. A. Wollenweber, M. A. Railsback, C. D. Cox, L. L. Stoll, and B. E. Britigan. 1998. Pseudomonas pyocyanin increases interleukin-8 expression by human airway epithelial cells. Infect. Immun. 66:5777-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fothergill, J. L., S. Panagea, C. A. Hart, M. J. Walshaw, T. L. Pitt, and C. Winstanley. 2007. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge, Y., X. Huang, S. Wang, X. Zhang, and Y. Xu. 2004. Phenazine-1-carboxylic acid is negatively regulated and pyoluteorin positively regulated by gacA in Pseudomonas sp. M18. FEMS Microbiol. Lett. 237:41-47. [DOI] [PubMed] [Google Scholar]
- 15.Ge, Y. H., D. L. Pei, P. Y. Feng, X. Q. Huang, and Y. Q. Xu. 2007. Autoinduction of RpoS biosynthesis in the biocontrol strain Pseudomonas sp. M18. Curr. Microbiol. 54:97-101. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, J., A. Sood, and D. A. Hogan. 2009. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 75:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 18.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 20.Hu, H., Y. Xu, F. Cheng, X. Zhang, and B. Hur. 2005. Isolation and characterization of a new fluorescent Pseudomonas strain producing both phenazine-1-carboxylic acid and pyoluteorin. J. Microbiol. Biotechnol. 15:86-90. [Google Scholar]
- 21.Huang, X., A. Yan, X. Zhang, and Y. Xu. 2006. Identification and characterization of a putative ABC transporter PltHIJKN required for pyoluteorin production in Pseudomonas sp. M18. Gene 376:68-78. [DOI] [PubMed] [Google Scholar]
- 22.Huang, X., X. Zhang, and Y. Xu. 2008. PltR expression modulated by the global regulators GacA, RsmA, LasI and RhlI in Pseudomonas sp. M18. Res. Microbiol. 159:128-136. [DOI] [PubMed] [Google Scholar]
- 23.Huang, X., D. Zhu, Y. Ge, H. Hu, X. Zhang, and Y. Xu. 2004. Identification and characterization of pltZ, a gene involved in the repression of pyoluteorin biosynthesis in Pseudomonas sp. M18. FEMS Microbiol. Lett. 232:197-202. [DOI] [PubMed] [Google Scholar]
- 24.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 25.Kumar, R. S., N. Ayyadurai, P. Pandiaraja, A. V. Reddy, Y. Venkateswarlu, O. Prakash, and N. Sakthivel. 2005. Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity and biofertilizing traits. J. Appl. Microbiol. 98:145-154. [DOI] [PubMed] [Google Scholar]
- 26.Lau, G. W., D. J. Hassett, H. Ran, and F. Kong. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599-606. [DOI] [PubMed] [Google Scholar]
- 27.Lau, G. W., H. Ran, F. Kong, D. J. Hassett, and D. Mavrodi. 2004. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 72:4275-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levitch, M. E., and R. E. Stadtman. 1964. A study of the biosynthesis of phenazine-1-carboxylic acid. Arch. Biochem. Biophys. 106:194-199. [DOI] [PubMed] [Google Scholar]
- 29.Li, L. L., J. E. Malone, and B. H. Iglewski. 2007. Regulation of the Pseudomonas aeruginosa quorum-sensing regulator VqsR. J. Bacteriol. 189:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y., H. Jiang, Y. Xu, and X. Zhang. 2008. Optimization of nutrient components for enhanced phenazine-1-carboxylic acid production by gacA-inactivated Pseudomonas sp. M18G using response surface method. Appl. Microbiol. Biotechnol. 77:1207-1217. [DOI] [PubMed] [Google Scholar]
- 31.Liang, H., L. Li, Z. Dong, M. G. Surette, and K. Duan. 2008. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 190:6217-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurhofer, M., C. Reimmann, P. Schmidli-Sacherer, S. Heeb, D. Haas, and G. Defago. 1998. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88:678-684. [DOI] [PubMed] [Google Scholar]
- 33.Mavrodi, D. V., W. Blankenfeldt, and L. S. Thomashow. 2006. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44:417-445. [DOI] [PubMed] [Google Scholar]
- 34.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mavrodi, D. V., V. N. Ksenzenko, R. F. Bonsall, R. J. Cook, A. M. Boronin, and L. S. Thomashow. 1998. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mavrodi, O. V., D. V. Mavrodi, D. M. Weller, and L. S. Thomashow. 2006. Role of ptsP, orfT, and sss recombinase genes in root colonization by Pseudomonas fluorescens Q8r1-96. Appl. Environ. Microbiol. 72:7111-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzola, M., R. J. Cook, L. S. Thomashow, D. M. Weller, and L. S. Pierson III. 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58:2616-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Morici, L. A., A. J. Carterson, V. E. Wagner, A. Frisk, J. R. Schurr, K. Honer zu Bentrup, D. J. Hassett, B. H. Iglewski, K. Sauer, and M. J. Schurr. 2007. Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol. 189:7752-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parsons, J. F., B. T. Greenhagen, K. Shi, K. Calabrese, H. Robinson, and J. E. Ladner. 2007. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry 46:1821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierson, L. S., III, V. D. Keppenne, and D. W. Wood. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J. Bacteriol. 176:3966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price-Whelan, A., L. E. Dietrich, and D. K. Newman. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189:6372-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price-Whelan, A., L. E. Dietrich, and D. K. Newman. 2006. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2:71-78. [DOI] [PubMed] [Google Scholar]
- 45.Rampioni, G., M. Schuster, E. P. Greenberg, I. Bertani, M. Grasso, V. Venturi, E. Zennaro, and L. Leoni. 2007. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 66:1557-1565. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73-81. [DOI] [PubMed] [Google Scholar]
- 48.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsenk, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 49.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomashow, L. S., D. M. Weller, R. F. Bonsall, and L. S. Pierson. 1990. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 56:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timms-Wilson, T. M., R. J. Ellis, A. Renwick, D. J. Rhodes, D. V. Mavrodi, D. M. Weller, L. S. Thomashow, and M. J. Bailey. 2000. Chromosomal insertion of phenazine-1-carboxylic acid biosynthetic pathway enhances efficacy of damping-off disease control by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 13:1293-1300. [DOI] [PubMed] [Google Scholar]
- 52.Troxler, J., P. Azelvandre, M. Zala, G. Defago, and D. Haas. 1997. Conjugative transfer of chromosomal genes between fluorescent pseudomonads in the rhizosphere of wheat. Appl. Environ. Microbiol. 63:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velázquez, F., K. Pflüger, I. Cases, L. I. De Eugenio, and V. de Lorenzo. 2007. The phosphotransferase system formed by PtsP, PtsO, and PtsN proteins controls production of polyhydroxyalkanoates in Pseudomonas putida. J. Bacteriol. 189:4529-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Y., X. Huang, H. Hu, X. Zhang, and Y. Xu. 2008. QscR acts as an intermediate in gacA-dependent regulation of PCA biosynthesis in Pseudomonas sp. M-18. Curr. Microbiol. 56:339-345. [DOI] [PubMed] [Google Scholar]
- 55.Waters, C. M., and B. L. Bassler. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 20:2754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, R., T. Pitt, G. Taylor, D. Watson, J. MacDermot, D. Sykes, D. Roberts, and P. Cole. 1987. Pyocyanin and 1-hydroxyphenazine produced by Pseudomonas aeruginosa inhibit the beating of human respiratory cilia in vitro. J. Clin. Investig. 79:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, H., W. Lin, H. Xia, S. Xu, Y. Li, H. Yao, F. Bai, X. Zhang, Y. Bai, P. Saris, and M. Qiao. 2005. Influence of ptsP gene on pyocyanin production in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 253:103-109. [DOI] [PubMed] [Google Scholar]
- 59.Yan, A., X. Huang, H. Liu, D. Dong, D. Zhang, X. Zhang, and Y. Xu. 2007. An rhl-like quorum-sensing system negatively regulates pyoluteorin production in Pseudomonas sp. M18. Microbiology 153:16-28. [DOI] [PubMed] [Google Scholar]
- 60.Yang, S. W., L. J. Lin, G. A. Cordell, P. Wang, and D. G. Corley. 1999. O- and N-methylation in the biosynthesis of staurosporine. J. Nat. Prod. 62:1551-1553. [DOI] [PubMed] [Google Scholar]
- 61.Yuan, L. L., Y. Q. Li, Y. Wang, X. H. Zhang, and Y. Q. Xu. 2008. Optimization of critical medium components using response surface methodology for phenazine-1-carboxylic acid production by Pseudomonas sp. M-18Q. J. Biosci. Bioeng. 105:232-237. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X. H., S. L. Wang, H. F. Gen, H. B. Hu, and Y. Q. Xu. 2005. Differential regulation of rsmA gene on biosynthesis of pyoluteorin and phenazine-1-carboxylic acid in Pseudomonas sp. M18. World J. Microbiol. Biotechnol. 21:883-889. [Google Scholar]