Abstract
DNA-based stable isotope probing in combination with terminal restriction fragment length polymorphism was used in order to identify members of the microbial community that metabolize biphenyl in the rhizosphere of horseradish (Armoracia rusticana) cultivated in soil contaminated with polychlorinated biphenyls (PCBs) compared to members of the microbial community in initial, uncultivated bulk soil. On the basis of early and recurrent detection of their 16S rRNA genes in clone libraries constructed from [13C]DNA, Hydrogenophaga spp. appeared to dominate biphenyl catabolism in the horseradish rhizosphere soil, whereas Paenibacillus spp. were the predominant biphenyl-utilizing bacteria in the initial bulk soil. Other bacteria found to derive carbon from biphenyl in this nutrient-amended microcosm-based study belonged mostly to the class Betaproteobacteria and were identified as Achromobacter spp., Variovorax spp., Methylovorus spp., or Methylophilus spp. Some bacteria that were unclassified at the genus level were also detected, and these bacteria may be members of undescribed genera. The deduced amino acid sequences of the biphenyl dioxygenase α subunits (BphA) from bacteria that incorporated [13C]into DNA in 3-day incubations of the soils with [13C]biphenyl are almost identical to that of Pseudomonas alcaligenes B-357. This suggests that the spectrum of the PCB congeners that can be degraded by these enzymes may be similar to that of strain B-357. These results demonstrate that altering the soil environment can result in the participation of different bacteria in the metabolism of biphenyl.
Polychlorinated biphenyls (PCBs) are very stable chloroorganic compounds with the general formula C12H10-xClx. Mixtures of PCBs have been used as coolants and lubricants in transformers, capacitors, and other electrical equipment as they do not burn easily and are good insulators. It is estimated that some 1.5 million tons of PCBs were produced up to 1988 worldwide (11; http://www.atsdr.cdc.gov/cercla; http://www.epa.gov/epawaste/hazard/tsd/pcbs/pubs/about.htm). Although production of these compounds was stopped, due to their long-term persistence, many sites all over the world are still contaminated with PCBs. Moreover, not only do PCBs threaten human health in the vicinity of the contaminated area, but lower PCB congeners volatilize and migrate to places far from where they were originally released (2, 3, 16). Also, their metabolic products have environmental significance; activities of both plants and microorganisms result in formation of different intermediates and final products whose toxicity can in some cases be even higher than that of the original toxicant (24, 26; http://www.atsdr.cdc.gov/cercla).
Physical-chemical methods used for the removal of PCBs often cause further natural disturbance and pollution; in contrast, biological methods of removal (i.e., bioremediation) are less expensive and more environmentally sound and thus have aroused much interest (7). These methods include the use of microorganisms and also exploitation of plants (i.e., phytoremediation) (19) and the cooperation of plants with microorganisms in the rhizosphere (i.e., rhizoremediation) (21). These bioremediation options also include the use of genetically modified bacteria (6) and/or plants (18, 23). PCBs were only recently introduced into the environment, and no completely efficient pathways for the aerobic bacterial degradation of all of these compounds have evolved (34); however, lower chlorinated PCB congeners can be degraded via the pathway that is used by aerobic bacteria to degrade biphenyl (35). Therefore, metabolism of biphenyl as a potential cometabolite of PCBs was the subject of this study.
The biphenyl degradation pathway is the same in all aerobic bacteria, and enzymes of this pathway degrade biphenyl in four steps into benzoate and 2-hydroxypenta-2,4-dienoate (21). The first enzyme of the pathway, biphenyl dioxygenase, has broad substrate specificity and thus permits degradation of biphenyl-related compounds (9). Substrates for biphenyl dioxygenase comprise, in addition to biphenyl itself, other diphenyl or benzene skeletons with several substituents, including halogens and bicyclic or tricyclic fused heterocyclic aromatics (35). These substrates also include certain natural compounds, including some plant flavonoids, phenols, or terpenes (10). Bacteria capable of metabolizing biphenyl are thus pervasive members of many microbial communities in vegetated soil.
As reported previously (20), there are two main problems with introduction of a new population of degrading or genetically modified microorganisms to enhance the biodegradation of PCBs in a contaminated environment: legislative barriers and the inability of strains added to the soil to survive. Therefore, the use of microorganisms for bioremediation of contaminated sites is not likely to be successful. Hence, understanding the biodegradative processes in the natural communities is necessary for planning remediation strategies. Identification of members of the community potentially responsible for the degradative process has recently been enabled by DNA-based stable isotope probing (SIP), as reviewed previously; therefore, this technique has become an efficient tool in microbial ecology (33). In this study, by tracking the transfer of 13C from [13C]biphenyl into bacterial DNA, it was possible to identify biphenyl-metabolizing bacteria in PCB-contaminated soil. To analyze how the bacterial diversity can be changed by introduction of a plant and subsequent cultivation in a greenhouse, bacteria in the rhizosphere of horseradish (Armoracia rusticana) cultivated in a contaminated soil were studied.
MATERIALS AND METHODS
Soil samples.
Soil was procured from a depth of approximately 0.5 m at a dumpsite for long-term PCB-contaminated soil with a PCB content of 153 mg kg−1 (dry weight) soil (25) located in Lhenice in the southern Czech Republic. The soil was homogenized and used for cultivation of 3-month-old horseradish plants. Before the plants were transferred to the contaminated soil, the roots were washed with tap water in order to remove the original soil. After 5 months of pot cultivation of the horseradish plants in 1-liter pots (three replicates) in stable conditions in a greenhouse, the roots had spread throughout the soil and the soil was harvested and designated the horseradish rhizosphere. Until the SIP experiment, initial bulk soil and horseradish rhizosphere samples were preserved at 4°C.
SIP microcosms.
Replicate microcosms were constructed in 250-ml sterilized glass Erlenmeyer flasks using 5 g of the rhizosphere and the initial bulk soil (referred to as soils below) and 45 ml of a basal mineral salt solution [1 g/liter (NH4)2SO4, 2.7 g/liter KH2PO4, 10.955 g/liter Na2HPO4·12H2O, 0.03 g/liter Ca(NO3)2, 0.01 g/liter FeSO4, 0.2 g/liter MgSO4] with 50 μg of biphenyl. Following a 5-day incubation on a rotary shaker at 28°C, SIP was begun by addition of 2.5 mg of [13C12]biphenyl (Sigma-Aldrich, United States) to four flasks containing each suspension; the remaining flaks were used as controls. SIP microcosms were harvested after 1, 3, 7, and 14 days of incubation at 28°C with shaking (130 rpm) by centrifugation at 2,500 × g for 30 min and were preserved at −80°C until DNA extraction.
DNA extraction.
DNA was extracted with a PowerMax soil DNA isolation kit (MoBio Laboratories Inc., United States) using the standard protocol, except that after the final elution the DNA was concentrated by adding 0.2 ml 5 M NaCl and 10.4 ml ethanol, incubated overnight at −20°C, and transferred gradually into 2-ml microtubes with 20 μg glycogen (Roche, Germany), which were centrifuged after each addition in order to obtain a single pellet. The pellet was then dissolved in 20 μl of water. The DNA concentration was evaluated by measuring the absorbance at 260 and 280 nm using a Bio-Rad Smart Spec Plus spectrophotometer (Bio-Rad, United States). All solutions were diluted to obtain a concentration of 0.6 μg·μl−1. Then 5 μl of each solution was mixed with 3.3 ml cesium trifluoroacetate (Amersham, United Kingdom) with a density of 1.6 g·ml−1 and subjected to isopycnic density gradient centrifugation.
Isopycnic centrifugation and gradient fractionation.
Isopycnic centrifugation was performed with a TL ultracentrifuge (Beckman Coulter, United States) at 145,000 × g for 70 h using a TLN 100 rotor and 3.3-ml OptiSeal cuvettes. Using a fraction recovery system (Beckman Coulter, United States) and Harvard Pump 11 Plus single syringe (Harvard Apparatus, United States), each gradient was fractionated into 80-μl fractions (with a flow rate of 240 μl·min−1). DNA was retrieved by adding 1 ml isopropanol and 20 μg glycogen, and after overnight incubation at 4°C, the DNA was centrifuged and the pellet was washed with 50 μl isopropanol, centrifuged again, and finally resuspended in 50 μl water. The DNA in each fraction was quantified by real-time PCR using primers 786f (5′-GATTAGATACCCTGGTAG-3′) and 939r (5′-CTTGTGCGGGCCCCCGTCAATTC-3′) targeting conserved regions of eubacterial 16S rRNA genes (1). DNA quantification was performed using the MiniOpticon real-time PCR detection system (Bio-Rad, United States), 15-μl reaction mixtures with iQ SYBR green Supermix (Bio-Rad, United States), 4.5 pmol each primer, and 2.5 μl template DNA, and the following program: 95°C for 5 min, followed by 30 cycles of 95°C for 40 s, 55°C for 40 s, and 72°C for 60 s and then a final extension at 72°C for 10 min. The gene copy number was determined using a standard curve constructed with Pseudomonas stutzeri JM300 genomic DNA as described previously (17). Fractions in which DNA was enriched with 13C were combined, and the resulting [13C]DNA was analyzed. Also, corresponding fractions from the control gradient were combined and analyzed so that the [12C]DNA that occurred throughout the whole gradient was not confused with 13C-enriched DNA (33).
Classification of metabolically active bacteria.
[13C]DNA from each time point, as well as total community DNA and control DNA (a mixture of DNA fractions from the initiation of the experiment corresponding to fractions in which [13C]DNA was located after [13C]biphenyl consumption), were used as templates for amplification of 16S rRNA genes. These DNA were amplified using primers 8f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 926r (5′-CCGTCAATTCCTTTRAGTTT-3′) targeting conserved regions of eubacterial 16S rRNA genes (29) with a Biometra TGradient thermocycler (Biometra, Germany) and a program consisting of 95°C for 5 min, followed by 25 cycles of 95°C for 45 s, 56°C for 45 s, and 72°C for 90 s and then a final extension at 72°C for 10 min. The 25-μl reaction mixtures contained a template, 5 pmol each primer (Generi Biotech, Czech Republic), 50 pmol deoxynucleoside triphosphates, 2.5 μg bovine serum albumin, and 0.5 U GoTaq DNA polymerase with the corresponding buffer (Promega, United States). Each PCR product was obtained in eight parallel experiments, and the resulting preparations were mixed, purified with a QIAquick PCR purification kit (Qiagen, Germany), and cloned using a TOPO-TA cloning kit for sequencing (Invitrogen, United States). Cultures of Escherichia coli transformed by appropriate inserts were plated on LB agar with ampicillin, and after overnight cultivation at 37°C, single colonies were transferred into a liquid medium and incubated overnight. Plasmids were isolated by alkaline lysis minipreparation using commercially available buffers P1 to P3 (Qiagen, Germany). Plasmid DNA sequencing was performed with a Beckman Coulter CEQ2000XL platform (Beckman Coulter, United States) using the program and parameters recommended by the manufacturer. Operational taxonomic units (OTUs) were defined at a level of sequence identity of 97%. Classification was performed by using RDPII Classifier (5) and an 80% confidence threshold. The web-based tool FastGroupII (36) was used to group similar sequences together (using a 97% sequence identity criterion). Phylogenetic trees were constructed using MEGA software (31) and the neighbor-joining method with the p-distance model and pairwise deletion of gaps or missing data.
Community profiling.
Fingerprinting analyses were performed using terminal restriction fragment length polymorphism (T-RFLP) of total community DNA, [13C]DNA from each time point, and control DNA. The templates were amplified by PCR (using the program described above) performed with primer 8f labeled at the 5′ end with 6-carboxyfluorescein and primer 926r in 25-μl reaction mixtures containing the template, 5 pmol each primer (Generi Biotech, Czech Republic), 50 pmol deoxynucleoside triphosphates (Promega, United States), 2.5 μg bovine serum albumin (Promega, United States), and 0.5 U DyNAzyme II DNA polymerase with the corresponding buffer (Finnzyme, Finland). The reconditioning step, purification, digestion with HhaI, and analyses were performed as described previously (17). In order to match the terminal restriction fragments (T-RFs) with the sequences in the 16S rRNA clone libraries, manual in silico digestion with HhaI was performed.
Analysis of genes responsible for biphenyl metabolism.
Portions of genes coding for the α subunit of biphenyl dioxygenase (bphA) were amplified from [13C]DNA isolated after 3 days of incubation of soils with [13C]biphenyl. The PCR conditions were the same as those used for amplification of 16S rRNA genes, and the previously described primers used (numbered based on positions in Burkholderia xenovorans LB400 bphA) were primers 352f (5′-TTCACCTGCASCTAYCACGGC-3′) and 1178r (5′-ACCCAGTTYTCDCCRTCGTCCTGC-3′) (27). The genes were cloned using a TOPO-TA cloning kit for sequencing (Invitrogen, United States). Ten clones from each library were sequenced with primers M13 forward and M13 reverse (TOPO-TA cloning kit for sequencing [Invitrogen, United States]), and in order to identify the closest sequence available, the sequences were subjected to a BLASTn search. The phylogenetic analysis was performed by using MEGA software (31) and the neighbor-joining method with the p-distance model and pairwise deletion of gaps or missing data.
Nucleotide sequence accession numbers.
Representative sequences determined in our analysis of the classification of metabolically active bacteria have been deposited in the GenBank database under accession no. FJ532316 to FJ532345. Representative sequences determined in our analysis of the genes responsible for biphenyl metabolism have been deposited in the GenBank database under accession no. FJ532314 and FJ532315.
RESULTS
Abundant members of bacterial communities.
T-RFLP profiles, as well as the libraries of total community DNA, show that the bacterial content in the horseradish rhizosphere differs from that in the bulk soil. In silico digestion of the sequences in the clone libraries was used to predict T-RFs that matched the peaks in the T-RFLP profiles (see Fig. S1 in the supplemental material). As the clone libraries did not provide complete community coverage according to rarefaction curves (data not shown), T-RFLP peak heights were considered more accurate indicators of relative abundance of microbial taxa than clone library detection frequency. In the bulk soil, the most abundant members of the community belonged to the Gammaproteobacteria, and Rhodanobacter was the predominant genus. In contrast, in the horseradish rhizosphere only one sequence was classified as a Rhodanobacter sequence (see Fig. S1 in the supplemental material); however, Gammaproteobacteria were also the most abundant organisms. Additional peaks in both profiles confirmed that 16S rRNA gene libraries covered only the more abundant members of the communities (see Fig. S1 in the supplemental material).
DNA labeling.
Real-time quantitative PCR analysis of all the fractions acquired by fractionation of unenriched DNA used as a control showed that the maximum quantity of DNA was in the 25th fraction. The DNA obtained after incubation of the soils with [13C]biphenyl was quantified for fractions 12 to 27. Enrichment with 13C was obvious in both soils after 3, 7, and 14 days of incubation with [13C]biphenyl; however, following 14 days of incubation, [13C]DNA was more dilute than it was after 3 or 7 days of incubation. After 1 day of incubation, no differences compared with unenriched control DNA were evident (see Fig. S2 in the supplemental material). Fractions 13 to 20 of the fractionated bulk soil DNA and fractions 14 to 20 of the horseradish rhizosphere DNA were combined and analyzed as [13C]DNA.
Classification of metabolically active bacteria based on 16S rRNA gene sequence analyses and community profiling.
16S rRNA gene amplicons acquired from total community DNA, [13C]DNA from each time point, and control DNA from both soils were cloned, and 50 clones in each library were sequenced with primers M13 forward and M13 reverse (TOPO-TA cloning kit for sequencing [Invitrogen, United States]). Sequences occurring in any of the [13C]DNA libraries that were grouped together with sequences in the control DNA by FastGroupII were omitted from further analyses. This resulted in libraries containing 19, 48, 48, and 29 sequences in the case of bulk soil DNA and 28, 47, 47, and 29 sequences in the case of horseradish rhizosphere DNA after 1, 3, 7, and 14 days of incubation, respectively.
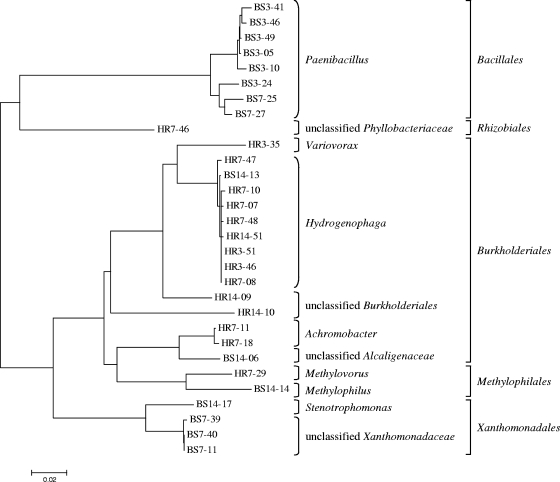
The microbial compositions of the clone libraries for 3-, 7-, and 14-day [13C]DNA based on RDP Classifier are shown in Table 1. Clone library analyses revealed that members of the genus Hydrogenophaga dominated biphenyl metabolism in the horseradish rhizosphere; sequences of members of this genus were detected in all of the [13C]DNA libraries (Fig. 1). Other sequences in the horseradish rhizosphere [13C]DNA libraries were classified as Variovorax (detected after 3 days of incubation), Achromobacter (the second most abundant taxon detected after 7 and 14 days of incubation), and Methylovorus (detected after 7 and 14 days of incubation). The other proteobacterial sequences occurring in the libraries after 7 and 14 days of incubation were not classified by RDPII Classifier at the genus level using the 80% confidence threshold (Fig. 1).
TABLE 1.
Numbers of 16S rRNA gene clones in clone libraries constructed from bulk soil and horseradish rhizosphere [13C]DNA obtained after 3, 7, and 14 days of incubation of the soils with [13C]biphenyl, as well as the number of unique OTUs (defined using 97% sequence identity) and the closest cultured relative(s) (according to Seqmatch at RDP-II) for each taxon
| Taxon | No. of OTUs | Closest cultured relative(s) according to RDP Seqmatcha | No. of clones in library
|
|||||
|---|---|---|---|---|---|---|---|---|
| Bulk soil
|
Horseradish rhizosphere
|
|||||||
| 3 days | 7 days | 14 days | 3 days | 7 days | 14 days | |||
| Paenibacillus | 8 | Paenibacillus validus (AF353697), Paenibacillus sp. strain Ao3 (EF208754) | 44 | 3 | ||||
| Unclassified Phyllobacteriaceae | 1 | Phyllobacterium myrsinacearum (D12789) | 1 | |||||
| Variovorax | 1 | Variovorax paradoxus (DQ256487) | 1 | 2 | ||||
| Hydrogenophaga | 9 | Hydrogenophaga palleronii (AF019073) | 4 | 31 | 26 | 45 | 37 | 23 |
| Unclassified Burkholderiales | 2 | Leptothrix mobilis (X97071) | 2 | |||||
| Achromobacter | 2 | Achromobacter xylosoxidans subsp. xylosoxidans (AJ491839) | 7 | 2 | ||||
| Unclassified Alcaligenaceae | 1 | Alcaligenes sp. strain L6 (X92415) | 3 | 1 | 1 | |||
| Methylovorus | 1 | Methylobacillus flagellatus (CP000284) | 2 | 1 | ||||
| Methylophilus | 1 | Methylophilus methylotrophus (AB193724) | 1 | |||||
| Stenotrophomonas | 1 | Stenotrophomonas dokdonensis (DQ178977) | 1 | |||||
| Unclassified Xanthomonadaceae | 3 | Pseudoxanthomonas spadix (AM418384) | 10 | |||||
| Total | 30 | 48 | 48 | 29 | 47 | 47 | 29 | |
The numbers in parentheses are accession numbers.
FIG. 1.
Phylogenetic closeness of OTUs (defined using 97% sequence identity) detected in 16S rRNA gene clone libraries from the 3-, 7-, and 14-day incubations of the soils with [13C]biphenyl grouped into genera and orders based on the RDP Classifier.
While only proteobacterial sequences were detected in the horseradish rhizosphere [13C]DNA clone libraries after 3, 7, and 14 days of incubation, the bulk soil [13C]DNA libraries also contained sequences of Firmicutes. The majority of the sequences in the library after 3 days of incubation were classified as Paenibacillus (Fig. 1), and similar sequences also appeared in the library after 7 days of incubation; however, in this library Hydrogenophaga sequences were the dominant sequences. Other members of the community deriving carbon from biphenyl were members of the genera Variovorax, Methylophilus, and Stenotrophomonas; also, some unclassified sequences belonging to members of the Xanthomonadaceae and Alcaligenaceae were detected (Fig. 1).
The [13C]DNA clone libraries after 1 day of incubation of both soils were more diverse than the other clone libraries. In both of these libraries, the dominant sequences were classified as Hydrogenophaga sequences (14 of 19 clones in the bulk soil DNA library and 22 of 28 clones in the horseradish rhizosphere DNA library), whereas only one copy of each of the other sequences occurred and these other sequences were not detected in the [13C]DNA libraries for the other time points, except for one Paenibacillus sequence in the bulk soil DNA library (data not shown).
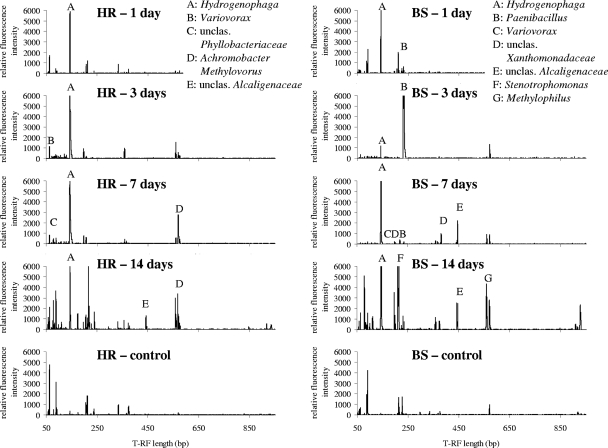
T-RFLP profiles of the [13C]DNA, together with the control profiles, are shown in Fig. 2. The control profiles contain peaks for the contaminating DNA that also occur in the other profiles, and thus these peaks are not considered peaks produced by [13C]DNA. In silico digestion of the sequences in clone libraries permitted identification of T-RFs and hence the relative abundance of community members (Fig. 2).
FIG. 2.
T-RFLP profiles of 16S rRNA gene amplicons of the [13C]DNA isolated from the horseradish rhizosphere (HR) and the bulk soil (BS) after 1, 3, 7, and 14 days of incubation with [13C]biphenyl. The control profiles are the profiles of unlabeled DNA isolated before [13C]biphenyl was added. Digestion was performed with HhaI. The origin of the T-RFs was predicted by in silico digestion of the sequences in the 16S rRNA gene clone libraries.
Functional genes involved in biphenyl metabolism.
Portions of bphA genes were amplified from [13C]DNA obtained after 3 days of incubation of the soils with [13C]biphenyl in order to identify functional genes that dominate biphenyl degradation. Use of primers 352f and 1178r for PCR amplification resulted in sequences that were 824 bp long. In both libraries, the sequences were only slightly different from each other. The nearest sequence in the databases matching the sequences obtained in both libraries was that of Pseudomonas alcaligenes B-357 (35). Using this sequence and other model sequences, a phylogenetic tree showing the clustering of deduced amino acid sequences was constructed (see Fig. S3 in the supplemental material).
DISCUSSION
The aim this work was to determine the identities of potential biphenyl-utilizing bacteria in the horseradish rhizosphere compared with the bacteria in bulk soil contaminated with PCBs. The SIP microcosms used were amended with nutrients; use of previously described unamended microcosms (17) did not result in detectable labeling of DNA after 14 days of incubation with [13C]biphenyl. Faster consumption of a substrate occurs in nutrient-amended microcosms, although the DNA of a less diverse population might be labeled (4, 33).
The results of quantitative PCR (see Fig. S2 in the supplemental material) show that 13C enrichment of DNA was insufficient after 1 day of incubation of the soils with [13C]biphenyl. This was confirmed by the 16S rRNA gene library analyses for this time point, in which 31 (bulk soil DNA) and 22 (horseradish rhizosphere DNA) of 50 sequences had to be ruled out because of their high levels of similarity to sequences detected in the control libraries. Moreover, except for 14 (bulk soil DNA) and 22 (horseradish rhizosphere DNA) sequences classified as Hydrogenophaga sequences, the sequences likely represented contaminating [12C]DNA. This can be illustrated by the T-RFLP profiles for this time point, in which there are no additional peaks compared to the control DNA T-RFLP profile, except for one matching Hydrogenophaga T-RF peak. Taking these findings into consideration, together with the fact that none of the sequences in the libraries for 1-day [13C]DNA except Hydrogenophaga and Paenibacillus sequences were in the libraries constructed using [13C]DNA from later time points, it is unlikely that these bacteria truly derived carbon from [13C]biphenyl.
The first time point at which there was sufficient labeling of DNA was after 3 days of incubation of soils with [13C]biphenyl (see Fig. S2 in the supplemental material). The 16S rRNA gene clone libraries and T-RF intensities for this time point might indicate the dominant role of Hydrogenophaga in the biphenyl metabolism in the horseradish rhizosphere (Table 1). In recent studies of Lambo and Patel (13-15), a member of Hydrogenophaga was characterized as a psychrotolerant biphenyl-utilizing bacterium that is able to cometabolize several mono-, di-, and trichlorobiphenyls at low temperatures and also a tetrachlorobiphenyl isomer at 30°C. Thus, this bacterium was put forward as an organism that could be useful for the bioremediation of PCBs in colder climates. Our results demonstrate the ability of Hydrogenophaga spp. to metabolize biphenyl in a complex community. Hydrogenophaga spp. appear to be invasive and fast growing, characteristics which are well illustrated by their increasing relative abundance in the clone libraries constructed using the 3-, 7-, and 14-day bulk soil [13C]DNA. This phenomenon, accompanied by the decreasing number of the Paenibacillus sequences over the time, is likely to provide evidence of the changing conditions in the microcosms and the response of the community to the changes. Therefore, the 3-day [13C]DNA is likely to most accurately represent potential biphenyl utilizers in the horseradish rhizosphere and in the initial bulk soil.
Bacteria belonging to the genus Paenibacillus seem to dominate biphenyl metabolism in the bulk soil, as shown by the frequent detection of Paenibacillus sequences in the 3-day bulk soil [13C]DNA sequences (Fig. 1). Because Paenibacillus sequences were also detected in the 16S rRNA gene library for the bulk soil total community DNA (see Fig. S1 in the supplemental material), Paenibacillus is likely to be the dominant genus of biphenyl-metabolizing bacteria in the bulk soil and might be quite abundant in this soil. On the other hand, the fact that it was not detected using the horseradish rhizosphere total community DNA or [13C]DNA indicates that horseradish plants may produce some compounds that make the environment unsuitable for Paenibacillus spp. or at least their biphenyl-metabolizing capabilities or that these species could be outcompeted by faster-growing organisms. Recently, Sakai et al. (28) isolated a bacterium that efficiently degraded di- to nonachlorobiphenyls and was identified as a Paenibacillus sp. A Paenibacillus strain capable of growing on biphenyl was also isolated in the laboratory of Michel Sylvestre (35).
Except for Paenibacillus, which belongs to the phylum Firmicutes (order Bacillales), all bacteria that were revealed by the 3-, 7-, and 14-day incubation with [13C]biphenyl were members of Proteobacteria that derived carbon either directly or indirectly from this substance. In addition to Hydrogenophaga spp., other betaproteobacterial sequences were also detected, including Variovorax and Achromobacter sequences, unclassified sequences of members of the family Alcaligenaceae, and unclassified sequences of members of the order Burkholderiales. Biphenyl-degrading bacteria are quite common in the Burkholderiales; isolates belonging to the genera Burkholderia (8), Pandoraea (30, 35), and Ralstonia (12) are well-investigated biphenyl-degrading strains. Variovorax was mentioned previously as one of the predominant genera that metabolize biphenyl in the root zone of pine growing in PCB-contaminated soil (17). Although DNA of Achromobacter spp. were detected in the total community DNA from the bulk soil (2 of 50 clones), they were detected only in the horseradish rhizosphere 7- and 14-day [13C]DNA. This finding suggests that horseradish roots exude some compounds that stimulate the enzymes of biphenyl catabolism or in some other way support the degradative abilities of these bacteria. Besides Achromobacter sequences, other sequences of members of the family Alcaligenaceae were detected in [13C]DNA from both soils. However, these sequences were not classified at the genus level and may represent so-far-undiscovered bacteria. In [13C]DNA from both soils, sequences of Methylophilales, namely members of the genera Methylovorus and Methylophilus, were also detected. Methylovorus spp. were detected repeatedly in 7- and 14-day [13C]DNA from the horseradish rhizosphere. Only single copies of DNA of Methylophilus spp. were detected in 14-day bulk soil [13C]DNA, but when the T-RFLP profiles were taken into account (Fig. 2), the peak that matched the Methylophilus predicted T-RF in 14-day [13C]DNA appeared at the same position in the profile of 7-day [13C]DNA. To the best of our knowledge, neither Methylovorus nor Methylophilus has been reported to metabolize biphenyl and/or PCBs. As these sequences did not appear in 3-day [13C]DNA, it is unclear whether these genera consumed [13C]biphenyl directly or via labeled metabolites. However, the presence of 13C-labeled Methylophilales might indicate that methanogens became active in the microcosms over the time, which may also be evidence of a decreasing amount of headspace oxygen and gradual transition to anoxic conditions. Some gammaproteobacterial sequences (family Xanthomonadaceae) occurred in the libraries from the 7- and 14-day incubations of the bulk soil with [13C]biphenyl, whereas an alphaproteobacterial sequence was detected in the 7-day horseradish rhizosphere [13C]DNA. Most of these sequences were not classified and may represent undescribed bacteria.
Deduced amino acid sequences of BphA (α subunit of biphenyl dioxygenase) of bacteria detected in the 3-day [13C]DNA from each of the soils clustered together. The finding that Paenibacillus and Hydrogenophaga sequences represented the vast majority of the sequences in 16S rRNA gene libraries constructed from the bulk soil and horseradish rhizosphere 3-day [13C]DNA, respectively, could possibly allow potential matching of the BphA amino acid sequences to these taxa, but the possibility of PCR biases cannot be ruled out. Both groups of sequences clustered with the BphA sequence of P. alcaligenes B-357 (see Fig. S3 in the supplemental material) and hence with the sequences of proteobacterial BphAs. If the BphAs from bulk soil originate from Paenibacillus spp., our results are in agreement with the findings of Vezina et al. (35); i.e., BphAs from Paenibacillus spp., which belong to the Firmicutes, cluster with proteobacterial BphAs and not with rhodococcal BphAs.
The amino acid sequence of a BphA is usually responsible for the specificity of the PCB congener degradation; in particular, region III, the sequence corresponding to B. xenovorans LB400 BphA positions 335, 336, 338, and 341, is known to influence substrate binding and specificity (22). The amino acids in this region in the sequences translated from 3-day [13C]DNA from each of the soils are Ala, Ile, Thr, and Ser, which are the amino acids in the P. alcaligenes B-357 sequence (35). The remainders of the sequences are almost identical to that of strain B-357, suggesting that the spectrum of the congeners that can be degraded by these BphAs is similar to that of P. alcaligenes B-357.
Our results demonstrate that the cultivation of a plant, such as horseradish, in contaminated soil may affect the spectrum of bacteria that are potentially responsible for the degradative process. The results obtained in this and two other studies that also utilized the SIP technique to identify members of the (polychlorinated) biphenyl-utilizing communities indicate that different microbial taxa may be active in different types of matrices. Tillmann et al. (32) used microcosm-based SIP to study active bacteria in a biofilm community grown directly on PCB droplets and found that Burkholderia spp. dominated aerobic degradation of PCBs. Leigh and colleagues (17) found that members of the genera Pseudonocardia, Kribella, Nocardioides, and Sphingomonas dominated biphenyl metabolism in unamended microcosms of PCB-contaminated soil from the root zone of pines in Czech soil. In the labeled populations detected in this study (17), members of the genera Variovorax and Hydrogenophaga, bacterial genera that appeared largely in [13C]DNA isolated from our soils, were also detected, suggesting that these two closely related genera might be quite abundant among biphenyl-metabolizing bacteria in PCB-contaminated soils in general.
Supplementary Material
Acknowledgments
This work was supported by grant NPVII 2B08031 from the Ministry of Education, Youth and Sports of the Czech Republic and by research projects MSM 6046137305 and Z 40550506 and grant 525/09/1058 from the Grant Agency of the Czech Republic.
We thank Sarka Pinkasova for her skillful technical assistance and Mary Beth Leigh for training in SIP.
Footnotes
Published ahead of print on 21 August 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard, M., M. J. Teil, E. Guigon, K. Larcher-Tiphagne, D. Ollivon, B. Garban, and M. Chevreuil. 2007. Persistent toxic substance inputs to the River Seine basin (France) via atmospheric deposition and urban sludge application. Sci. Total Environ. 375:232-243. [DOI] [PubMed] [Google Scholar]
- 3.Braune, B. M., M. L. Mallory, G. H. Grant, R. J. Letcher, and K. G. Drouillard. 2007. Levels and trends of organochlorines and brominated flame retardants in ivory gull eggs from the Canadian Arctic, 1976 to 2004. Sci. Total Environ. 378:403-417. [DOI] [PubMed] [Google Scholar]
- 4.Cebron, A., L. Bodrossy, N. Stralis-Pavese, A. C. Singer, I. P. Thompson, J. I. Prosser, and J. C. Murrell. 2007. Nutrient amendments in soil DNA stable isotope probing experiments reduce the observed methanotroph diversity. Appl. Environ. Microbiol. 73:798-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Carcer, D. A., M. Martin, M. Mackova, T. Macek, U. Karlson, and R. Rivilla. 2007. The introduction of genetically modified microorganisms designed for rhizoremediation induces changes on native bacteria in the rhizosphere but not in the surrounding soil. ISME J. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 7.Demnerova, K., M. Mackova, V. Spevakova, K. Beranova, L. Kochankova, P. Lovecka, E. Ryslava, and T. Macek. 2005. Two approaches to biological decontamination of groundwater and soil polluted by aromatics—characterization of microbial populations. Int. Microbiol. 8:205-211. [PubMed] [Google Scholar]
- 8.Erickson, B. D., and F. J. Mondello. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa, K., H. Suenaga, and M. Goto. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez, B. S., S. C. Koh, M. Chial, and D. D. Focht. 1997. Terpene-utilizing isolates and their relevance to enhanced biotransformation of polychlorinated biphenyls in soil. Biodegradation 8:153-158. [Google Scholar]
- 11.Holoubek, I. 2001. Polychlorinated biphenyl (PCB) contaminated sites worldwide, p. 17-26. In L. W. Robertson and L. G. Hansen (ed.), PCBs. Recent advances in environmental toxicology and health effects. University of Kentucky Press, Lexington.
- 12.Kahl, S., and B. Hofer. 2003. A genetic system for the rapid isolation of aromatic-ring-hydroxylating dioxygenase activities. Microbiology 149:1475-1481. [DOI] [PubMed] [Google Scholar]
- 13.Lambo, A. J., and T. R. Patel. 2006. Cometabolic degradation of polychlorinated biphenyls at low temperature by psychrotolerant bacterium Hydrogenophaga sp. IA3-A. Curr. Microbiol. 53:48-52. [DOI] [PubMed] [Google Scholar]
- 14.Lambo, A. J., and T. R. Patel. 2006. Isolation and characterization of a biphenyl-utilizing psychrotrophic bacterium, Hydrogenophaga taeniospiralis IA3-A, that cometabolizes dichlorobiphenyls and polychlorinated biphenyl congeners in Aroclor 1221. J. Basic Microbiol. 46:94-107. [DOI] [PubMed] [Google Scholar]
- 15.Lambo, A. J., and T. R. Patel. 2007. Biodegradation of polychlorinated biphenyls in Aroclor 1232 and production of metabolites from 2,4,4′-trichlorobiphenyl at low temperature by psychrotolerant Hydrogenophaga sp. strain IA3-A. J. Appl. Microbiol. 102:1318-1329. [DOI] [PubMed] [Google Scholar]
- 16.Lebeuf, M., M. Noel, S. Trottier, and L. Measures. 2007. Temporal trends (1987-2002) of persistent, bioaccumulative and toxic (PBT) chemicals in beluga whales (Delphinapterus leucas) from the St. Lawrence Estuary, Canada. Sci. Total Environ. 383:216-231. [DOI] [PubMed] [Google Scholar]
- 17.Leigh, M. B., V. H. Pellizari, O. Uhlik, R. Sutka, J. Rodrigues, N. E. Ostrom, J. Zhou, and J. M. Tiedje. 2007. Biphenyl-utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs). ISME J. 1:134-148. [DOI] [PubMed] [Google Scholar]
- 18.Macek, T., P. Kotrba, A. Svatos, M. Novakova, K. Demnerova, and M. Mackova. 2008. Novel roles for genetically modified plants in environmental protection. Trends Biotechnol. 26:146-152. [DOI] [PubMed] [Google Scholar]
- 19.Macek, T., M. Mackova, and J. Kas. 2000. Exploitation of plants for the removal of organics in environmental remediation. Biotechnol. Adv. 18:23-34. [DOI] [PubMed] [Google Scholar]
- 20.Macek, T., O. Uhlik, K. Jecna, M. Novakova, P. Lovecka, J. Rezek, V. Dudkova, P. Stursa, B. Vrchotova, D. Pavlikova, K. Demnerova, and M. Mackova. 2009. Advances in phytoremediation and rhizoremediation, p. 257-277. In A. Singh, R. C. Kuhad, and O. P. Ward (ed.), Advances in applied bioremediation. Springer, Berlin, Germany.
- 21.Mackova, M., D. Dowling, and T. Macek. 2006. Phytoremediation and rhizoremediation. Theoretical background. Springer, Dordrecht, The Netherlands.
- 22.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novakova, M., M. Mackova, Z. Chrastilova, J. Viktorova, M. Szekeres, K. Demnerova, and T. Macek. 2009. Cloning the bacterial bphC gene into Nicotiana tabacum to improve the efficiency of PCB phytoremediation. Biotechnol. Bioeng. 102:29-37. [DOI] [PubMed] [Google Scholar]
- 24.Park, J. S., L. Linderholm, M. J. Charles, M. Athanasiadou, J. Petrik, A. Kocan, B. Drobna, T. Trnovec, A. Bergman, and I. Hertz-Picciotto. 2007. Polychlorinated biphenyls and their hydroxylated metabolites (OH-PCBs) in pregnant women from eastern Slovakia. Environ. Health Perspect. 115:20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavlikova, D., T. Macek, M. Mackova, and M. Pavlik. 2007. Monitoring native vegetation on a dumpsite of PCB-contaminated soil. Int. J. Phytoremediat. 9:71-78. [DOI] [PubMed] [Google Scholar]
- 26.Rezek, J., T. Macek, M. Mackova, J. Triska, and K. Ruzickova. 2008. Hydroxy-PCBs, methoxy-PCBs and hydroxy-methoxy-PCBs: metabolites of polychlorinated biphenyls formed in vitro by tobacco cells. Environ. Sci. Technol. 42:5746-5751. [DOI] [PubMed] [Google Scholar]
- 27.Ryslava, E., T. Macek, M. Mackova, and Z. Krejcik. 2004. Detection of polychlorinated biphenyl-degrading bacteria in soil, p. 839-842. In W. Verstraete (ed.), Environmental biotechnology. Taylor & Francis Group plc, London, United Kingdom.
- 28.Sakai, M., S. Ezaki, N. Suzuki, and R. Kurane. 2005. Isolation and characterization of a novel polychlorinated biphenyl-degrading bacterium, Paenibacillus sp. KBC101. Appl. Microbiol. Biotechnol. 68:111-116. [DOI] [PubMed] [Google Scholar]
- 29.Shyu, C., T. Soule, S. J. Bent, J. A. Foster, and L. J. Forney. 2007. MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. Microb. Ecol. 53:562-570. [DOI] [PubMed] [Google Scholar]
- 30.Sylvestre, M., M. Sirois, Y. Hurtubise, J. Bergeron, D. Ahmad, F. Shareck, D. Barriault, I. Guillemette, and J. M. Juteau. 1996. Sequencing of Comamonas testosteroni strain B-356-biphenyl/chlorobiphenyl dioxygenase genes: evolutionary relationships among Gram-negative bacterial biphenyl dioxygenases. Gene 174:195-202. [DOI] [PubMed] [Google Scholar]
- 31.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 32.Tillmann, S., C. Strompl, K. N. Timmis, and W. R. Abraham. 2005. Stable isotope probing reveals the dominant role of Burkholderia species in aerobic degradation of PCBs. FEMS Microbiol. Ecol. 52:207-217. [DOI] [PubMed] [Google Scholar]
- 33.Uhlik, O., K. Jecna, M. B. Leigh, M. Mackova, and T. Macek. 2009. DNA-based stable isotope probing: a link between community structure and function. Sci. Total Environ. 407:3611-3619. [DOI] [PubMed] [Google Scholar]
- 34.Vaillancourt, F. H., M. A. Haro, N. M. Drouin, Z. Karim, H. Maaroufi, and L. D. Eltis. 2003. Characterization of extradiol dioxygenases from a polychlorinated biphenyl-degrading strain that possess higher specificities for chlorinated metabolites. J. Bacteriol. 185:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vezina, J., D. Barriault, and M. Sylvestre. 2008. Diversity of the C-terminal portion of the biphenyl dioxygenase large subunit. J. Mol. Microbiol. Biotechnol. 15:139-151. [DOI] [PubMed] [Google Scholar]
- 36.Yu, Y., M. Breitbart, P. McNairnie, and F. Rohwer. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.