Summary
Microorganisms have evolved an impressive array of mechanisms to adapt to stress induced by reactive oxygen species (ROS) of virtually any kind. One such regulator is B. subtilis PerR, a member of the ubiquitous Fur (Ferric uptake regulator) family of metalloregulatory repressors, which senses hydrogen peroxide. In this issue of Molecular Microbiology, Duarte, Latour and colleagues report the structure of the Mn(II)-bound form of PerR, a first for the Fe/Mn-selective members of the Fur family. The structure reveals how a regulatory metal drives a quaternary structural switch that allosterically activates the PerR dimer to bind its DNA operator, while also providing detailed insight into the mechanism of metal-catalyzed ligand oxidation and transcriptional derepression that uniquely characterizes PerR.
The formation of intracellular reactive oxygen species (ROS) is a potentially deleterious byproduct of metabolism in an aerobic environment (Imlay, 2008). ROS can take on many forms, including superoxide anion, hydrogen peroxide (H2O2), organic peroxides (ROOH) and the highly reactive hydroxyl radical (HO•), all of which are capable of damaging virtually all biopolymers in the cell. Misregulation of the homeostasis of transition metals can also lead to oxidative stress, where H2O2 feeds the autocatalytic production of OH• from reduced Fe(II) and Cu(I) via redox cycling by cellular reductants. The ability of microorganisms to detoxify ROS continuously is therefore critical for their survival.
Under normal conditions, many cells constitutively produce low levels of enzymes, e.g., peroxidases and superoxide dismutases, which protect against low or ambient levels of ROS. Under conditions of high ROS or oxidative stress, the transcription of genes for these and other proteins is strongly upregulated. This is accomplished by what are collectively termed “redox sensing” transcriptional regulators. One common feature of virtually all redox sensors specific for ROS, with the notable exception of B. subtilis PerR discussed here (Jacquamet et al., 2009) and E. coli SoxR (Watanabe et al., 2008), is that they possess at least one redox-active cysteine, derivatization of which drives “redox-dependent” transcriptional control (Paget & Buttner, 2003). For example, a highly reactive Cys thiolate in OxyR, the primary hydrogen peroxide sensor in E. coli, reduces H2O2 to form a sulphenic acid adduct, which is then resolved by formation of a disulfide bond with a nearby Cys, a low-molecular cellular thiol, e.g., cysteine or glutathione, or other oxidation products (Lee et al., 2004).
In B. subtilis, the transcriptional reprogramming that occurs in response to peroxide stress is carried out by a general stress response sigma factor, σB, and two transcriptional repressors, OhrR and PerR (Helmann et al., 2003). B. subtilis OhrR specifically senses organic peroxides and is a repressor of a gene encoding a thiol peroxidase (OhrA) (Hong et al., 2005). Derepression occurs via inactivation of the operator DNA binding activity by derivatization of the lone Cys in the DNA binding domain, first to a Cys sulfenate and then to a number of reaction products that leads to OhrR dissociation from the operator to allow RNA polymerase access to the promoter. Although the mechanistic details might vary, the vast majority of ubiquitous so-called “single-Cys” and “two-Cys” redox sensors are known or projected to function essentially in this way.
B. subtilis PerR is a striking and significant departure from this model. PerR was first discovered by Helmann’s group in their search for functional orthologs of the iron regulatory protein, E. coli Fur (Ferric uptake regulator), in B. subtilis (Bsat et al., 1998). Fur, a global regulator of Fe and oxidative stress regulons in many bacteria, turns off the transcription of genes for Fe uptake and siderophore biosynthesis under Fe-replete conditions to avoid Fe toxicity and the oxidative stress that would ensue (Lee & Helmann, 2007). In Fur, Fe(II) functions as a co-repressor and activates the binding of Fur to the Fur-box; in the absence of Fe, Fur has an undetectable affinity for DNA. Fur is now known to be representative of a large family of metalloregulatory proteins that sense other transition metal ions, including Zur (zinc uptake regulator), Nur (nickel uptake regulator), and Mur (manganese uptake regulator) as well as heme availability (Irr) (Lee & Helmann, 2007).
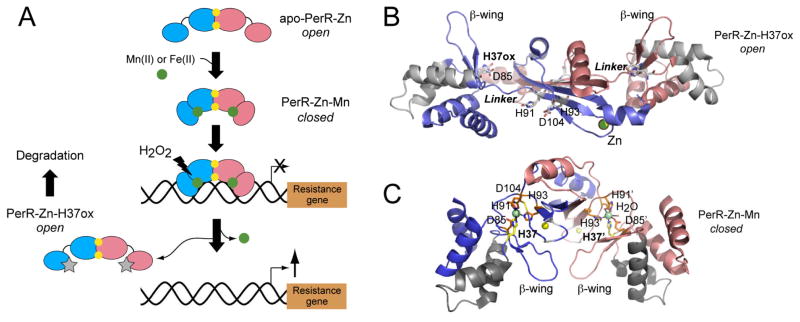
In contrast to these Fur orthologs, PerR functions primarily as a hydrogen peroxide sensor and is therefore the functional equivalent of OxyR in B. subtilis. Like Fur family proteins however, PerR employs a metal, Fe(II) or Mn(II), to activate operator DNA binding. The striking departure from Fur is that this Fe(II)-bound PerR is effectively a substrate for Fe(II)-catalyzed H2O2-dependent oxidation of PerR itself, which in turn leads to derepression of the PerR regulon (Fig. 1A). Purified PerR incubated with H2O2, as well as PerR isolated from H2O2-treated cells, is oxidized exclusively at His37 and to a lesser extent His91 to 2-oxo-His, with both histidines predicted to be ligands to the regulatory Fe atom in PerR (Lee & Helmann, 2006b, Traore et al., 2009). The bound Fe(II) is proposed to coordinate H2O2 and generate HO•, which then reacts directly with nearby His ligands in a non-repairable, “sacrificial” metal-catalyzed oxidation (MCO) reaction, leading to dissociation of the metal and the DNA (Lee & Helmann, 2006b).
Fig. 1.
Functional states of B. subtilis PerR. (A) Schematic rendering of the conformational changes in PerR that result from Mn(II) or Fe(II) binding (green spheres) to the PerR dimer, which activates operator binding and represses the transcription of a resistance gene, e.g., katG (catalase). Subsequent metal-catalyzed oxidation of H37, results in dissociation of the regulatory metal, and dissociation from the DNA operator, leading to derepression of transcription. One subunit is blue, and the other is shaded salmon (small ellipse, N-terminal DBD; larger ellipse, C-terminal regulatory domain, connected to one another by a linker); structural Zn(II) ions (yellow spheres); 2-oxo-His37 (H37ox) (gray stars) also indicated. (B) Ribbon representation of oxidized PerR, with H37ox indicated (Traore et al., 2009); the structure of apo-PerR-Zn is substantially identical (Traore et al., 2006). One subunit shaded blue, and the other shaded salmon, and the extended segment that connects the N-terminal and C-terminal domains in each subunit is labeled Linker. The metal ligands are indicated in stick representation (labeled), and the structural Zn(II) ions are shaded yellow. The helix-turn-helix (shaded gray ribbon) and the β-wing of each the winged helix domain of each subunit are highlighted. (C) Ribbon representation of the Mn(II)-activated PerR dimer (Jacquamet et al., 2009), with His37 from the DBD is in yellow stick; Zn(II) ions are in yellow; Mn(II) ions in green.
Now, enter three recent crystallographic structures of B. subtilis PerR solved by Duarte, Latour and coworkers, including the one reported in this issue of the journal (Traore et al., 2006, Traore et al., 2009, Jacquamet et al., 2009). Key unresolved issues addressed by the structural work center around two questions: (1) How is PerR selective for hydrogen peroxide as an oxidant in a way that leads to oxidation of primarily one histidine (His37)? and, (2) How does Fe(II) binding to the regulatory sites allosterically activate the binding of PerR to the operator (a question of general significance to the entire Fur family), which is then essentially unraveled by 2-oxo-His37 formation?
The structure of “apo” PerR, termed apo-PerR-Zn, gave us our first glimpse of a bona fide Fur family repressor (Traore et al., 2006). The structure reveals an “open” conformation in which the N-terminal DNA-binding winged helical domains (DBDs) are “splayed out” from the C-terminal dimerization domain, connected by a short tether (residues 86–91) (see Fig. 1B). This C-terminal domain contains a tetrathiolate Zn(II) coordination complex, found in some Fur proteins, which might stabilize the dimer (Lee & Helmann, 2006a). The apo-PerR-Zn structure suggests that the DBDs will adopt a wide range of orientations relative to the C-terminal domain in the absence of Fe(II) or Mn(II) in solution, but this is not yet proven. In any case, this conformation(s) would appear poorly suited to bind DNA, with the anticipated regulatory metal site ligands, His37 from the DBD, Asp85, His91 and His93 from the connecting linker, and Asp104 from the C-terminal domain (Lee & Helmann, 2006b) clearly not in a position to bind a metal (Fig. 1B). Strikingly, the structure of oxidized apo-PerR-Zn in which 2-oxo-His is unambiguously confirmed by crystallographic methods, reveals exactly the same “open” conformation as apo-PerR-Zn (Traore et al., 2009) and, thus, immediately suggests how MCO leads to inactivation of PerR as a repressor (Fig. 1B).
The new structure of the PerR-Zn-Mn complex reported in this issue of the journal provides new insights into what is anticipated to be the DNA-bound conformation of PerR on which MCO works, as well as metalloregulation of DNA binding by other metal-sensing Fur family members (Jacquamet et al., 2009) (Fig. 1C). First, the binding of Mn(II) to the pair of regulatory sites on the dimer leads to a caliper-like closure of the dimer due primarily to recruitment of His37 from the DBD; this effectively “freezes out” any PerR conformations that are open and not directly suited for high affinity DNA binding (see Fig. 1A). The result is formation of a pentacoordinate, square pyramidal Mn(II) complex, where His37 and His91 are ligands in the equatorial plane, with an axially coordinated His93 essentially opposite a solvent accessible open coordination site to which an H2O2 can make close approach to the metal (Fig. 1C). Provided the structure of the PerR-Zn-Mn complex is a close mimic of the redox-active PerR-Zn-Fe complex (see below), it immediately explains why 2-oxo-His37 and 2-oxo-His91 are the only observed reaction products (Lee & Helmann, 2006b), since the side chain of His93 is completely inaccessible to the locally generated OH•; furthermore, the structure readily rationalizes the exquisite selectivity for H2O2 over bulkier organoperoxides on steric grounds. Finally, since the Fe(II) complex is far more reactive than the Mn(II) chelate, the authors went to great lengths, using x-ray absorption spectroscopy, to establish that Fe(II) adopts a coordination complex that is similar, if not identical, to that formed by Mn(II).
Thus, this study (Jacquamet et al., 2009) firmly establishes that Fe(II)-activation and subsequent inactivation of PerR by its inducer hydrogen peroxide is explicable by a ordered closing and opening of a caliper, with a key allosteric role played in both steps by the distal metal ligand His37 (Fig. 1A). The structural features that distinguish a bona fide Fur from a PerR are not yet known, but they presumably attenuate the reactivity of the regulatory Fe(II) in Fur, likely via formation of a coordinately saturated Fe(II)-(N/O)6 complex. It is also of interest to compare the structures of Zn(II)-bound Zur (FurB) from M. tuberculosis (Lucarelli et al., 2007) and Ni(II)-bound Nur from S. coelicolor (An et al., 2009) with PerR to gain insight into metal selectivity of Fur proteins and general mechanisms of metalloregulation of operator DNA binding. It is striking that a domain-bridging Zn(II) complex like that in PerR is also formed in Zur, but one that assumes tetrahedral metal coordination, the most common coordination geometry for Zn(II). In contrast to PerR, the Zn(II)-Zur structure is rather open, and a major conformational change seems likely on DNA binding (Lucarelli et al., 2007).
There are many similarities in the Nur and PerR structures, including a “closed” conformation, in which the β-wings, like in PerR and P. aureginosa Fur (Pohl et al., 2003), are modeled to pack into the minor groove in the middle of the operator (An et al., 2009) (Fig. 1C). Two pairs of bound Ni(II) ions are found in the Nur structure. Strikingly, each Ni(II) ion of one pair forms a square planar complex, a common coordination geometry for Ni(II) with biological ligands. These Ni(II) sites largely overlap the regulatory Fe(II)/Mn(II) sites in PerR, including metal coordination by a distal histidine from the DBD. However, the authors of that study argue that the other pair of coordinately unsaturated, octahedrally bound Ni(II) ions in the β-wing region not previously observed in any Fur family regulator, are the primary Ni(II)-sensing sites in Nur, a proposal with parallels to the Ni(II) sensor E. coli NikR (Leitch et al., 2007). Taken collectively, these studies underscore the need for DNA-bound structures of metal-activated Fur family repressors, while emphasizing the challenge in assigning regulatory functions for metal ions in Fur proteins that bind multiple metals on the basis of structures alone. The compelling work on the Mn(II)- and Fe(II)-activated conformations of PerR reported by Duarte, Latour and coworkers in this issue represents a significant advance in our understanding of PerR function and Fur repressor family structure in general (Jacquamet et al., 2009).
References
- An YJ, Ahn BE, Han AR, Kim HM, Chung KM, Shin JH, Cho YB, Roe JH, Cha SS. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp198. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Helmann JD, Wu MF, Gaballa A, Kobel PA, Morshedi MM, Fawcett P, Paddon C. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J Bacteriol. 2003;185:243–253. doi: 10.1128/JB.185.1.243-253.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquamet L, Traoré D, Ferrer J-L, Proux O, Hazemann J-L, Nazarenko E, Ghazouni AE, Caux-Thang C, Duarte V, Latour J-M. Structural characetrization of the active form of PerR: Insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol Microbiol. 2009;72 doi: 10.1111/j.1365-2958.2009.06753.x. in press. [DOI] [PubMed] [Google Scholar]
- Lee C, Lee SM, Mukhopadhyay P, Kim SJ, Lee SC, Ahn WS, Yu MH, Storz G, Ryu SE. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J Biol Chem. 2006a;281:23567–23578. doi: 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006b;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. Nickel-Specific Response in the Transcriptional Regulator, Escherichia coli NikR. J Am Chem Soc. 2007;129:5085–5095. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J Biol Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- Paget MS, Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- Traore DA, El Ghazouani A, Ilango S, Dupuy J, Jacquamet L, Ferrer JL, Caux-Thang C, Duarte V, Latour JM. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol Microbiol. 2006;61:1211–1219. doi: 10.1111/j.1365-2958.2006.05313.x. [DOI] [PubMed] [Google Scholar]
- Traore DAK, Ghazouani AE, Jacquamet L, Borel F, Ferrer JL, Lascoux D, Ravanat JL, Jaquinod M, Blondin G, Caux-Thang C, Duarte V, Latour JM. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat Chem Biol. 2009;5:53–59. doi: 10.1038/nchembio.133. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Nat Acad Sci USA. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]