Abstract
microRNAs (miRNAs) were originally discovered as regulators of developmental in C. elegans. Recent results have revealed that miRNAs also regulate developmental timing in plants, and have provided a long-awaited molecular connection between the juvenile-to-adult transition and flowering. Specifically, the transition from juvenile to adult development in flowering plants is regulated by two temporally-expressed microRNAs, miR156 and miR172. These miRNAs target two families of plant-specific transcription factors (respectively, SBP-box and AP2-like factors), that cooperate to regulate phase-specific vegetative traits, as well as genes involved in flowering. Small RNAs have also been shown to play a role in the transition between different stages of gametophyte development in the moss Physcomitrella patens. The use of small RNAs for temporal regulation is therefore quite ancient in plants.
Introduction
Plants undergo several major changes in body plan during the course of their life cycle. The most significant of these changes occur during the switch between the haploid, gamete-producing (gametophyte) and diploid, spore-producing, (sporophyte) phases of the life cycle. Additional, sometimes quite dramatic, transformations occur during the development of both the gametophyte and the sporophyte. These include changes in vegetative morphology, an increase in reproductive competence and, finally, the production of structures involved in sexual reproduction. Goebel [1] termed the early stage of the development of the gametophyte and the sporophyte the “juvenile” phase, and the later phase the “adult” phase, and drew attention to the similarity between the juvenile-to-adult transition in plants and developmental transitions in animals, such as metamorphosis. Recent studies suggest that this similarity may extend from the overt phenomenology of these events to their molecular mechanisms.
It is now apparent that in plants, as in animals [2], small RNAs play central roles in the timing of developmental transitions. In higher plants, interest has focused on two temporally-regulated miRNAs, miR156 and miR172. Both of these miRNAs target plant-specific families of transcription factors, and recent studies have provided new insights into the function of these transcription factors in both the juvenile-to-adult transition and floral induction. Small RNAs have also been found to regulate the transition between different stages of gametophyte development in moss, demonstrating that they have an ancient role in developmental regulation in plants.
Phase change during sporophytic development: the function of miR156
The shoot of a flowering plant begins development in the juvenile phase. This phase is characterized by a variety of morphological traits including the shape, size, and pattern of epidermal differentiation of the leaf blade, and by the insensitivity of the shoot to floral stimuli [3]. The transition to the adult phase (vegetative phase change) is marked by changes in leaf morphology and by an increase in reproductive competence. Flower production begins during the adult phase, at a time that is dependent on endogenous signals as well as environmental factors, such as mineral nutrition, temperature, and photoperiod [4].
It is now apparent that miR156 is a key regulator of vegetative phase change in both Arabidopsis, maize, and rice. In Arabidopsis, this discovery was the result of molecular analyses of HASTY. Loss-of-function mutations of HASTY were originally identified in screens for mutations that accelerated vegetative phase change [5]. Given that HASTY is the Arabidopsis ortholog of the miRNA nuclear export receptor Exportin5 [6], we hypothesized that miRNA export must be important for the juvenile-to-adult transition in Arabidopis [7]. Analysis of the levels of various miRNAs in hst revealed that miR156 is expressed at a significantly higher level in juvenile shoots than in adult shoots. Furthermore, miR156 is strongly reduced at both stages in hst mutants, leading to the hypothesis that miR156 promotes juvenile development in Arabidopsis. The discovery that constitutive expression of mR156 significantly delayed both vegetative phase change and flowering demonstrated that miR156 is sufficient for the expression of the juvenile phase, and provided the first direct evidence for its role in phase change (Figure 1A)[8].
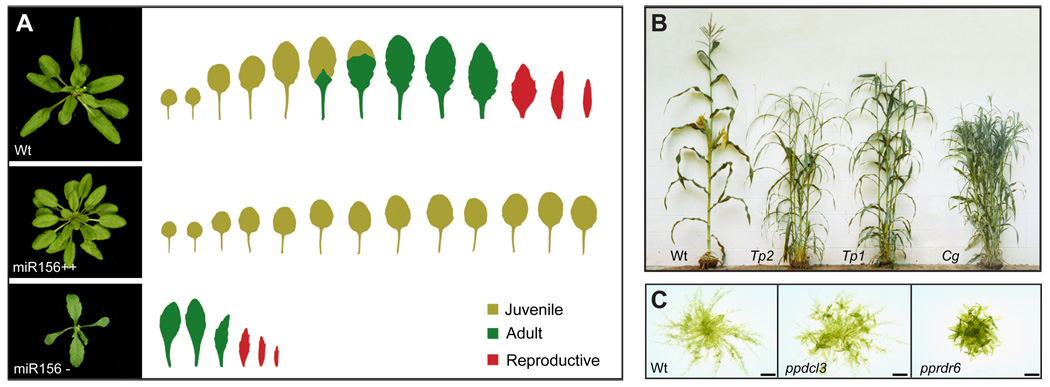
Figure 1.
The phenotypes of genes involved in phase change in Arabidopsis, maize and P. patens. A) The phenotypes of wild-type Arabidopsis (Wt), and a transgenic plant constitutively expressing miR156a under the regulation of the CaMV 35S promoter (miR156++), and a plant constitutively expressing a miR156 target site mimic under the regulation of the 35S promoter (miR156- ); this mimic reduces the activity of miR156 (18). Over-expression of miR156 prolongs the expression of the juvenile phase, while reducing the acitivity of miR156 has the opposite effect. B) The phenotype of wild type maize, and dominant mutations of three different miR156 loci that cause miR156 to be over-expressed. All three mutations prolong the expression of the juvenile phase and transform reproductive structures into leaves. C) A wild type P. patens gametophyte in the juvenile stage of development, and mutants lacking ppDCL3 and ppRDR6. Both of these mutants have begun to produce leafy gametophores; figure modified with permission from (51).
Evidence that miR156 is also required for the juvenile phase in Arabidopsis comes from an analysis of SQUINT (SQN)--the Arabidopsis ortholog of the co-chaperone Cyclophilin-40 [9]--and from the phenotype of transgenic plants in which miR156 is inactive. sqn mutations produce a precocious phenotype that is associated with a decrease in the activity of miR156 and an increase in the expression of its SPL targets [10]. The observation that this phenotype can be nearly completely suppressed by a transgene that over-expresses miR156 indicates that this phenotype is attributable to the decreased activity of miR156. Additional evidence for the importance of miR156 is provided by the observation that plants over-expressing a miR156 “sponge” sequence lack the juvenile phase and flower early [11]. These results demonstrate that miR156 is both necessary and sufficient for the juvenile phase.
In maize, the route to miR156 was less circuitous. The starting point was a classic mutation, Corngrass (Cg), that has a striking effect on vegetative and reproductive morphology (Figure 1B) [12]. Cg has a similar phenotype to dominant gain-of-function mutations of two other genes in maize, teopod1(tp1) and teopod2 (tp2) (Figure 1B)[13]. All of these mutations prolong the expression of the juvenile phase, delay the transition from vegetative to reproductive development, and transform floral organs into leaves [13–15]. Chuck and colleagues recently demonstrated that cg corresponds to a polycistronic gene encoding miR156 b/c, which is over-expressed in the Cg mutant [16]. The Tp1 and Tp2 mutations have since been found to cause over-expression of other miR156 genes in maize (M. Y. Park, M. de la luz Gutierrez-Nava and R. S. Poethig, unpublished results). Over-expression of miR156 in rice [17] produces a branching and inflorescence phenotype similar to that of of Cg, Tp1 and Tp2, implying that miR156 also promotes juvenile development in this species. However, this hypothesis is difficult to test because the juvenile and adult phases of vegetative development are poorly differentiated in rice. Comparative sequence analysis of the miR156 b/c locus in cultivated and wild accessions of rice suggests that this locus played an important role in the evolution of cereal crops [18], as originally predicted by Singleton [12].
Targets of miR156
miR156 directly represses the expression of members of the squamosa promoter binding protein family of transcription factors [19]. These plant-specific proteins were first identified in Antirrhinum majus by their ability to bind to the promoter of the floral meristem identity gene SQUAMOSA [20], and share a highly conserved sequence (the SBP-box) that is necessary and sufficient for DNA binding [21,22]. SBP-box genes are present in every major plant taxon and include genes with and without a miR156 target site [23–27]. This observation suggests that miR156 and its SBP-box targets play an important role in vegetative phase change throughout the plant kingdom.
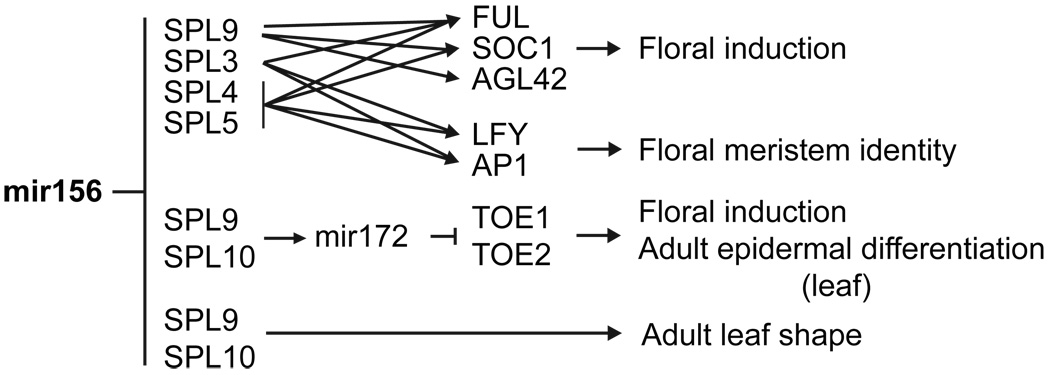
SBP-box genes can be grouped into 7 major clades in flowering plants. Four of these clades include genes regulated by miR156 [25]. Loss-of-function and/or gain-of-function phenotypes have been described for genes in 6 of the 7 clades in Arabidopsis [8,28–34], and for two SBP-box genes in maize [35,36] and one gene in tomato [37]. In Arabidopsis, the miR156-regulated clades are SPL3/4/5, SPL9/15, SPL2/10/11, and SPL6/SPL13a/b [25,31]. Although technical issues--such as the tight linkage between paralogous genes--and functional redundancy have made it difficult to determine the precise function of many of the SPL genes in Arabidopsis, the gain-of-function phenotypes of plants expressing miR156-resistant forms of these genes has provided insights into the processes they control. SPL3, SPL4 and SPL5 promote an adult pattern of epidermal differentiation in leaves, but have little or no effect on leaf shape [8, 19]. Over-expression of all three genes also causes early flowering [8,28]. SPL9, SPL15, SPL10 and SPL11 promote all adult phase-specific aspects of leaf morphology; these genes also decrease the rate of leaf production, and promote flowering [11,32,33]. However, SPL10 and SPL11 have different effects on these traits than SPL9 and SPL15 [11]. These results suggest that SPL genes operate in several functionally distinct pathways that promote adult phase vegetative traits and flowering and miR156 functions to coordinately repress these pathways early in shoot development (Figure 2).
Figure 2.
The function of miR156 and its targets in Arabidopsis thaliana.
Although the mechanism by which SPL genes regulate vegetative morphology is unknown, two recent studies have provided insights into their role in flower development (Figure 2) [38,39]. Analyses of gene expression and chromatin immunoprecipitation experiments demonstrate that SPL3, SPL4, SPL5 and SPL9 promote flowering and floral meristem identity by directly promoting the transcription of key regulators of these traits. SPL9 promotes the transcription of FUL, SOC1 and AGL42 [39], SPL3 promotes the transcription of FUL, LFY, and AP1 [38,39], and SPL4/SPL5 promote the transcription of FUL, SOC1, LFY, AP1, and FT [38]‥ Interestingly, these studies also indicate that SPL3 and SPL9 act in parallel to the floral inducer, FT. These results provide an explanation for the increase in reproductive competence that accompanies vegetative phase change, and suggest that these SPL genes operate in a novel pathway for the control of flowering in Arabidopsis.
miR172 acts downstream of miR156
In contrast to miR156, miR172 promotes flowering [40–42] and adult patterns of epidermal differentiation in leaves [11]. miR172 is expressed in an inverse pattern to miR156, increasing in abundance during shoot development [11,40,42,43]. In Arabidopsis, the temporal expression pattern of miR172 is regulated by miR156 through its effect on SPL genes that promote the transcription of miR172, one of which (SPL9) directly regulates the transcription of miR172b [11]. miR156 also regulates miR172 expression in maize [16], but the SPL genes that mediate this function remain to be determined.
miR172 specifically targets AP2, TOE1, TOE2, TOE3, SMZ, and SNZ in Arabidopsis [40,41,44] and 6 AP2-like genes in maize, including gl15 [45]. Epistasis experiments indicate that TOE1, TOE2 and Gl15 are required for the effect of miR156 on epidermal identity, implying that they act downstream of mIR156 [11,46,47]. Over-expression of these genes delays flowering, whereas loss-of-function mutations in TOE1 and TOE2 cause early flowering [40,42,43]. It is therefore likely that TOE1, TOE2 and Gl15 contribute to the effect of miR156 on flowering time. miR156 acts by regulating the expression of SPB-box genes. Although existing data are consistent with the idea that SBP-box factors regulate the expression of TOE1, TOE2 and Gl15 via their effect on the expression of miR172 (Figure 2), the possibility that SBP-box genes directly promote the transcription of these AP2-like genes has not been excluded.
Phase change during gametophyte development
In non-vascular plants, such as the moss P patens, the gametophyte is typically larger and more complex than the sporophyte, so phase change is most readily observed during this stage of the life cycle. The P. patens gametophyte initially grows as an irregularly branched monofilament known as the protonema. The protonema eventually gives rise to larger leaf-bearing shoots (gametophores) which produce a series of increasingly more complex leaves before producing gamete-producing structures, the antheridia and archegonia.
P. patens produces several different classes of small RNAs, including miRNAs [48] trans-acting siRNAs (tasiRNAs) [49,50], and 23 nt siRNAs derived from transposon-rich regions of the genome [51]. Although there is still no evidence for the involvement of miRNAs in phase change in P. patens, two types of siRNAs appear to play a role in this phenomenon. Gametophytes lacking the RNA dependent RNA polymerase PpRDR6 [49] or the Dicer-like gene PpDCL3 [51] produce gametophores unusually early (Figure 1C), implying that the siRNAs produced by these genes promote the juvenile, protonemal phase of development. pprdr6 mutants completely lack tasiRNAs derived from the ppTAS3a–d loci [49,51]. ppTAS3a–d have no sequence similarity to the TAS3 locus in Arabidopsis, but are produced by the same mechanism as TAS3 and regulate the same targets as TAS3--the transcription factors ARF3 and ARF4 [50,52]. Because Arabidopsis mutants that specifically lack TAS3 tasiRNAs have a precious phenotype [53,54], it has been suggested that TAS3a–d may regulate the juvenile-to-adult transition in moss [49]. This hypothesis is difficult to test because it is technically challenging to mutate all four paralogs of TAS3 in P. patens. Approaching this question by mutating the targets of TAS3 is also a difficult proposition because in addition to ppARF3 and ppARF4, TAS3a–d target several AP2-like genes [49,52]. ppdcl3 mutants have a weaker precocious phentoype than pprdr6 (Figure 1C). These mutants lack 23 nt siRNAs produced by 48 loci in P. patens [51]; determining which of these loci is responsible for the developmental phenotype of ppdcl3 is also a major challenge.
Conclusion
The use of small RNAs for the regulation of developmental timing originated early in plant evolution and has been conserved. Whether this reflects the general importance of small RNAs in gene regulation, or is a consequence of their special regulatory properties is still unknown; however, it is intriguing that they were also selected for this purpose in animals. Genetic analyses of vegetative phase change in Arabidopsis and maize have revealed that the same genes regulate this process in both species. It will be important to determine if these genes also regulate this process in other major plant lineages (pteridophytes and gymnosperms) and in woody plants, where the regulation of vegetative phase change is a major issue and has significant economic implications.
Acknowledgements
Stewart Gillmor, Keith Earley and Matthew Willmann provided helpful comments on this manuscript. Research on developmental timing the author’s laboratory is supported by grants from the NIH and NSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goebel K. In: Organography of plants Part I. General organography. Balfour IB, translator. Oxford: Clarendon Press; 1900. [Google Scholar]
- 2.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 3.Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–336. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- 4.Wilkie JD, Sedgley M, Olesen T. Regulation of floral initiation in horticultural trees. J Exp Bot. 2008;59:3215–3228. doi: 10.1093/jxb/ern188. [DOI] [PubMed] [Google Scholar]
- 5.Telfer A, Poethig RS. HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development. 1998;125:1889–1898. doi: 10.1242/dev.125.10.1889. [DOI] [PubMed] [Google Scholar]
- 6.Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003;130:1493–1504. doi: 10.1242/dev.00362. [DOI] [PubMed] [Google Scholar]
- 7.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521.This paper provides the first evidence that miR156 regulates developmental timing, and demonstrates that SPL3, SPL4 and SPL5 are targets of this miRNA, and regulate flowering time.
- 9.Berardini TZ, Bollman K, Sun H, Poethig RS. Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science. 2001;291:2405–2407. doi: 10.1126/science.1057144. [DOI] [PubMed] [Google Scholar]
- 10.Smith MR, Willmann MR, Wu G, Berardini TZ, Möller B, Weiers D, Poethig RS. Cyclophilin40 is required for miRNA activity in Arabidopsis. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0812729106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;137 doi: 10.1016/j.cell.2009.06.031. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singleton WR. Inheritance of Corngrass as a macromutation in maize, and its possible significance as an ancestral type. Am. Nat. 1951;85:81–96. [Google Scholar]
- 13.Poethig RS. Heterochronic mutations affecting shoot development in maize. Genetics. 1988;119:959–973. doi: 10.1093/genetics/119.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galinat WC. Corn grass. II. Effect of the corn grass gene on the development of the maize inflorescence. Am. J. Bot. 1954;41:803–806. [Google Scholar]
- 15.Galinat WC. The corn grass and teopod loci involve phase change. Maize Genet Coop. News Lett. 1966;40:102–103. [Google Scholar]
- 16.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001.Corngrass, a classic mutation in maize and a potential target of selection during the evolution of maize, is shown to correspond to miR156a/c.
- 17.Xie K, Wu C, Xiong L. Genomic organization, differential expression and interaction of SPL transcription factors and microRNA156 in rice. Plant Physiol. 2006 doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Zhu QH, Guo X, Gui Y, Bao J, Helliwell C, Fan L. Molecular evolution and selection of a gene encoding two tandem microRNAs in rice. FEBS Lett. 2007;581:4789–4793. doi: 10.1016/j.febslet.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet. 1996;250:7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Nunokawa E, et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol. 2004;337:49–63. doi: 10.1016/j.jmb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Birkenbihl RP, Jach G, Saedler H, Huijser P. Functional dissection of the plant-specificSBP-domain overlap of the DNA-binding and nuclear localization domains. J Mol Biol. 2005;352:585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Cardon G, Hohmann S, Klein J, Nettesheim K, Saedler H, Huijser P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/s0378-1119(99)00308-x. [DOI] [PubMed] [Google Scholar]
- 24.Riese M, Hohmann S, Saedler H, Munster T, Huijser P. Comparative analysis of the SBP- box gene families in P. patens and seed plants. Gene. 2007;401:28–37. doi: 10.1016/j.gene.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Wang X, Gu S, Hu Z, Xu H, Xu C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene. 2008;407:1–11. doi: 10.1016/j.gene.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 27.Hultquist JF, Dorweiler JE. Feminized tassels of maize mop1 and ts1 mutants exhibit altered levels of miR156 and specific SBP-box genes. Planta. 2008;229:99–113. doi: 10.1007/s00425-008-0813-2. [DOI] [PubMed] [Google Scholar]
- 28.Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 1997;12:367–377. doi: 10.1046/j.1365-313x.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- 29.Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003;15:1009–1019. doi: 10.1105/tpc.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone JM, Liang X, Nekl ER, Stiers JJ. Arabidopsis AtSPL14, a plant-specific SBP- domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005;41:744–754. doi: 10.1111/j.1365-313X.2005.02334.x. [DOI] [PubMed] [Google Scholar]
- 31.Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP-box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP- box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell. 2009;21:347–361. doi: 10.1105/tpc.108.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 1997;11:616–628. doi: 10.1101/gad.11.5.616. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. The microRNA-regulated SBP-box transcription factor SPL3 is a direct transcriptional activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell. 2009;16 doi: 10.1016/j.devcel.2009.06.007. in press.This is one of two papers first describing a molecular link between genes involved in vegetative phase and genes with well-described roles in flowering time and floral meristem identity
- 39.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define and endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;137 doi: 10.1016/j.cell.2009.06.014. in press.This is one of two papers first describing a molecular link between genes involved in vegetative phase and genes with well-described roles in flowering time and floral meristem identity
- 40.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238.This paper describes a role for miR172 and its targets TOE1 and TOE2 in flowering time. It is the first demonstration of a role for miRNAs in developmental timing in plants.
- 41.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. The GIGANTEA- regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci U S A. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 45.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- 46.Evans MM, Passas HJ, Poethig RS. Heterochronic effects of glossy15 mutations on epidermal cell identity in maize. Development. 1994;120:1971–1981. doi: 10.1242/dev.120.7.1971. [DOI] [PubMed] [Google Scholar]
- 47.Moose SP, Sisco PH. Glossy15 controls the epidermal juvenile-to-adult phase transition in maize. Plant Cell. 1994;6:1343–1355. doi: 10.1105/tpc.6.10.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 49.Talmor-Neiman M, Stav R, Klipcan L, Buxdorf K, Baulcombe DC, Arazi T. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 2006;48:511–521. doi: 10.1111/j.1365-313X.2006.02895.x.This paper demonstrates that siRNAs regulate the transition from juvenile to adult development in moss, and provides the first evidence that siRNAs have a function in non-vascular plants.
- 50.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 51.Cho SH, Addo-Quaye C, Coruh C, Arif MA, Ma Z, Frank W, Axtell MJ. Physcomitrella patens DCL3 is required for 22–24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 2008;4:e1000314. doi: 10.1371/journal.pgen.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hunter C, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol. 2003;13:1734–1739. doi: 10.1016/j.cub.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]