Abstract
We present a particle-based nonlinear filtering scheme, related to recent work on chainless Monte Carlo, designed to focus particle paths sharply so that fewer particles are required. The main features of the scheme are a representation of each new probability density function by means of a set of functions of Gaussian variables (a distinct function for each particle and step) and a resampling based on normalization factors and Jacobians. The construction is demonstrated on a standard, ill-conditioned test problem.
Keywords: pseudo-Gaussian, Jacobian, chainless sampling
There are many problems in science in which the state of a system must be identified from an uncertain equation supplemented by a stream of noisy data (ref. 1). A natural model of this situation consists of a stochastic differential equation (SDE)
where x = (x 1,x 2,…,x m) is an m-dimensional vector, w is an m-dimensional Brownian motion, f is an m-dimensional vector function, and g(x,t) is an m by m diagonal matrix. The Brownian motion encapsulates all the uncertainty in this equation. The initial state x(0) is assumed given and may be random as well.
As the experiment unfolds, it is observed, and the values b n of a measurement process are recorded at times t n. For simplicity, assume t n = nδ, where δ is a fixed time interval and n is an integer. The measurements are related to the evolving state x(t) by
where h is a k-dimensional, generally nonlinear, vector function with k ≤ m, G is a diagonal matrix, x n = x(nδ), and W n is a vector whose components are independent Gaussian variables of mean 0 and variance 1, independent also of the Brownian motion in Eq. 1. The task is to estimate x on the basis of Eq. 1 and the observations in Eq. 2.
If the system in Eq. 1 and Eq. 2 are linear and the data are Gaussian, the solution can be found via the Kalman–Bucy filter (2). In the general case, it is natural to try to estimate x via its evolving probability density. The initial state x is known and so is its probability density; all one has to do is evaluate sequentially the density P n+1 of x n+1 given the probability densities P k of x k for k ≤ n and the data b n+1. This evaluation can be done by following “particles” (replicas of the system) whose empirical distribution approximates P n. In a Bayesian filter (3,9–10), one uses the probability density function (PDF) P n and Eq. 1 to generate a prior density, and then one uses the new data b n+1 to generate a posterior density P n+1. In addition, one has to sample backward to take into account the information each measurement provides about the past as well as to avoid having too many identical particles. This can be very expensive, in particular because the number of particles needed can grow catastrophically (11, 12).
In this paper, we offer an alternative to the standard approach in which P n+1 is sampled more directly and backward sampling is done without chains (13). Our direct sampling is based on a pseudo-Gaussian representation of a variable with density P n+1, i.e. a representation by a collection of functions of Gaussian variables with sample-dependent parameters. The construction is related to chainless sampling as described in ref. 13. The idea in chainless sampling is to produce a sample of a large set of variables by sequentially sampling a growing sequence of nested, conditionally independent subsets, with discrepancies balanced by sampling weights. As observed in refs. 14 and 15, chainless sampling for a SDE reduces to interpolatory sampling, as explained below. Our construction will be explained in the following sections through an example where the position of a ship is deduced from the measurements of an azimuth, already used as a test bed in a number of previous papers (7, 16, 17). We call our sampling “implicit” by analogy with implicit schemes for solving differential equations, where the determination of a next value requires the solution of algebraic equations.
If the SDE in Eq. 1 and the observation function in Eq. 2 are linear, our construction becomes a reformulation of sequential importance sampling with an optimal importance function, see refs. 5 and 6.
Sampling by Interpolation and Iteration
First, we explain how to sample via interpolation and iteration in a simple problem, related to the example and the construction in ref. 14. Consider the scalar SDE
where β is a constant. We want to find sample paths x = x(t) , 0 ≤ t ≤ 1, subject to the conditions x(0) = 0, x(1) = X.
Let N(a, v) denote a Gaussian variable with mean a and variance v. We first discretize Eq. 3 on a regular mesh t 0,t 1,…,t N, where t n = nδ, δ = 1/N, 0 ≤ n ≤ N, with x n = x(t n), and, following ref. 14, use a balanced, implicit discretization (18, 19):
where and Wn+1 is N(0,β/N). The joint probability density of the variables x 1,…,x N−1 is Z −1 exp(−∑0N−1 V n), where Z is the normalization constant and
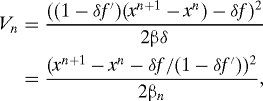 |
where f,f′ are functions of the x n,t n, and βn = βδ/(1 − δf′)2 (20). One can obtain sample solutions by sampling this density, e.g. by Markov chain Monte Carlo, or one can obtain them by interpolation (chainless sampling), as follows.
Let a n = f(x n,t n)δ/(1 − δf′(x n,t n)). Consider first the special case f(x,t) = f(t), so that in particular f′ = 0; we recover a version of a Brownian bridge (21). Each increment x n+1 − x n is now a N(a n,β/N) variable, with the a n = f(t n)δ known explicitly. Let N be a power of 2. Consider the variable x N/2. On the one hand,
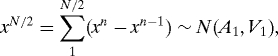 |
where A 1 = ∑1N/2 a n, V 1 = β/2. On the other hand,
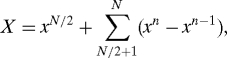 |
so that
with
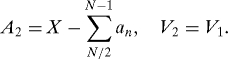 |
The PDF of x N/2 is the product of the two PDFs; one can check that
where , , and ; e −ϕ is the probability of getting from the origin to X, up to a normalization constant.
Pick a sample ξ1 from the N(0,1) density; one obtains a sample of x N/2, by setting . Given a sample of x N/2, one can similarly sample x N/4,x 3N/4, then x N/8, x 3N/8, etc., until all the x j have been sampled. If we define ξ = (ξ1,ξ2,…,ξN−1), then for each choice of ξ we find a sample x = (x 1,…,x N−1), such that
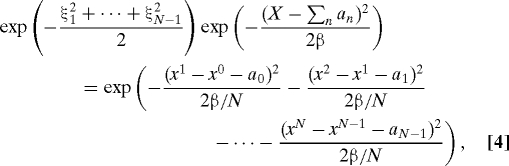 |
where the factor on the left is the probability of the fixed end value X up to a normalization constant. In this linear problem, this factor is the same for all the samples and therefore harmless. The Jacobian J of the variables x 1,…,x N−1 with respect to the variables ξ1,…,ξN−1 can be seen to be a constant independent of the sample and is also immaterial. One can repeat this sampling process for multiple choices of the variables ξ; each sample of the corresponding set of x n is independent of any previous samples of this set.
Now return to the general case. The functions f, f′ are now functions of the x j. We obtain a sample of the probability density we want by iteration. The simplest iteration proceeds as follows. First, pick ξ = (ξ1,ξ2,…,ξN−1), where each ξl, l = 1,…,N − 1 is drawn independently from the N (0,1) density (this vector remains fixed during the iteration). Make a first guess x 0 = (x 01,x 02,…,x 0N−1) (for example, if X ≠ 0, pick x 0 = 0). Evaluate the functions f,f′ at x j (note that now f′ ≠ 0, and therefore the variances of the various displacements are no longer constants). We are back in the previous case and can find values of the increments x j+1n+1 − x j+1n corresponding to the values of f,f′ we have. Repeat the process starting with the new iterate. If the vectors x j converge to a vector x = (x 1,…,x N−1), we obtain, in the limit, Eq. 4, where now on the right side β depends on n so that β = βn, and both a n,βn are functions of the final x. The left hand side of Eq. 4 becomes
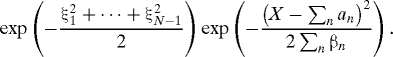 |
The factor is now different from sample to sample and changes the relative weights of the different samples. The Jacobian J of the x variables with respect to the ξ variables, is now also a function of the sample. It can be evaluated step by step the last time the iteration is carried out, either by an implicit differentiation or by repeating the iteration for a slightly different value of the relevant ξ and differencing. In averaging, one should take the product F|J| as weight or resample as described at the end of the following section. In order to obtain more uniform weights, one also can use the strategies in refs. 13 and 14.
One can readily see that this iteration converges if
where K is the Lipshitz constant of f, and L is the length of the interval on which one works (here L = 1). If this iteration fails to converge, more sophisticated iterations are available. One should of course choose N large enough so that the results are converged in N. We do not provide more details here because they are extraneous to our purpose, which is to explain chainless/interpolatory sampling and the use of reference variables in a simple context.
Finally, we chose the reference density to be a product of independent N(0,1) variables, which is a convenient but not mandatory choice. In applications, one may well want to choose other variances or make the variables be dependent.
The Ship Azimuth Problem
The problem we focus on is discussed in refs. 7, 16 and 17, where it is used to demonstrate the capabilities of particular filters. A ship sets out from a point (x 0,y 0) in the plane and undergoes a random walk,
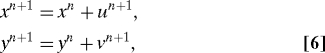 |
for n ≥ 0, u n+1 = N(u n,β), v n+1 = N(v n,β), i.e., each displacement is a sample of a Gaussian random variable whose variance β does not change from step to step and whose mean is the value of the previous displacement. An observer makes noisy measurements of the azimuth arctan(y n/x n) (for the sake of definiteness, we take the branch in −[π/2,π/2)), recording
where the variance s is also fixed; here, the observed quantity b n is scalar and is not denoted by a boldfaced letter. The problem is to reconstruct the positions x n = (x n,y n) from Eqs. 6 and 7. We take the same parameters as ref. 7: x 0 = 0.01, y 0 = 20, u 1 = 0.002, v 1 = −0.06, β = 1 · 10−6, s = 25 · 10−6. We follow numerically M particles, all starting from X i0 = x 0, Y i0 = y 0, as described in the following sections, and we estimate the ship's position at time nδ as the mean of the locations X in = (X in, Y in),i = 1,…,M of the particles at that time. The authors of ref. 7 also show numerical results for runs with varying data and constants; we discuss those refinements in the numerical results section below.
Forward Step
Assume that we have a collection of M particles X n at time t n = nδ whose empirical density approximates P n; now we find displacements U n+1 = (U n+1,V n+1) such that the empirical density of X n+1 = X n + U n+1 approximates P n+1. P n+1 is known implicitly: It is the product of the density that can be deduced from the dynamics and the one that comes from the observations, with the appropriate normalization. If one is given sample displacements, their probabilities p (the densities P n+1 evaluated at the resulting positions X n+1) can be evaluated, so p is a function of U n+1, p = p(U n+1). For each particle i, we are going to sample a reference density, obtain a reference sample of probability ρ, and then attempt to solve (by iteration) the equation
to obtain U in+1.
Define f(x,y) = arctan(y/x) and f n = f(X n,Y n). We are working on one particle at a time, so the index i can be temporarily suppressed. Pick two independent samples ξx, ξy from a N(0,1) density (the reference density in the present calculation), and set ; the variables ξx, ξy remain unchanged until the end of the iteration. We are looking for displacements U n+1, V n+1, and parameters a x,a y,v x,v y,ϕ, such that:
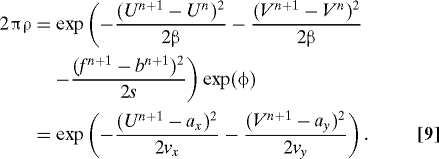 |
The first equality states what we wish to accomplish. Indeed, divide this first equality by exp(ϕ). The equality now defines implicitly new displacements U n+1, V n+1, functions of ξx,ξy, with the probability of these displacements with respect to P n+1 given (up to an unknown normalization constant). The second equality in Eq. 9 defines parameters a x,a y,v x,v y, (all functions of X n and ξx,ξy) that will be used to actually find the displacements U n+1, V n+1. One should remember that in our example, the mean of U n+1 before the observation is taken into account is U n, with a similar statement for V n+1.
We use the second equality in Eq. 9 to set up an iteration for vectors U n+1,j(= U j for brevity) that converges to U n+1. Start with U 0 = 0. We now explain how to compute U j+1 given U j.
Approximate the observation in Eq. 7 by
where the derivatives f x,f y are, like f, evaluated at X j = X n + U j, i.e., approximate the observation equation by its Taylor series expansion around the previous iterate. Define a variable ηj+1 = (f x · U j+1 + f y · V j+1)/. The approximate observation equation says that ηj+1 is a N(a 1,v 1) variable, with
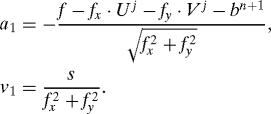 |
On the other hand, from the equations of motion one finds that ηj+1 is N(a 2,v 2), with a 2 = (f x · U n + f y · V n)/ and v 2 = β. Hence the PDF of ηj+1 is, up to normalization factors,
where , , .
We can also define a variable η+j+1 that is a linear combination of U j+1, V j+1 and is uncorrelated with ηj+1:
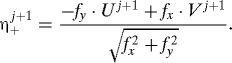 |
The observations do not affect η+j+1, so its mean and variance are known. Given the means and variances of ηj+1, η+j+1 one can easily invert the orthogonal matrix that connects them to U j+1, V j+1 and find the means and variances a x,v x of U j+1 and a y,v y of V j+1 after their modification by the observation (the subscripts on a,v are labels, not differentiations). Now one can produce values for U j+1,V j+1:
where ξx, ξy are the samples from N(0,1), chosen at the beginning of the iteration. This completes the iteration.
This iteration converges to X n+1, such that f(X n+1) = b n+1 + N(0,s), and the phases ϕj converge to a limit ϕ = ϕi, where the particle index i has been restored. The time interval over which the solution is updated in each step is short, and there are no problems with convergence, either here or in the next section (see Eq. 5); in all cases, the iteration converges in a small number of steps.
We now calculate the Jacobian J of the U n+1 variables with respect to ξx,ξy. The relation between these variables is laid out in the first equality of Eq. 9. Take the log of this equation, partition it into a part parallel to the direction in which the observation is made (i.e., parallel to the vector (f x,f y)) and a part orthogonal to that direction. Because the increment U n+1, V n+1 is now known, the evaluation of J is merely an exercise in implicit differentiation. J can also be evaluated numerically by finding the increment U n+1,V n+1 that corresponds to nearby values of ξx, ξy, and differencing.
Do this for all the particles and obtain new positions with weights W j = exp(−ϕj)|J j|, where J j is the Jacobian for the jth particle. One can get rid of the weights by resampling, i.e., for each of M random numbers θk,k = 1,…,M drawn from the uniform distribution on [0,1], choose a new kn+1 = Xin+1, such that A−1 ∑j=1i−1Wj < θk ≤ A−1 ∑j=1i W j (where A = ∑j=1M W j), and then suppress the hat. We have traded the usual Bayesian resampling based on the posterior probabilities of the samples for a resampling based on the normalizing factors of the several Gaussian densities; this is a worthwhile trade because in a Bayesian filter one gets a set of samples, many of which may have low probability with respect to P n+1, and here we have a set of samples, each one of which has high probability with respect to a PDF close to P n+1 (see Numerical Results and Conclusion).
Note also that the resampling does not have to be done at every step—for example, one can add up the phases for a given particle and resample only when the ratio of the largest cumulative weight exp(−∑(ϕi − log|J i|)) to the smallest such weight exceeds some limit L (the summation is over the weights accrued to a particular particle i since the last resampling). If one is worried by too many particles being close to each other (“depletion” in the Bayesian terminology), one can divide the set of particles into subsets of small size and resample only inside those subsets, creating a greater diversity. As will be seen in the numerical results section, none of these strategies will be used here, and we will resample fully at every step.
Finally, note that if the SDE in Eq. 1 and the observation in Eq. 2 are linear, and if at time nδ one is given the means and the covariance matrix of a Gaussian x, then our algorithm produces, in one iteration, the means and the covariance matrix of a standard Kalman filter.
Backward Sampling
The algorithm of the previous section is sufficient to create a filter, but accuracy, when the problem is not Gaussian, may require an additional step. Every observation provides information not only about the future but also about the past—it may, for example, tag as improbable earlier states that had seemed probable before the observation was made; in general, one has to go back and correct the past after every observation (this backward sampling is often misleadingly motivated solely by the need to create greater diversity among the particles in a Bayesian filter). As will be seen below, this backward sampling does not provide a significant boost to accuracy in the present problem, but it must be described for the filter to be of general use as well as be generalizable to problems involving smoothing.
Given a set of particles at time (n + 1)δ, after a forward step and maybe a subsequent resampling, one can figure out where each particle i was in the previous two steps and have a partial history for each particle i: X in−1,X in,X in+1 (if resamples had occurred, some parts of that history may be shared among several current particles). Knowing the first and the last members of this sequence, one can interpolate for the middle term as in the first example above, thus projecting information backward. This requires that one recompute U n.
Let U tot = U n + U n+1; in the present section, this quantity is assumed known and remains fixed. In the azimuth problem discussed here, one has to deal with the slight complication due to the fact that the mean of each displacement is the value of the previous one, so that two successive displacements are related in a slightly more complicated way than usual. The displacement U n is a N(U n−1,β) variable, and U n+1 is a N(U n,β) variable, so that one goes from X n−1 to X n+1 by sampling first a (2U n−1,4β) variable that takes us from X n−1 to an intermediate point P, with a correction by the observation half-way up this first leg, and then one samples a N(U tot,β) variable to reach X n+1, and this is done similarly for Y. Let the variable that connects X n−1 to P be U new, so that what replaces U n is U new/2. Accordingly, we are looking for a new displacement U new = (U new, V new) and for parameters a xnew,a ynew,v xnew,v ynew, such that
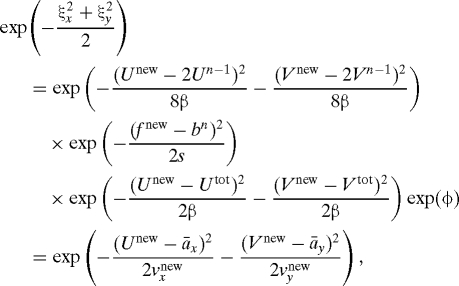 |
where f new = f(X n−1 + U new/2,Y n−1 + V new/2) and ξx, ξy are independent N(0,1) Gaussian variables. As in Eq. 9, the first equality embodies what we wish to accomplish—find displacements, which are functions of the reference variables that sample the new PDF at time nδt. The new PDF is defined by the forward motion, by the constraint imposed by the observation, and by knowledge of the position at time (n + 1)δt. The second equality states that this is done by finding particle-dependent parameters for a Gaussian density.
We again find these parameters as well as the displacements by iteration. Much of the work is separate for the X and Y components of the equations of motion, so we write some of the equations for the X component only. Again set up an iteration for variables U new,j = U j, which converge to U new. Start with U 0 = 0. To find U j+1 given U j, approximate the observation in Eq. 7, as before, by Eq. 10; define again variables ηj+1,η+j+1, one in the direction of the approximate constraint and one orthogonal to it. In the direction of the constraint, multiply the PDFs as in the previous section; construct new means a x1,a y1 and new variances v x1,v y1 at time n, taking into account the observation at time n as before. This also produces a phase ϕ = ϕ0.
Now take into account that the location of the ship at time n + 1 is known; this creates a new mean āx, a new variance v̄x, and a new phase ϕx, by , , , where a 1 = 2a 1,v 1 = 4v x1,a 2 = X tot,v 2 = β. Finally, find a new interpolated position (the calculation for V j+1 is similar, with a phase ϕy), and we are done. The total phase for this iteration is ϕ = ϕ0 + ϕx + ϕy. As the iterates U j converge to U new, the phases converge to a limit ϕ = ϕi. One also has to compute Jacobians and set up a resampling. After one has values for X new, a forward step gives corrected values of X n+1; one can use this interpolation process to correct estimates of X k by subsequent observations for k = n − 1, k = n − 2, …, or as many as are useful.
Numerical Results
Before presenting examples of numerical results for the azimuth problem, we discuss the accuracy one can expect. We run the ship once and record synthetic observations (with the appropriate noise), which will remain fixed. Then we find other ship trajectories compatible with these fixed observations as follows. We have 160 observations. We note that the maximum likelihood estimate of s given 160 observations is a random variable with mean zero and variance .11s. Then we make other runs of the ship, record the azimuths along the path and calculate the differences between these azimuths and the fixed observations. If the set of these differences in any run is a likely set of 160 samples of a N(0,s) variable (which is what the noise is supposed to be), then we declare that the new run is compatible with the fixed observations. We view the set of differences as likely if their empirical variance is within one standard variation (.11s) of the nominal variance s of the observation noise. One can impose further requirements (for example, one may demand that the empirical covariance of two successive noises be within a standard deviation of zero), but these turn out to be weaker requirements. To lighten the burden of computation, we make the new ship runs have fixed displacements in the observed direction (equal to those that the first ship experienced) and sample new displacements only in the direction orthogonal to the observed direction. We use the variability of the compatible runs as an estimate of the lower bound on the possible accuracy of the reconstruction.
In Table 1, we display the standard deviations of the differences between the resulting paths and the original path that produced the observations after the number of steps indicated there (the means of these differences are statistically indistinguishable from zero). This table provides an estimate of the accuracy we can expect. It is fair to assume that these standard deviations are underestimates of the uncertainty—a maximum variation of a single standard deviation in s is a strict requirement, and we allowed no variability in β. In particular, our construction, together with the particular set of directions in the linearized observation equations that arises with our data, conspire to make the error estimate in the x component unrealistically small.
Table 1.
Intrinsic uncertainty in the azimuth problem
| Step | x component | y component |
|---|---|---|
| 40 | 0.0005 | 0.21 |
| 80 | 0.004 | 0.58 |
| 120 | 0.010 | 0.88 |
| 160 | 0.017 | 0.95 |
If one wants reliable information about the performance of the filter, it is not sufficient to run the ship once, record observations, and then use the filter to reconstruct the ship's path, because the difference between the true path and the reconstruction is a random variable that may be accidentally small or large. We have therefore run a large number of such reconstructions and computed the means and standard deviations of the discrepancies between path and reconstruction as a function of the number of steps and of other parameters. In Tables 2 and 3, we display the means and standard deviations of these discrepancies (not of their mean!) in the the x and y components of the paths with 2,000 runs, at the steps and numbers of particles indicated, with no backward sampling. (ref. (7) used 100 particles). On average, the error is zero so that the filter is unbiased and the standard deviation of the discrepancies cannot be expected to be better than the lower bound of Table 1, and in fact it is compatible to that lower bound. The standard deviation of the discrepancy is not catastrophically larger with one particle (and no resampling at all!) than with 100—the main source of the discrepancy is the insufficiency of the data for accurate estimation of the trajectories. The more-sophisticated resampling strategies discussed above make no discernible difference here because they are unable to remedy the limitations of the dataset. One can check to see that backward sampling also does not make much difference for this problem, where the underlying motion is Gaussian and the variance of the observation noise is much larger than the variance in the model.
Table 2.
Mean and standard variation of the discrepancy between synthetic data and their reconstruction, 2,000 runs, no back step, 100 particles
| Number of steps |
x component |
y component |
||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| 40 | .0004 | 0.04 | 0.001 | 0.17 |
| 80 | −0.001 | 0.04 | −0.01 | 0.54 |
| 120 | −0.0008 | 0.07 | −0.03 | 1.02 |
| 160 | −0.002 | 0.18 | −0.05 | 1.62 |
Table 3.
Mean and standard variation of the discrepancy between synthetic data and their reconstruction, 2,000 runs, no back step, one particle
| Number of steps |
x component |
y component |
||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| 40 | −0.003 | 0.20 | −0.0008 | 0.19 |
| 80 | −0.01 | 0.54 | −0.019 | 0.56 |
| 120 | −0.01 | 0.84 | −0.027 | 1.04 |
| 160 | −0.016 | 0.94 | −0.02 | 1.62 |
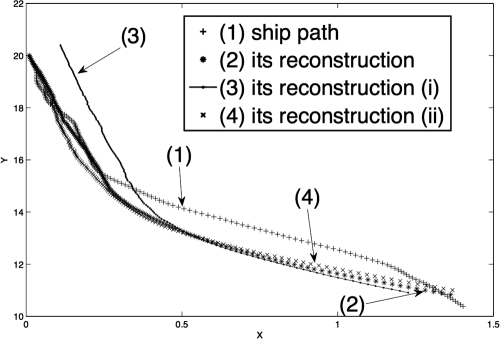
In Fig. 1 we plot a sample ship path, its reconstruction, and the reconstructions obtained (i) when the initial data for the reconstruction are strongly perturbed (here, the initial data for x,y were perturbed initially by, respectively, 0.1 and 0.4), and (ii) when the value of β assumed in the reconstruction is random: β = N(β0,∈β0), where β0 is the constant value used until now and ∈ = 0.4 but the calculation is otherwise identical. This produces variations in β of the order of 40%; any larger variance in the perturbations produced here a negative value of β. The differences between the reconstructions and the true path remain within the acceptable range of errors. These graphs show that the filter has little sensitivity to perturbations (we did not calculate statistics here because the insensitivity holds for each individual run).
Fig. 1.
Some ship trajectories (as explained in the text).
We now show that the parameter β can be estimated from the data. The filter needs an estimate of β to function; call this estimate βassumed. If βassumed ≠ β, the other assumptions used to produce the dataset (e.g. independence of the displacements and of the observations) are also false, and all one has to do is detect the fallacy. We do it by picking a trajectory of a particle and computing the quantity
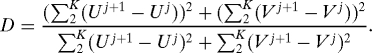 |
If the displacements are independent, then on the average D = 1; we will try to find the real β by finding a value of βassumed for which this happens. We chose K = 40 (the early part of a trajectory is less noisy than the later parts).
As we already know, a single run cannot provide an accurate estimate of β, and accuracy in the reconstruction depends on how many runs are used. In Table 4 we display some values of D averaged over 200 and over 3,000 runs as a function of the ratio of βassumed to the value of β used to generate the data. From the longer computation, one can find the correct value of β with an error of about 3%, whereas with 200 runs the uncertainty is about 10%. The limited accuracy reported in previous work can of course be achieved with a single run. A detailed discussion of parameter estimation using our algorithm will be presented elsewhere.
Table 4.
The mean of the discriminant D as a function of σassumed/σ, 30 particles
| σassumed/σ | 3,000 runs | 200 runs |
|---|---|---|
| 0.5 | 1.15 ± 0.01 | 1.15 ± 0.06 |
| 0.6 | 1.07 ± 0.01 | 1.07 ± 0.06 |
| 0.7 | 1.07 ± 0.01 | 1.07 ± 0.05 |
| 0.8 | 1.04 ± 0.01 | 1.04 ± 0.05 |
| 0.9 | 1.02 ± 0.01 | 1.02 ± 0.05 |
| 1.0 | 1.01 ± 0.01 | 1.00 ± 0.05 |
| 1.1 | 0.95 ± 0.01 | 1.01 ± 0.05 |
| 1.2 | 0.95 ± 0.01 | 0.95 ± 0.04 |
| 1.3 | 0.94 ± 0.01 | 0.96 ± 0.05 |
| 1.4 | 0.90 ± 0.01 | 0.88 ± 0.04 |
| 1.5 | 0.89 ± 0.01 | 0.88 ± 0.04 |
| 2.0 | 0.85 ± 0.01 | 0.83 ± 0.04 |
Conclusions
The numerical results for the test problem are comparable with those produced by other filters. What should be noted is that our filter behaves well as the number of particles decreases (down to a single particle in the test problem). There is no linearization or other uncontrollable approximation. This good behavior persists as the number of variables increases. The difficulty encountered by Bayesian particle filters when the number of variables increase is due to the fact that the relative size of that part of space that the data designate as probable decreases, so that it is harder for a Bayesian filter to produce probable samples. This situation can be modeled as the limit of a problem where both the variance β of the model and the variance s of the observation noise tend to zero; it is easy to see that in this limit, our iteration produces the correct trajectories without difficulty. In refs. 11 and 12 Snyder, Bickel, et al. produced a simple, many-dimensional problem where a Bayesian filter collapses because a single particle hogs all the probability; one can see that in that problem our filter produces the same weights for all the particles with any number of variables.
Acknowledgments.
We thank Prof. G.I. Barenblatt, Prof. R. Kupferman, Prof. R. Miller, and Dr. J. Weare for asking searching questions and providing good advice, and most particularly, Prof. J. Goodman, who read the manuscript carefully and pointed out areas that needed work. This work was supported in part by U.S. Department of Energy Contract No. DE-AC02-05CH11231 and by the National Science Foundation Grant DMS-0705910.
Footnotes
The authors declare no conflict of interest.
References
- 1.Doucet A, de Freitas N, Gordong N, editors. Sequential Monte Carlo Methods in Practice. New York: Springer; 2001. [Google Scholar]
- 2.Bozic S. Digital and Kalman Filtering. Oxford: Butterworth-Heinemann; 1994. [Google Scholar]
- 3.Maceachern S, Clyde M, Liu J. Sequential importance sampling for nonparametric Bayes models: the next generation. Can J Stat. 1999;27:251–267. [Google Scholar]
- 4.Liu J, Sabatti C. Generalized Gibbs sampler and multigrid Monte Carlo for Bayesian computation. Biometrika. 2000;87:353–369. [Google Scholar]
- 5.Doucet A, Godsill S, Andrieu C. On sequential Monte Carlo sampling methods for Bayesian filtering. Stat Comp. 2000;10:197–208. [Google Scholar]
- 6.Arulampalam M, Maskell S, Gordon N, Clapp T. A tutorial on particle filters for online nonlinear/nongaussian Bayesian tracking. IEEE Trans Sig Proc. 2002;50:174–188. [Google Scholar]
- 7.Gilks W, Berzuini C. Following a moving target—Monte Carlo inference for dynamic Bayesian models. J R Stat Soc B. 2001;63:127–146. [Google Scholar]
- 8.Chorin AJ, Krause P. Dimensional reduction for a Bayesian filter. Proc Natl Acad Sci USA. 2004;101:15013–15017. doi: 10.1073/pnas.0406222101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowd M. A sequential Monte Carlo approach for marine ecological prediction. Environmetrics. 2006;17:435–455. [Google Scholar]
- 10.Doucet A, Johansen A. Particle filtering and smoothing: Fifteen years later. In: Crisan D, Rozovsky B, editors. Handbook of Nonlinear Filtering. Oxford: Oxford Univ Press; 2009. preprint. [Google Scholar]
- 11.Snyder C, Bengtsson T, Bickel P, Anderson J. Obstacles to high-dimensional particle filtering. Monthly Weather Rev. 2008;136:4629–4640. [Google Scholar]
- 12.Bickel P, Li B, Bengtsson T. IMS Collections: Pushing the Limits of Contemporary Statistics: Contributions in Honor of Jayanta K Ghosh. Vol. 3. Beachwood, OH: Institute of Math Stat; 2008. Sharp failure rates for the bootstrap particle filter in high dimensions; pp. 318–329. [Google Scholar]
- 13.Chorin AJ. Monte Carlo without chains. Commun Appl Math Comput Sci. 2008;3:77–93. [Google Scholar]
- 14.Weare J. Efficient Monte Carlo sampling by parallel marginalization. Proc Natl Acad Sci USA. 2007;104:12657–12662. doi: 10.1073/pnas.0705418104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weare J. Particle filtering with path sampling and an application to a bimodal ocean current model. J Comput Phys. 2009;228:4321–4331. [Google Scholar]
- 16.Gordon N, Salmond D, Smith A. Novel approach to nonlinear/non-Gaussian Bayesian state estimation. IEEE Proc F. 1993;140:107–113. [Google Scholar]
- 17.Carpenter J, Clifford P, Fearnhead P. An improved particle fiter for nonlinear problems. IEEE Proc Radar Sonar Navig. 1999;146:2–7. [Google Scholar]
- 18.Milstein G, Platen E, Schurz H. Balanced implicit methods for stiff stochastic systems. SIAM J Num Anal. 1998;35:1010–1019. [Google Scholar]
- 19.Kloeden P, Platen E. Numerical Solution of Stochastic Differential Equations. Berlin: Springer; 1992. [Google Scholar]
- 20.Stuart A, Voss J, Wilberg P. Conditional path sampling of SDEs and the Langevin MCMC method. Commun Math Sci. 2004;4:685–697. [Google Scholar]
- 21.Levy P. Gauthiers-Villars: Paris; 1954. Le Mouvement Brownien. in French. [Google Scholar]