Abstract
Schistosomiasis, caused by infections by human blood flukes (Trematoda), continues to disrupt the lives of over 200,000,000 people in over 70 countries, inflicting misery and precluding the individuals’ otherwise reasonable expectations of productive lives. Infection requires contact with freshwater in which infected snails (the intermediate hosts of schistosomes) have released cercariae larvae. Habitats suitable for the host snails continue to expand as a consequence of water resource development. No vaccine is available, and the emergence and spread of resistance to the single licensed schistosomicide drug would be devastating. Since human infections would cease if parasite infections in snails were prevented, efforts are being made to discover requirements of intra-molluscan development of these parasites. Wherever blood flukes occur, naturally resistant conspecific snails are present. To understand the mechanisms used by parasites to ensure their survival in immunocompetent hosts, one must comprehend the internal defense mechanisms that are available to the host. For one intermediate host snail (Biomphalaria glabrata) and trematodes for which it serves as vector, molecular genetic and proteomic surveys for genes and proteins influencing the outcomes on infections are yielding lists of candidates. A comparative approach drawing on data from studies in divergent species provides a robust basis for hypothesis generation to drive decisions as to which candidates merit detailed further investigation. For example, reactive oxygen and nitrogen species are known mediators or effectors in battles between infectious agents and their hosts. An approach targeting genes involved in relevant pathways has been fruitful in the Schistosoma mansoni-B. glabrata parasitism, leading to discovery of a functionally relevant gene set (encoding enzymes responsible for the leukocyte respiratory burst) that associates significantly with host resistance phenotype. This review summarizes advances in the understanding of strategies used by both this trematode parasite and its molluscan host to ensure their survival.
Keywords: Biomphalaria glabrata, blood fluke, S. mansoni, schistosomiasis, immunoparasitology, oxidative burst
1. Introduction
“Each time a schistosome miracidium enters a snail, the outcome of the encounter is determined by the level of concordance of genetically determined characters”
Paraphrased from Paul Basch (1975 [1]).
Why the topic is important and a review is timely
The roughly 200,000,000 people in 74 countries infected with schistosomes [2] all share the fact that they contacted freshwater harboring infected snails. Starting about a month after being infected by one or more miracidia larvae of a compatible schistosome, a susceptible snail typically sheds (over several weeks) thousands of cercariae larvae into the water where they seek human skin and, if successful, penetrate to establish an infection, eventually taking up residence and maturing in blood vessels of the small intestine (Schistosoma mansoni) or bladder wall (S. haematobium). Obligatory parasitism of the snail, without which no-one can be infected, is the reason why this schistosome-snail interaction is a biomedically relevant target for research. An important snail host of human intestinal schistosomes is Biomphalaria glabrata. Expanding knowledge of this host-parasite system not only inspires hope of reducing the human costs of this insidious disease [2, 3], but holds the promise of broadening and deepening our grasp of the origins and evolutionary histories of the strategies exploited by different hosts and parasites to sustain other symbioses. With this grasp can come a deeper understanding of the origins and evolution of immune systems [4] and of the forces that influence the co-evolution of host and parasite [5, 6].
The last thorough assessment of schistosome-snail interactions was a multi-author work [7]. Big advances have characterized the intervening years 2001-2009, due in large part to exploratory studies in proteomics [8-12], functional genomics [13-29], and population genetics [30-37]. These have yielded floods of new data, and several novel hypotheses have emerged. In the meantime, high throughput protocols are giving way to even more productive technologies, with possibilities that challenge the imagination.
Molluscs have impressive capacities to protect themselves against infections by potential pathogens. For example, those rare microbes known to be pathogenic in any snail, limpet, slug or oyster are restricted to marine species [38, 39]. This implies versatility in both the recognition and the destruction of microbes by the molluscan immune (or internal defense) system. Molluscan responses to immune challenge are lymphoid-independent, mediated by phagocytic hemocytes in cooperation with humoral components [40]. That is not to say that the innate immune system relies exclusively on constitutive (pre-formed) elements: microbes entering the body cavity of a mollusc can elicit a variety of responses [23]. When a foreign object is encountered by a hemocyte in the body of a gastropod mollusc, soluble opsonins may have already marked the object for attention [40-41], plasma-borne enzymes may have altered its surface chemistry [42, 43], non-self receptors on the cell surface will signal through kinases and phosphatases to activate or deactivate intracellular enzymes and transcription factors [44-46], gene transcription is selectively enhanced or reduced [13-29 see Table 1], specific plasma proteins are synthesized and secreted [47, 48], and cell behaviors (migration, phagocytosis or encapsulation and degranulation) alter in orchestrated ways that reflect the innate immune repertoire of the mollusc. Resistance to infection is the norm; susceptibility – as in the case of compatible schistosomes - is the exception.
Table 1.
Studies (2001-present) designed to discover genes and proteins with roles in snail-trematode interactions. Bold underlined species names are the producers. For full lists of ‘hits’, consult the original papers. Many studies discovered genes for unknown proteins (not listed here). Studies which targeted specific genes (e.g. FREPs, SOD1) are covered elsewhere in the text. For brevity, the suffix ‘-like’ is omitted. ‘Targeted’ means that, rather than doing a study of ‘discovery’, the authors selected genes to study. With apologies for any missing entries, the publications cited in this Table are representative rather than inclusive of all relevant studies.
| Putative identification [names assigned by authors]. |
Genome / Transcriptome / Proteome. |
Compared susceptible / resistant? |
Compared challenge d / naïve? |
Cell/tissue | Host species | Parasite species |
Reference 1st author, number. |
Year | Technical approach |
|---|---|---|---|---|---|---|---|---|---|
| Cu/Zn SOD, glutathione-S-transferase, aldo-keto-reductase, triose-phosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase, aldolase, enolase, MICAL-like, calreticulin, NADH dehydrogenase, phosphoenol pyruvate hydratase, actin, acetyl coA transferase, RNA recognition, histones H1 and H4, ubiquinone cyt C chaperone, hemoglobin-like, hypothetical protein. | Proteome. | N.A. | N.A. | Secreted and excreted proteins from primary sporocysts (SEP). | N.A. | S. mansoni, E. caproni. | Guillou 8 | 2007 | Proteomic. |
| Twelve, incl. aldolase, intermediate filament protein, cytidine deaminase, ribosomal protein P1, histone H4. | Proteome. | Yes. | Yes. | Hemocytes. | B. glabrata | E. caproni. | Bouchut 9 | 2006 | Proteomic. |
| Thirteen, incl. glycolytic enzymes, calcium-binding proteins, cysteine-protease inhibitor. | Proteome. | Yes. | Yes. | Plasma (and albumen gland). | B. glabrata. | E. caproni. | Vergote 10 | 2005 | Proteomic, PCR. |
| Twenty three peptides, incl. ROS scavengers, components of primary metabolism, mucin-like and unidentified proteins. | Proteome. | N.A. | N.A. | ESP | B. glabrata | S. mansoni | Roger 11 | 2008 | Proteomic, PCR. |
| Thirty one named, incl. reverse transcriptase, aggregation factor, possible lectin, transposase, chitinase, others. | Transcriptome. | Yes. | Yes. | Hemocytes, Bge cells, body regions. | B. glabrata | S. mansoni | Raghavan 13 | 2003 | EST libraries, enzymology. |
| Over 860, (31 immune-relevant) incl. Dual oxidase, Prx, LBP/BPI, glucanase, theromacin, coagulation factor, proteinases, cystatin, FREPs, peptidoglycan recognition protein, dormapontin, matrilin, galectin, MIF, allograft inflammatory factor, others. | Transcriptome. | Yes. | Yes. | Hemocytes. | B. glabrata. | E. caproni. | Mita 14 | 2005 | EST |
| 23 cDNAs including cytochrome C, methyl-binding proteins, gluatmine synthetases, protease inhibitors. | Transcriptome. | N.A. | Yes [ESPs] | Bge cell line. | B. glabrata. | S. mansoni, E. caproni. | Coustau 15. | 2003 | DD-RT-PCR, RT-PCR |
| Detox. enzymes (GST, SOD), antimicrobial proteins (LBI/BPI), protease inhibitors (cystatins), calcium-binding proteins, C-type lectins, disulfide isomerase, HSP10, dermatopontin, cyclophilin, cabin, CEBP, S/TP kinase. Total 116 unique sequences. | Transcriptome. | Resistant only. | Yes. | Hemocytes, albumen gland. | B. glabrata. | E. caproni. | Guillou 16 | 2007 | SSH for ESTs; qPCR; ISH. |
| Bg selectin. Targeted. | Transcriptome. | Yes. | Yes. | Whole snail bodies. | B. glabrata. | E. caproni. | Guillou 17 | 2004 | PCR. |
| Antioxidant (peroxidasin, dual oxidase, Fe-dependent peroxidase, peroxinectin), >6 signaling pathways, transcriptional regulation, immune-related (incl. α2-mac, ABCF1, ferritin), glycan biosynthesis. 1,843 non-redundant clones. | Transcriptome. | No. | Yes. | Selected tissues, incl. hemocytes. | B. glabrata. | S. mansoni. | Lockyer 18 | 2007 | ORESTES. |
| 11 unique, incl. oxidative stress (killing of parasite), ferritin, HtrA2 (serine protease), FREP, possible MAPK. | Transcriptome. | Yes. | Yes. | Hemocytes; hemopoietic organ. | B. glabrata. | S. mansoni. | Lockyer 19 | 2007 | SSH for ESTs. |
| 10 incl. MINT1 (neurotransmission), HSP70, and a globin-containing peptide. | Transcriptome. | Yes. | Yes. | Brain, Hemopoietic organ, kidney. | B. glabrata. | S. mansoni. | Lockyer 20 | 2004 | Differential display; PCR. |
| 170 unique: SOD, calcium-binding proteins, collagen, carbohydrases, proteases, protease inhibitors, lectin, lysozyme, aplysianin-like molecule, cell adhesion molecules, hemoglobin, coaggulation factor, dermatopontin. | Transcriptome. | Yes. | No. | Whole snail bodies; hemocytes. | B. glabrata | N.A. | Bouchut 21 | 2007 | SSH, qPCR. |
| Dermatopontin, matrilin, cadherin, thrombospondin, von Willebrand domain, chitin-binding, EGF/TSP domain, FREP3. Targetted. | Transcriptome. | Yes. | Yes. | Hemocytes; whole snails. | B. glabrata. | E. caproni. | Bouchut 22 | 2006 | Database search; Q-PCR. |
| 2,129 unique: 175 named putative immune-related genes, incl. pattern recognition, cell adhesion, signal transduction, antioxidant-related, apoptosis, stress response, metal-binding serpins, proteases, inflammatory response, other putative immune factors. | Transcriptome. | No. | Yes. | Whole bodies. | B. glabrata. | S. mansoni; also E. coli, M. luteus. | Hanelt 23 | 2008 | ORESTES. |
| 8: SmTCP 1-C (chaperonin), α-tubulin, β-tubulin, cytochrome oxidase, glutaminyl-tRNA synthetase. | Transcriptome. | N.A. | Yes. | Sporocysts. | B. glabrata (Bge). | S. mansoni. | Coppin 24 | 2003 | Differential display, PCR. |
| FREPs 2, 3, 4, 7. Targeted. | Transcriptome. | Yes. | Yes. | Whole snails. | B. glabrata. | S. mansoni, E. paraensei. | Hertel 25 | 2005 | PCR. |
| 88 unique: cutochrome C oxidase subunit, FREP, cystatin B, serpin 6, ferritin, α-B crystalin, novel ESTs. | Transcriptome. | Yes. | Yes. | Snail head-feet. | B. glabrata. | S. mansoni. | Nowak 26 | 2004 | SSH, q-PCR. |
| 22: adhesion molecule, defensin, serine/threonine kinase, myoglobin, glycosidase. | Transcriptome. | Yes. | No. | Hemocytes | B. glabrata. | S. mansoni. | Schneider 27 | 2001 | DDRT-PCR, SSCP. |
| 65: transposase, histidine-rich glycoprotein, others. | Transcriptome. | Yes. | Yes. | Hemocytes. | B. glabrata. | S. mansoni. | Miller 28 | 2001 | DDRT-PCR |
| Nimbus (BgI non-LTR retrotransposon). Targeted. | Genome. | Yes. | Yes. | Whole snails, hemocytes. | B. glabrata. | N.A. | Raghavan 29 | 2007 | Sequence. |
| Cathepsin B. Targeted. | Proteome, transcriptome. | Yes. | Yes. | Hepatopancreas | B. glabrata | S. mansoni. | Myers 101 | 2008 | Enzymology, qPCR. |
This review is timely because, predominantly from research on the Schistosoma mansoni- Biomphalaria glabrata and Echinostoma spp-B. glabrata systems, the years since the last review [7] have yielded both a torrent of information [reports cited above, with the addition of the S. mansoni genome [49], (http://www.genedb.org/genedb/smansoni/, http://schistodb.net/schistodb20/, http://www.sanger.ac.uk/Projects/S_mansoni/), an expansive database of the S. japonicum transcriptome [50], and progress in sequencing the B. glabrata genome (http://genome.wustl.edu/genome.cgi?GENOME=Biomphalaria%20glabrata)], but powerful new tools: a BAC library for B. glabrata ([51], http://biology.unm.edu/Biomphalaria-genome/index.html), oligonucleotide microarrays for both parasite [52, 53] and host [Loker ES, University of New Mexico, and C. Jones, University of Aberdeen, personal communications], and the means to suppress gene expression in both the snail [54] and the trematode [55]. Furthermore, it has become apparent that epigenetic phenomena may regulate schistosome gene transcription [C. Cosseau et al, presented at Xth EMOP in Paris, France, August 2008]. On the host side, studies that have targeted molecules falling within the sphere of oxidative and nitrative stress have provided a solid basis for appreciating mechanisms by which snail hemocytes (phagocytic leukocytes) kill parasites [56,57].
In the past 10 years, resistance to the anti-schistosome ‘drug of choice’ (Praziquantel) has been observed [58] and just one new potential drug has emerged [59]. At the same time, human activities such as dam construction that expand habitats for vector snails and facilitate transmission have run ahead of efforts to short-circuit parasite transmission through the provision of piped water and sanitary systems. It is widely hoped that a vaccine may be developed with the potential to reduce morbidity and mortality, but even if vaccine development succeeds, it is not seen as a likely panacea [3, 58]. In short, we are not yet providing the tools for public health personnel to lessen suffering and mortality due to human blood fluke infections, and a multipronged approach is needed to lessen their human impacts.
The challenge we now face is to separate the grain from the chaff in the process of ascertaining what molecules are relevant to the success of the parasite and to the failure of susceptible hosts leading to the shed of cercariae from snails. This is a noble challenge because such knowledge may help in the effort to reduce blood fluke burdens in afflicted human populations. However, if the complete account of schistosome-snail interactions were to take the form of a painting by Van Gogh, no more than 1/10th of the canvass would have paint, and the strokes remain mostly broad. This review will focus on the painted regions of the canvass to see what they suggest or reveal, and to gain a sense of direction in choosing where to invest future efforts in the hope of gaining a comprehensive picture of snail-schistosome relationships.
Key attributes adding value to this model host-parasite system
Species of Biomphalaria (B. pfeifferi [31, 32] and B. glabrata [33]) and their trematode parasites [34, 35] are surprisingly genetically diverse. Furthermore, in natural populations only some schistosome individuals are compatible with any snail individual [1, 7, 30, 60-61]. Throughout the geographic ranges of schistosome species parasitizing humans (mostly the humid tropics), such compatibility polymorphisms are a hallmark of the trematode-mollusc relationship [11, 62, 63]. These facts were first documented decades ago [60-63], and the underlying temporal and spatial heterogeneities in compatibility phenotypes have, with rare exceptions (e.g. B. glabrata strain BS90 individuals are invariably resistant to S. mansoni), stymied efforts to describe susceptibility/resistance phenotypes using stable quantitative values. We seem to have a moving target, consistent with rapid co-evolution. Key concepts about which we must be clear are shown in Box 1. In Nature, a range of compatibilities exist, and the fates of schistosome larvae that penetrate snails of the host species vary from (i) destruction within hours to (ii) productive infections that yield human-infective cercariae several weeks later. Subtle additional phenotypes exist, such as infections that persist long term in a developmentally stalled state, and that may or may not regain developmental momentum for reasons that no-one knows.
Box 1. Key concepts of infectivity, susceptibility, resistance, compatibility, incompatibility, unsuitability: what these mean in this system.
Infectivity: the capacity of individuals in a parasite population to establish viable infections in individuals within a host population.
Susceptibility: condition in which a host fails to inactivate or kill a parasite, resulting in an infection that progresses to patency, requiring physiological suitability of the host’s milieu interieur for parasite development.
Resistance: condition in which a host prevents the development of the parasite by killing it.
Compatibility: a quality of a pair of species in which an individual parasite infects an individual host in which the parasite completes the part of its life cycle appropriate to the host.
Incompatibility: a quality of a pair of species in which an individual parasite infects an individual host in which the parasite fails to complete the part of its life cycle appropriate to the host.
Unsuitability: host condition in which the needs of the parasite are not adequately met.
Laboratory models of this host-parasite system [65] are readily available (e.g. in the USA, http://www.schisto-resource.org/), relatively easily maintained in the laboratory, and widely exploited by investigators interested in gaining an understanding of this host-parasite system.
Infectivity, susceptibility, resistance, compatibility, incompatibility, unsuitability: what does it all mean?
Schistosome infections are typified by their very high degree of host snail specificity: a miracidium is capable of infecting only some individuals of any one strain of the host species. Given the impressive efficacy of the snail’s defenses, some very special circumstances must account for the success of these parasites in exploiting the snail as a host. Classical genetics showed, unsurprisingly, that genotypes of both partners determine the outcome of each infection [1, 62]. With snails, they implied a multi-gene control of resistance in juveniles, and a single dominant gene controlling resistance in adults. With schistosomes [1, 6, 65], they revealed that the capacity to infect one strain of snail can be independent of the capacity to infect another strain of snail. The challenges we now face are to identify genes controlling the different S/R compatibility phenotypes and to use this knowledge to reduce parasite burdens in human populations.
Aspects of schistosome and snail biology most relevant to the topic of this review are shown in Box 2. The echinostomes are closely related trematodes. Some parasitize Biomphalaria snails, but appear to be more proactive than schistosomes in immune suppression/interference [15], and we lack the space here to describe the differences.
Box 2. The schistosome’s survival mechanisms that need to be understood, and Questions which need to be addressed concerning the snail’s mechanisms, with answers when available.
The schistosome’s survival mechanisms that we need to understand.
Locating a host snail.
Penetrating a host snail.
Minimizing the furor of the attack by the host’s internal defense (immune) system.
Protecting self and counter-defending against host immune effector pathways.
Metamorphosing from miracidium to mother sporocyst.
Migration in the host.
Nutrition, growth and differentiation, including asexual proliferation.
Redirecting host resources to the benefit of the schistosome.
Ensuring the survival of the host (in order to sustain parasite development).
Escaping (birth) at the cercarial stage, and emigrating from the snail fit for survival in freshwater.
Locating and penetrating human skin (and subsequent activities within the definitive host)
Questions which need to be addressed concerning the snail’s mechanisms, and answers when available.
Do snails release clues that can be used by the miracidium to locate a host? YES [98].
Are there barriers to penetration of the miracidium? NOT EFFECTIVE.
How is the presence of the schistosome recognized? NOT KNOWN.
What mechanisms are marshaled to kill the invader? These are humoral and cell-mediated, with behavioral, membrane-dependent and granule-dependent components. There are both constitutive changes (not evidenced by changes in the transcriptome) and induced changes (evidenced as altered levels of mRNAs and proteins).
What else is known of responsible immune mediators and effectors? SEE TEXT.
What is the process of dealing with killed parasites? SEE TEXT.
How are resources partitioned to sustain both symbionts when a snail is unable to get rid of the invader? NOT KNOWN.
What, then, are the mechanisms that govern the variable outcomes of schistosome-snail encounters and for which of those that are theoretically possible do we have empirical data supporting or devaluing a model that incorporates that mechanism?
Others [66] have argued that the success or failure of the host’s recognition system plays the dominant role in determining the outcomes of schistosome-snail encounters. We prefer to think that the fate of a parasite is determined by several factors including recognition or its failure: a complex interplay of the recognition capacities of the host, evasion capacities of parasites, effectiveness of killing activities of hosts, and counter-defense capacities of parasites seems a more realistic scenario. Some elements of the system will be active acutely (within minutes to hours of the infection) while others act chronically, even for the duration of the infection (up to >2 months).
Recognition and means to escape it
We have explained why - in the absence of special circumstances - snails would be expected to resist infections. However, before unleashing the furor of its defense system, the snail needs first to recognize the presence of something to be attacked. Trematodes do not ‘lie low’ when they enter a snail; to the contrary, they release a veritable cloud of molecules that have come to be known as their excretory-secretory products (ESP) [8, 12]. Many are released from the penetration glands, and it is possible that others derive from ciliated plates that are shed when the miracidia metamorphose to sporocysts, exuding tegumental components in doing so. These ESP molecules include enzymes (cysteine proteases [12], superoxide dismutase [8, 67]) along with a variety of polymorphic mucins, the so-called SmPoMuc family members [11]. Research at the University of Perpignan, France [68] suggests that these mucins become diversified within the life of each individual and that their expression is regulated to some degree by epigenetic changes that could be different between individual parasites. These polymorphic mucins are released by schistosomes as they penetrate a snail, and could be recognized by reciprocally polymorphic receptors in host plasma and on host hemocytes [25, 47, 48], an idea that has been independently suggested by E. Roger (personal communication).
Knowledge of receptors that recognize foreign (non-self) determinants in molluscs is disproportionately comprehensive for one family, the FREPs [25, 47, 48, 70-75, 77], and fragmentary for others. However, most, if not all, non-self receptors described in molluscs are lectins. This holds for cell receptors that bind sporocyst molecules [69] and for plasma proteins which bind parasite ESPs [see FREPs papers]. Other candidates for molluscan immune-activating non-self receptors include mannan- and laminarin-binding molecules [76], and peptidoglycan-binding and Gram-negative-bacteria-binding proteins [77], but others exist [78]. Members of the FREP family of plasma proteins (14 family members are now known [75]; these are lectins but not known to be cell membrane receptors; [79] is a recent review) appear or increase in the plasma following trematode infections. By recognizing sugars, they precipitate glycan-bearing molecules released by trematodes, and will bind to sporocysts, bacteria and yeast [75]. The FREPs are compelling because, in addition to these properties, they are mosaic proteins with 1 or 2 immunoglobulin-like (Ig) domains and a fibrinogen domain, and the Ig domains are diversified within individual snails [70]. This discovery has bolstered a paradigm shift in comparative immunology, changing from the old view that immune recognition in invertebrates relies entirely on invariant, genome-encoded receptors. Insects have been reported to exploit diversification of receptors within the lifetime of the individual [80], while the genomes of echinoderms [81] and protochordates [82] encode the proteins to diversify receptors, a signature event of the adaptive immune systems of jawed vertebrates. The presence in snail plasma of the Ig superfamily FREP proteins and of the related FReM protein [71] is not predictive of resistance. Despite the variable Ig domains in FREPs, evidence that they trigger effective immune-type responses remains elusive. One might conjecture that their function is to scavenge excretory-secretory products (ESPs) from developing parasites. Do they (like Ig antibody receptors on lymphocytes) occur, even ephemerally, as integral membrane receptors? It will be intriguing to discover the proximate stimuli for their hypersynthesis (might this be akin to antigen-antibody binding?) and to learn if their ligands include or are related to the polymorphic schistosome mucins reported by others [68] or to other components of trematode ESPs.
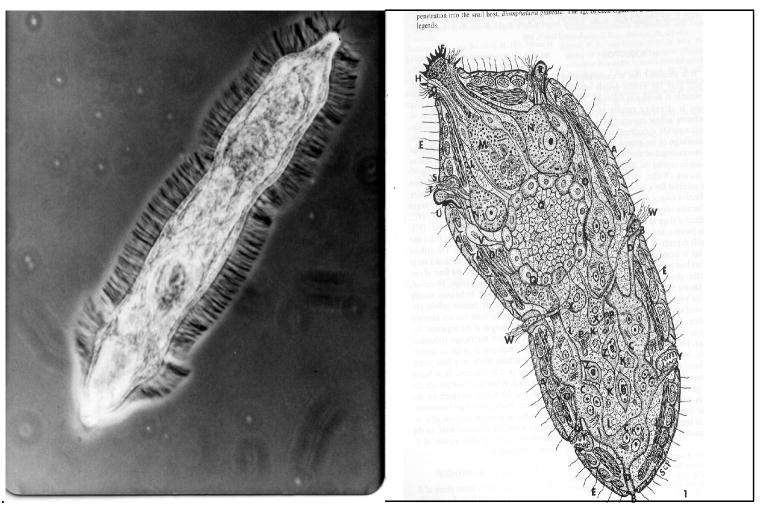
On contacting the soft (Latin ‘mollis’ means soft) body surface of a snail, a miracidium (a ~200 μm long, ciliated larva of ~ 200-300 cells; Fig 2) uses the contents of its penetration glands, along with muscular squirming, to penetrate the body wall. Within minutes of arriving in the body cavity, it sheds its ciliated surface ‘plates’ and exudes new cytoplasm and membrane lipids from cells in its interior. This exudate spreads over the surface, forming a tegument: the miracidium has metamorphosed into a sporocyst [64, 83] within which germinal cells begin to develop into a generation of daughter sporocysts, a process that takes weeks. The body cavity of the snail where these events occur is an open circulatory system in which blood (hemolymph) containing numerous plasma proteins [43] and ameboid leukocytes (called hemocytes) is continually circulated. Parasites that have penetrated have nowhere to hide.
Figure 2.
Left: a miracidium of Schistosoma mansoni photographed under phase contrast microscopy. The surface cilia, with which the larva swims, are evident as a fuzzy surface.
Right: an artist’s reconstruction of the internal anatomy of a miracidium, based on electron micrographs (from Pan CT, 1980 [64]).
How, then, might a schistosome entering the hemocoel of a snail avoid eliciting an attack? One suggestion for which there is increasing evidence is that compatible parasites avoid the wrath of the host snail by mimicking the snail’s own self markers. The case for this has been reasoned [40, 84], and recent evidence is supportive [85]. In a nutshell, it must be the case that any and all immunocompetent organisms have means to avoid inflicting unintentional damage on self. The means to achieve this is probably ancient though it was first described within gnathostome vertebrates (mice and humans): receptors (on immune cells) that recognize self determinants effectively shut off signaling pathways that otherwise elicit aggressive responses. This is how our own natural killer cells are normally prevented from attacking our tissues [reviewed in 86]. Since mammals injected with schistosome antigens produce antibodies that recognize host snail antigens, and those injected with snail antigens produce antibodies that recognize schistosome antigens, we can not escape the inference that schistosomes mimic snail (host) epitopes [84, 87]. It is also clear that sporocysts quickly coat themselves with host plasma proteins, and display these over the long term [43]. Why would parasites both mimic and acquire-display host determinants unless there were functions for the epitopes? We conjecture that these epitopes are recognized by negative-signaling ‘self’ receptors on snail hemocytes. These putative receptors would be functional analogs of those on natural killer cells that express the immunoreceptor tyrosine-based inhibitory motifs [ITIMs, 86] with the ability to counteract kinase-based activation pathways. Parasites such as blood flukes which develop without access to immunologically privileged sites (they live in the blood streams of both molluscan and mammalian hosts) are especially likely to derive protection from mechanisms such as this. Though the idea remains speculative, reports that sporocysts and their products interfere with ERK signaling in snail (Lymnaea stagnalis) hemocytes from susceptible but not resistant snails [84], and that glycopeptides in trematode (echinostome) ESPs interfere with defenses of susceptible strain hemocytes, and not resistant hemocytes [85], underscore its merit. Also the sharing of specific carbohydrate structure by B. glabrata and S. mansoni [87] provides the chemical basis of the observed mimicry. It will be valuable to characterize the responsible receptor(s) upstream of ERK in snail hemocytes and to assess the possibility that it/they recognize trematode mucins and reduce phosphorylation of ERK.
The parasite - host interplay is, of course, dynamic. Much may be determined by constitutive components of the two partners, including the parasite’s abilities to appear not much different from the host and to mop up potentially destructive host molecules. But to these first lines of defense, the parasite may add others as the infection progresses to its chronic phase [7].
The ciliated plates shed after miracidial penetration of a snail are anucleate and are quickly phagocytosed by hemocytes. Further, the capacity of sporocysts to prevent recognition is a property that they loose the moment that they die (unpublished observations). The work of staying alive in an immunologically hostile environment appears to require housekeeping that communicates identity and health, but how this is done and why the signals decay quickly remain mysterious.
The host’s capacity to kill and the means a schistosome may use to resist it
If, when a miracidium enters a snail, it fails to avoid being recognized, it faces an ugly predicament. The body cavity of a gastropod provides no places in which to hide: without exception, hemocytes will locate the metamorphosing miracidium-sporocyst. The hemocytes are circulating, phagocytic leukocytes. They are capable of chemotaxis, can extend long pseudopods, and will avidly engulf or encapsulate (‘frustrated phagocytosis’) foreign objects that are too large to be phagocytosed – with the exception of compatible parasites. The hemocytes contain lysosomes with proteolytic enzymes, and upon appropriate stimulation will degranulate these, releasing their contents both extracellularly and into phagosomes. Receptor-mediated signaling appears to conform to some degree with pathways documented in other species [44 - 46, 84]. They also are armed with the biochemical machinery to produce reactive oxygen (ROS) and reactive nitrogen species (RNS), and do so when appropriately stimulated [91-94]. Hemocytes discover parasites within minutes and can inflict significant damage within a few hours [11, 94]. The capacity to generate ROS, known also by the term ‘leukocyte respiratory burst’ due to its rapid consumption of O2, is central to the cytotoxic capacities of leukocytes in organisms across the evolutionary spectrum. Once generated, the first ROS of the burst (superoxide, O2-) can be metabolized through alternative pathways, some leading to other toxic ROS while others detoxify the products (Fig. 4).
Figure 4.
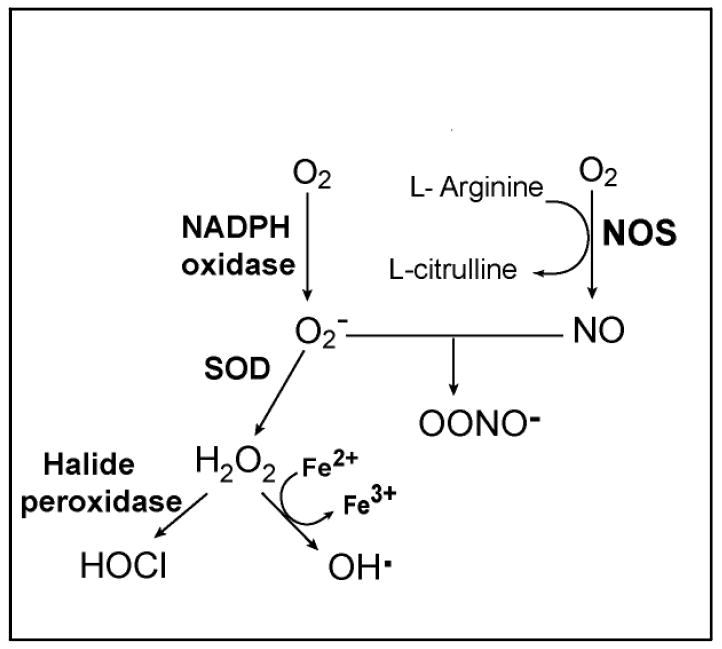
A simplified schematic of the pathways that generate ROS and RNS in phagocytic cells. NADPH oxidase converts molecular oxygen (O2) into superoxide (O2-). The dismutation of O2- to hydrogen peroxide (H2O2) is catalyzed by superoxide dismutase (SOD). In the presence of chloride ion, H2O2 can be converted to hypochlorous acid (HOCl) by halide peroxidases. H2O2 reacts with ferrous ions (Fe2+) to yield hydroxyl radical (OH·). The enzyme nitric oxide synthase (NOS) utilizes O2 and arginine to produce nitric oxide (NO), which can react with O2- to form the highly reactive peroxynitrite (ONOO-).
The inference that the respiratory burst is likely to be an important component of molluscan cellular defences led us to target the burst using an in vitro model of schistosome sporocyst encapsulation by snail hemocytes [91-93] (Fig 3). These experiments revealed that the capacity of hemocytes to kill sporocysts was compromised by inhibitors of enzymes in the respiratory burst pathway and by molecules that scavenge ROS. In addition, the knowledge that nitric oxide can react with O2- to yield peroxynitrite (Fig. 4), possibly enhancing the overall cytotoxic potential, led us to ask if an inhibitor of nitric oxide synthase would alter the killing of sporocysts in our in vitro model; it did [93].
Figure 3.
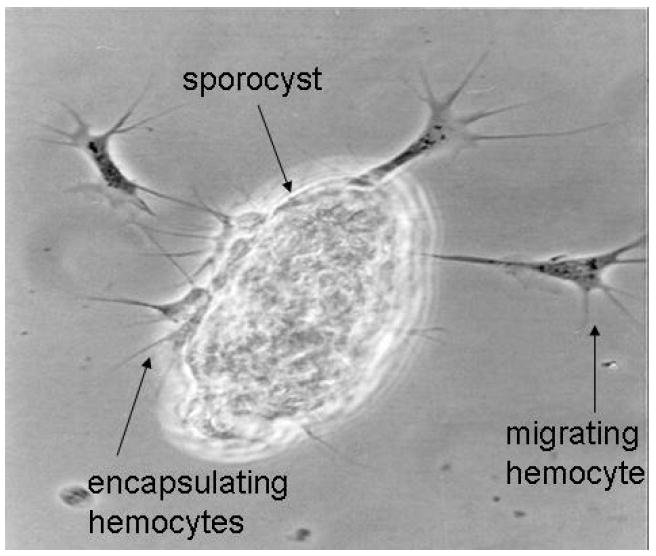
A sporocyst of S. mansoni encapsulated by B. glabrata hemocytes, with additional migrating hemocytes. This in vitro model replicates events that occur in vivo each time a schistosome enters a snail. When (in vivo) the snail is resistant to the parasite and when (in vitro) the hemocytes are from such a snail, the consequence of encapsulation is death of the sporocyst [94]. The kinetics of events in vivo and in vitro are the same. In the process of killing the parasite, hemocytes produce H2O2 and a product of arginine metabolism, possibly nitric oxide [91-93].
This potential of ROS and RNS to inflict damage in the B. glabrata – S. mansoni parasitism implicates enzymes at all stages of the respiratory burst. As H2O2 is pivotal to the effector pathways of the burst, we measured its production by hemocytes from a strain that is more resistant (R) and another that is more susceptible (S) to our strain of S. mansoni. Consistent with a role in determining the S/R phenotype, S hemocytes produced less H2O2 than did hemocytes more able to kill sporocysts [95].
The generation of H2O2 is accelerated by the enzyme superoxide dismutase (SOD). So we asked if the activity levels of SOD differ in S and R hemocytes, and found that R hemocytes had about twice the activity seen in S hemocytes [96]. This difference was found to be due to the intracellular Cu/Zn SOD and not to a Mn SOD which is characteristically located in mitochondria.
The early work of identifying and characterizing these enzymes at the levels of DNA, RNA and protein has resulted in the first association of a functionally relevant gene with the R (resistant)/S (susceptible) phenotype of a snail to a trematode parasite. The cytoplasmic Cu/Zn SOD of B. glabrata (SOD1) is encoded in our lab populations by three or more alleles, A, B and C (DQ239577, DQ239578 and DQ239579). In a strain of B. glabrata that is variable in S/R phenotype, one of the alleles has been found to be significantly associated with resistance [97]. This association of a functionally relevant locus with the S/R phenotype in B. glabrata is unprecedented. It begs the question, is the R-associated SOD1 allele also associated with higher numbers of transcripts? Quantification of allele-specific mRNAs in individuals revealed that snails carrying the allele in question had significantly higher levels of SOD1 transcript than individuals without this allele [98]. Using data on the average percent R of each genotype, we asked if this correlated with the mean level of SOD1 expression of each of the six genotypes (AA, AB, AC, BB, BC, CC): there is indeed a significant correlation, consistent with the view that SOD1 expression is directly involved in R/S to S. mansoni [98].
It would be premature to conclude that oxygen and/or nitrogen radicals directly kill S. mansoni; ROS are also responsible for intracellular signaling, so peroxide may not be the effector but rather an upstream mediator of parasite damage. However, underscoring the relevance of oxidative stress genes to the S/R phenotype in this model, peroxiredoxins (Prx) also appear to be involved. These enzymes have two known functions: one is to scavenge H2O2 while the other involves immune-relevant intracellular signaling [99]. There appear to be at least four Prx loci in B. glabrata [Knight et al, unpublished data]. In lab populations, we see 3 allelic variants for one of these – the apparent ortholog of vertebrate Prx4. We have found (unpublished observations) that Prx4 transcript abundance is a highly variable trait, but the locus appears to be transcriptionally activated in snails as a result of infection, with more rapid activation in a resistant strain than a susceptible one [Knight et al., unpublished data].
As we mentioned with the canvass analogy, there are as yet just limited areas where the image is clear. We posit, however, that variation in the expression or activity of any hemocyte factor contributing to the production or to the consumption of H2O2 is likely to influence the potency of resistance to S. mansoni in B. glabrata.
The targeting of loci involved in the generation of ROS and NO is a rational approach that is validated by data coming from more global approaches. Among genes that differentiate susceptible and resistant strains of snail, oxidative stress genes appear repeatedly [see Table 1]. Furthermore, SOD is the most highly expressed gene in the outermost layer (tegument) of larval and adult schistosomes [100].
Fruits of the more global approaches for discovery of genes and proteins that influence the outcomes of trematode parasite-snail host encounters
There are undoubtedly additional (ROS- and RNS-independent) means, both humoral and cell-mediated, used by snails to injure and kill schistosomes. Just as in the world of human affairs, reciprocal antagonists evolve supplementary mechanisms for attack, counter-attack and for self-defense. The wide variety of snail genes that are transcriptionally altered in response to parasitism include many whose products have functions that are not known to be related to oxidative stress (see Table 1). Many others code for proteins of unknown identity. Some are likely responsive to offensive signals from the trematodes, serving to protect the host even if not harming the parasite, while others (especially those with rapid – hours – increases in expression) may contribute to either cell-mediated or humoral aggression against the sporocyst.
Table 1 illustrates the extent of efforts that have been expended to discover genes and proteins which may account for the phenotypes in the B. glabrata – S. mansoni and the B. glabrata - Echinostoma spp parasitisms. The predictable rewards have been lists of genes and proteins that differentiate susceptible from resistant hosts, and infective/compatible from non-infective/incompatible parasites. Some are constitutively different while others are diagnostic of host or parasite responses to encounter with their partner. The majority of these papers include exhortations to conduct functional studies aimed at clarifying the roles of named genes in the host-parasite interaction [e.g. 21]. However, the large numbers of genes implicated impose a need to decide which leads are worthy of further experimentation, and what approaches are likely to best reward our efforts. While all such deliberations must be speculative, reasonable cases can be made for the roles of certain molecules in the causation of phenotypes. For example, there is an hypothetical framework (see earlier) that is consistent with roles for cell receptors, and another consistent with roles of oxidative stress enzymes, so that SOD, ROS scavengers, G3PDH, oxidases, peroxidasin and other products ‘fit’ a model, and suggestions as to functionality can be considered reasonable. Similarly, there are precedents for speculating that proteases, protease inhibitors, cell adhesion factors, coagulation factors, stress proteins, signal transduction proteins, enzymes involved in both the biosynthesis and the degradation of glycans, lectins and a variety of other genes and proteins in the lists play roles that contribute to the outcomes of parasite-host encounters. Many of the papers cited in Table 1 (see, for example, refs. 11, 14, 16) present well reasoned scenarios as to how the discovered molecules may contribute. Actually demonstrating the role(s) of each presents significant challenges, as is well illustrated by the evolving FREPs story [79]. Further, it is surely the case that, among the numerous unidentified genes and unknown proteins in the discovery studies, there are more that influence outcomes. The involvement of each may be made evident by gene knock-down approaches [54], but discovering specific mechanisms and actions of each will take time. However, if the development of fully protective vaccines remains insurmountable, and other methods of controlling schistosomiasis continue to fall short, the goal of breaking transmission should sustain these efforts to understand the basis for trematode propagation in their intermediate hosts.
Some further considerations
The speed of new technologies and their enormous capacities to discover genomic features that distinguish compared phenotypes have made them appealing. But their positive attributes come at a cost that is not obvious: minor differences in sequence can be missed (for example in microarray hybridizations), yet may have real phenotypic consequences (e.g. enzyme kinetics). Even when not missed, distinct alleles can influence phenotypes in subtle yet consequential ways such as the quantities of protein in the cell, and the efficiencies with which receptors and enzymes execute their functions.
While knowledge of Biomphalaria glabrata and Schistosoma mansoni genomes and transcriptomes has been increasing, related data have been accumulating for many additional species. Given the universality of many pathways in the biosphere, comparative approaches that draw on the broad datasets are increasingly rewarding as we seek to formulate and test new hypotheses.
Figure 1.
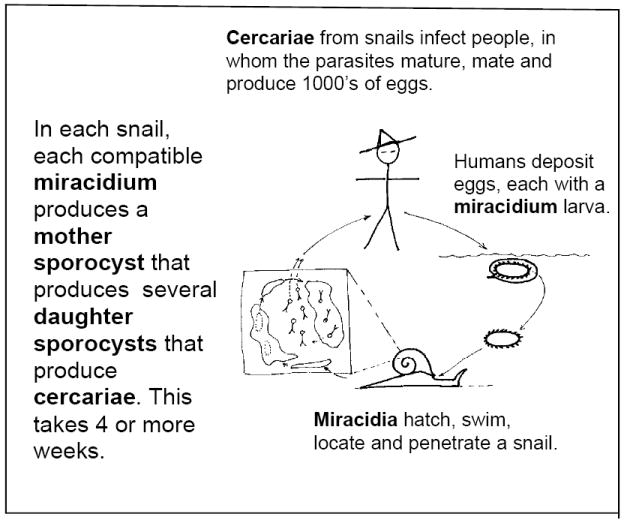
The life cycle of a schistosome that parasitizes humans and is responsible for schistosomiasis (blood fluke).
Acknowledgments
The author’s research has been supported by NIH award AI-16137. I thank each of those individuals who have spent, and continue to spend, time in my lab, those who have had me in theirs, and those who, at professional meetings, have taken the time to discuss comparative immunology with me. I am also indebted to Mike Blouin, Virginia Weis, Randy Bender and two anonymous reviewers who gave me feedback on an earlier version of this paper.
Abbreviations
- S
susceptible/susceptibility
- R
resistant/resistance
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- ESP
excretory-secretory products
- FREP
fibrinogen-related protein
- SOD
superoxide dismutase
- Prx
peroxiredoxin
- ERK
extracellular signal-regulated kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Basch PF. Intermediate host specificity in Schistosoma mansoni. Exper Parasitol. 1976;30:150–69. doi: 10.1016/0014-4894(76)90022-9. [DOI] [PubMed] [Google Scholar]
- 2.Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6:411–25. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- 3.Bergquist R, Utzinger J, McManus DP. Trick or treat: the role of vaccines in integrated schistosomiasis control. PLoS Negl Trop DiS. 2008;2(6):e244. doi: 10.1371/journal.pntd.0000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayne CJ. Origins and evolutionary relationships between the innate and adaptive arms of immune systems. Integr Comp Biol. 2003;43:293–9. doi: 10.1093/icb/43.2.293. [DOI] [PubMed] [Google Scholar]
- 5.Gower CM, Webster JP. Intraspecific competition and the evolution of virulence in a parasitic trematode. Evolution. 2005;59(3):544–53. [PubMed] [Google Scholar]
- 6.Webster JP, Gower CM, Blair L. Do hosts and parasites coevolve? Empirical support from the Schistosoma system. Am Nat. 2004;164(Suppl 5):S33–51. doi: 10.1086/424607. [DOI] [PubMed] [Google Scholar]
- 7.Rollinson D. Flukes and snails revisited – Preface. Parasitology. 2001;123 Supplement, containing 19 relevant review papers. [Google Scholar]
- 8.Guillou F, Roger E, Moné Y, Rognon A, Grunau C, Théron A, Mitta G, Coustau C, Gourbal BE. Excretory-secretory proteome of larval Schistosoma mansoni and Echinostoma caproni, two parasites of Biomphalaria glabrata. Mol Biochem Parasitol. 2007;155(1):45–56. doi: 10.1016/j.molbiopara.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Bouchut A, Sautiere PE, Coustau C, Mitta G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: Potential involvement of proteins from hemocytes revealed by a proteomic approach. Acta Trop. 2006;98(3):234–46. doi: 10.1016/j.actatropica.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Vergote D, Bouchut A, Sautière PE, Roger E, Galinier R, Rognon A, Coustau C, Salzet M, Mitta G. Characterisation of proteins differentially present in the plasma of Biomphalaria glabrata susceptible or resistant to Echinostoma caproni. Int J Parasitol. 2005;35(2):215–24. doi: 10.1016/j.ijpara.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Roger E, Mitta G, Moné Y, Bouchut A, Rognon A, Grunau C, Boissier J, Théron A, Gourbal BE. Molecular determinants of compatibility polymorphism in the Biomphalaria glabrata/Schistosoma mansoni model: new candidates identified by a global comparative proteomics approach. Mol Biochem Parasitol. 2008;157(2):205–16. doi: 10.1016/j.molbiopara.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Lodes MJ, Yoshino TP. Characterization of excretory-secretory proteins synthesized in vitro by Schistosoma mansoni primary sporocysts. J Parasitol. 1989;75:853–62. [PubMed] [Google Scholar]
- 13.Raghavan N, Miller AN, Gardner M, FitzGerald PC, Kerlavage AR, Johnston DA, Lewis FA, Knight M. Comparative gene analysis of Biomphalaria glabrata hemocytes pre- and post-exposure to miracidia of Schistosoma mansoni. Mol Biochem Parasitol. 2003;126(2):181–91. doi: 10.1016/s0166-6851(02)00272-4. [DOI] [PubMed] [Google Scholar]
- 14.Mitta G, Galinier R, Tisseyre P, Allienne JF, Girerd-Chambaz Y, Guillou F, Bouchut A, Coustau C. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. 2005;29(5):393–407. doi: 10.1016/j.dci.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Coustau C, Mitta G, Dissous C, Guillou F, Galinier R, Allienne JF, Modat S. Schistosoma mansoni and Echinostoma caproni excretory-secretory products differentially affect gene expression in Biomphalaria glabrata embryonic cells. Parasitology. 2003;127(6):533–42. doi: 10.1017/s0031182003004049. [DOI] [PubMed] [Google Scholar]
- 16.Guillou F, Mitta G, Galinier R, Coustau C. Identification and expression of gene transcripts generated during an anti-parasitic response in Biomphalaria glabrata. Dev Comp Immunol. 2007;31(7):657–71. doi: 10.1016/j.dci.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Guillou F, Mitta G, Dissous C, Pierce R, Coustau C. Use of individual polymorphism to validate potential functional markers: case of a candidate lectin (BgSel) differentially expressed in susceptible and resistant strains of Biomphalaria glabrata. Comp Biochem Physiol B Biochem Mol Biol. 2004;138(2):175–81. doi: 10.1016/j.cbpc.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Lockyer AE, Spinks JN, Walker AJ, Kane RA, Noble LR, Rollinson D, Dias-Neto E, Jones CS. Biomphalaria glabrata transcriptome: identification of cell-signalling, transcriptional control and immune-related genes from open reading frame expressed sequence tags (ORESTES) Dev Comp Immunol. 2007;31(8):763–82. doi: 10.1016/j.dci.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockyer AE, Spinks J, Noble LR, Rollinson D, Jones CS. Identification of genes involved in interactions between Biomphalaria glabrata and Schistosoma mansoni by suppression subtractive hybridization. Mol Biochem Parasitol. 2007;151(1):18–27. doi: 10.1016/j.molbiopara.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockyer AE, Noble LR, Rollinson D, Jones CS. Schistosoma mansoni: resistant specific infection-induced gene expression in Biomphalaria glabrata identified by fluorescent-based differential display. Exp Parasitol. 2004;107(12):97–104. doi: 10.1016/j.exppara.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Bouchut A, Coustau C, Gourbal B, Mitta G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: new candidate genes evidenced by a suppressive subtractive hybridization approach. Parasitology. 2007;134(4):575–88. doi: 10.1017/S0031182006001673. [DOI] [PubMed] [Google Scholar]
- 22.Bouchut A, Roger E, Coustau C, Gourbal B, Mitta G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: potential involvement of adhesion genes. Int J Parasitol. 2006;36(2):175–84. doi: 10.1016/j.ijpara.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Hanelt B, Lun CM, Adema CM. Comparative ORESTES-sampling of transcriptomes of immune-challenged Biomphalaria glabrata snails. J Invertebr Pathol. 2008;99(2):192–203. doi: 10.1016/j.jip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coppin JF, Lefebvre C, Caby S, Cocquerelle C, Vicogne J, Coustau C, Dissous C. Gene expression changes in Schistosoma mansoni sporocysts induced by Biomphalaria glabrata embryonic cells. Parasitol Res. 2003;89(2):113–9. doi: 10.1007/s00436-002-0643-2. [DOI] [PubMed] [Google Scholar]
- 25.Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei. Dev Comp Immunol. 2005;29(4):295–303. doi: 10.1016/j.dci.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Nowak TS, Woodards AC, Jung Y, Adema CM, Loker ES. Identification of transcripts generated during the response of resistant Biomphalaria glabrata to Schistosoma mansoni infection using suppression subtractive hybridization. J Parasitol. 2004;90(5):1034–40. doi: 10.1645/GE-193R1. [DOI] [PubMed] [Google Scholar]
- 27.Schneider O, Zelck UE. Differential display analysis of hemocytes from schistosome-resistant and schistosome-susceptible intermediate hosts. Parasitol Res. 2001;87(6):489–91. doi: 10.1007/s004360100394. [DOI] [PubMed] [Google Scholar]
- 28.Miller AN, Raghavan N, FitzGerald PC, Lewis FA, Knight M. Differential gene expression in haemocytes of the snail Biomphalaria glabrata: effects of Schistosoma mansoni infection. Int J Parasitol. 2001;31(7):687–96. doi: 10.1016/s0020-7519(01)00133-3. [DOI] [PubMed] [Google Scholar]
- 29.Raghavan N, Tettelin H, Miller A, Hostetler J, Tallon L, Knight M. Nimbus (BgI): an active non-LTR retrotransposon of the Schistosoma mansoni snail host Biomphalaria glabrata. Int J Parasitol. 2007;37(12):1307–18. doi: 10.1016/j.ijpara.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira AL, Da Silva D, Zanotti-Magalhaes EM, Abdel-Hamid AZ, Ribeiro-Paes JT. Schistosome/mollusk: genetic compatibility. Genet Mol Res. 2008;7(2):518–26. doi: 10.4238/vol7-2gmr444. [DOI] [PubMed] [Google Scholar]
- 31.Webster JP, Davies CM, Ndamba J, Noble LR, Jones CS, Woolhouse ME. Spatio-temporal genetic variability in the schistosome intermediate host Biomphalaria pfeifferi. Ann Trop Med Parasitol. 2001;95(5):515–27. doi: 10.1080/00034980120072239. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman JI, Webster JP, Ndamba J, Woolhouse ME. Extensive genetic variation revealed in adjacent populations of the schistosome intermediate host Biomphalaria pfeifferi from a single river system. Ann Trop Med Parasitol. 1998;92(6):693–8. doi: 10.1080/00034989859159. [DOI] [PubMed] [Google Scholar]
- 33.Zavodna M, Sandland GJ, Minchella DJ. Effects of intermediate host genetic background on parasite transmission dynamics: a case study using Schistosoma mansoni. Exp Parasitol. 2008;120(1):57–61. doi: 10.1016/j.exppara.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiele EA, Sorensen RE, Gazzinelli A, Minchella DJ. Genetic diversity and population structuring of Schistosoma mansoni in a Brazilian village. Int J Parasitol. 2008;38(34):389–99. doi: 10.1016/j.ijpara.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandland GJ, Foster AV, Zavodna M, Minchella DJ. Interplay between host genetic variation and parasite transmission in the Biomphalaria glabrata-Schistosoma mansoni system. Parasitol Res. 2007;101(4):1083–9. doi: 10.1007/s00436-007-0593-9. [DOI] [PubMed] [Google Scholar]
- 36.Prugnolle F, de Meeûs T, Pointier JP, Durand P, Rognon A, Théron A. Geographical variations in infectivity and susceptibility in the host-parasite system Schistosoma mansoni/Biomphalaria glabrata: no evidence for local adaptation. Parasitology. 2006;133(3):313–9. doi: 10.1017/S0031182006000412. [DOI] [PubMed] [Google Scholar]
- 37.Theron A, Sire C, Rognon A, Prugnolle F, Durand P. Molecular ecology of Schistosoma mansoni transmission inferred from the genetic composition of larval and adult infrapopulations within intermediate and definitive hosts. Parasitology. 2004;129(5):571–85. doi: 10.1017/s0031182004005943. [DOI] [PubMed] [Google Scholar]
- 38.Cai J, Wang Z, Cai C, Zhou Y. Characterization and identification of virulent Klebsiella oxytoca isolated from abalone (Haliotis diversicolor supertexta) postlarvae with mass mortality in Fujian, China. J Invert Pathol. 2008;97:70–5. doi: 10.1016/j.jip.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Ford SE, Chintala MM, Bushek D. Comparison of in vitro-cultured and wild-type Perkinsus marinus. I. Pathogen virulence. Dis Aquat Organ. 2002;51(3):187–201. doi: 10.3354/dao051187. [DOI] [PubMed] [Google Scholar]
- 40.Loker ES, Bayne CJ. Molecular Studies of the Molluscan Response to Digenean Infection. In: Beck G, Sugumaran M, Cooper EL, editors. Advances in Experimental Medicine and Biology. Vol. 484 “ Phylogenetic perspectives on the vertebrate immune system”. Kluwer Academic/Plenum; New York, NY 20001: 2001. pp. 209–22. [DOI] [PubMed] [Google Scholar]
- 41.Bayne CJ, Fryer SE. Phagocytosis and invertebrate opsonins in relation to parasitism. Ann N Y Acad Sci. 1994;712:162–77. doi: 10.1111/j.1749-6632.1994.tb33571.x. [DOI] [PubMed] [Google Scholar]
- 42.Zelck UE. Glycosidase activities in plasma of naive and schistosome-infected Biomphalaria glabrata (Gastropoda) Parasitology. 1999;119(6):563–8. doi: 10.1017/s0031182099005028. [DOI] [PubMed] [Google Scholar]
- 43.Bayne CJ, Loker ES, Yui MA. Interactions between the plasma proteins of Biomphalaria glabrata (Gastropoda) and the sporocyst tegument of Schistosoma mansoni (Trematoda) Parasitology. 1986;92:653–64. doi: 10.1017/s0031182000065513. [DOI] [PubMed] [Google Scholar]
- 44.Plows LD, Cook RT, Davies AJ, Walker AJ. Integrin engagement modulates the phosphorylation of focal adhesion kinase, phagocytosis, and cell spreading in molluscan defence cells. Biochim Biophys Acta. 2006;1763(8):779–86. doi: 10.1016/j.bbamcr.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Zelck UE, Gege BE, Schmid S. Specific inhibitors of mitogen-activated protein kinase and PI3-K pathways impair immune responses by hemocytes of trematode intermediate host snails. Dev Comp Immunol. 2007;31(4):321–31. doi: 10.1016/j.dci.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Humphries JE, Yoshino TP. Regulation of hydrogen peroxide release in circulating hemocytes of the planorbid snail Biomphalaria glabrata. Dev Comp Immunol. 2008;32(5):554–62. doi: 10.1016/j.dci.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci U S A. 1997;94(16):8691–6. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang SM, Léonard PM, Adema CM, Loker ES. Parasite-responsive IgSF members in the snail Biomphalaria glabrata: characterization of novel genes with tandemly arranged IgSF domains and a fibrinogen domain. Immunogenetics. 2001;53(8):684–94. doi: 10.1007/s00251-001-0386-8. Erratum in: Immunogenetics 2002;53(10-11):992. [DOI] [PubMed] [Google Scholar]
- 49.Haas BJ, Berriman M, Hirai H, Cerqueira GG, Loverde PT, El-Sayed NM. Schistosoma mansoni genome: closing in on a final gene set. Exp Parasitol. 2007;117(3):225–8. doi: 10.1016/j.exppara.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, Song HD, Xu XR, Wang ZJ, Rong YP, Zeng LC, Wu J, Zhang X, Wang JJ, Xu XN, Wang SY, Fu G, Zhang XL, Wang ZQ, Brindley PJ, McManus DP, Xue CL, Feng Z, Chen Z, Han ZG. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nat Genet. 2003;35(2):139–47. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- 51.Adema CM, Luo MZ, Hanelt B, Hertel LA, Marshall JJ, Zhang SM, DeJong RJ, Kim HR, Kudrna D, Wing RA, Soderlund C, Knight M, Lewis FA, Caldeira RL, Jannotti-Passos LK, Carvalho Odos S, Loker ES. A bacterial artificial chromosome library for Biomphalaria glabrata, intermediate snail host of Schistosoma mansoni. Mem Inst Oswaldo Cruz. 2006;101(Suppl 1):167–77. doi: 10.1590/s0074-02762006000900027. [DOI] [PubMed] [Google Scholar]
- 52.Fitzpatrick JM, Johnston DA, Williams GW, Williams DJ, Freeman TC, Dunne DW, Hoffmann KF. An oligonucleotide microarray for transcriptome analysis of Schistosoma mansoni and its application/use to investigate gender-associated gene expression. Mol Biochem Parasitol. 2005;141:1–13. doi: 10.1016/j.molbiopara.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Gobert GN, McInnes R, Moertel L, Nelson C, Jones MK, Hu W, McManus DP. Transcriptomics tool for the human Schistosoma blood flukes using microarray gene expression profiling. Exp Parasitol. 2006;114(3):160–72. doi: 10.1016/j.exppara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 54.Jiang Y, Loker ES, Zhang SM. In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference. Dev Comp Immunol. 2006;30(10):855–66. doi: 10.1016/j.dci.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyle JP, Wu XJ, Shoemaker CB, Yoshino TP. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Mol Biochem Parasitol. 2003 May;128(2):205–15. doi: 10.1016/s0166-6851(03)00078-1. [DOI] [PubMed] [Google Scholar]
- 56.Goodall CP, Bender RC, Broderick EJ, Bayne CJ. Constitutive differences in Cu/Zn superoxide dismutase mRNA levels and activity in hemocytes of Biomphalaria glabrata (Mollusca) that are either susceptible or resistant to Schistosoma mansoni (Trematoda) Mol Biochem Parasitol. 2004;137(2):321–8. doi: 10.1016/j.molbiopara.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Bender RC, Broderick EJ, Goodall CP, Bayne CJ. Respiratory burst of Biomphalaria glabrata hemocytes: Schistosoma mansoni-resistant snails produce more extracellular H2O2 than susceptible snails. J Parasitol. 2005;91(2):275–9. doi: 10.1645/GE-415R. [DOI] [PubMed] [Google Scholar]
- 58.McManus DP, Loukas A. Current status of vaccines for schistosomiasis. Clin Microbiol Rev. 2008;21(1):225–42. doi: 10.1128/CMR.00046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sayed AA, Simeonov A, Thomas CJ, Inglese J, Austin CP, Williams DL. Identification of oxadiazoles as new drug leads for the control of schistosomiasis. Nat Med. 2008;14(4):407–12. doi: 10.1038/nm1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richards CS, Merritt JW., Jr Genetic factors in the susceptibility of juvenile Biomphalaria glabrata to Schistosoma mansoni infection. Am J Trop Med Hyg. 1972;21:425–34. doi: 10.4269/ajtmh.1972.21.425. [DOI] [PubMed] [Google Scholar]
- 61.Richards CS. Genetic factors in susceptibility of Biomphalaria glabrata for different strains of Schistosoma mansoni. Parasitology. 1975;70:231–41. doi: 10.1017/s0031182000049696. [DOI] [PubMed] [Google Scholar]
- 62.Richards CS, Shade PC. The genetic variation of compatibility in Biomphalaria glabrata and Schistosoma mansoni. J Parasitol. 1987;73(6):1146–51. [PubMed] [Google Scholar]
- 63.Michelson EH, DuBois L. Susceptibility of Bahian populations of Biomphalaria glabrata to an allopatric strain of Schistosoma mansoni. Am J Trop Med Hyg. 1978;27:782–6. doi: 10.4269/ajtmh.1978.27.782. [DOI] [PubMed] [Google Scholar]
- 64.Pan SC. The fine structure of the miracidium of Schistosoma mansoni. J Invert Pathol. 1980;36:307–72. doi: 10.1016/0022-2011(80)90040-3. [DOI] [PubMed] [Google Scholar]
- 65.Lewis FA, Stirewalt MA, Souza CP, Gazzinelli G. Large-scale laboratory maintenance of Schistosoma mansoni, with observations on three schistosome/snail host combinations. J Parasitol. 1986;72(6):813–29. [PubMed] [Google Scholar]
- 66.Théron A, Coustau C. Are Biomphalaria snails resistant to Schistosoma mansoni? J Helminthol. 2005;79(3):187–91. doi: 10.1079/joh2005299. [DOI] [PubMed] [Google Scholar]
- 67.Zelck UE, Von Janowsky B. Antioxidant enzymes in intramolluscan Schistosoma mansoni and ROS-induced changes in expression. Parasitology. 2004;128(5):493–501. doi: 10.1017/s0031182004004895. [DOI] [PubMed] [Google Scholar]
- 68.Roger E, Grunau C, Pierce RJ, Hirai H, Gourbal B, Galinier R, Emans R, Cesari IM, Cosseau C, Mitta G. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata) PLoS Negl Trop Dis. 2008;2(11):e330. doi: 10.1371/journal.pntd.0000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnston LA, Yoshino TP. Larval Schistosoma mansoni excretory-secretory glycoproteins (ESPs) bind to hemocytes of Biomphalaria glabrata (Gastropoda) via surface carbohydrate binding receptors. J Parasitol. 2001;87(4):786–93. doi: 10.1645/0022-3395(2001)087[0786:LSMESG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 70.Léonard PM, Adema CM, Zhang SM, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269(12):155–65. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]
- 71.Zhang SM, Nian H, Zeng Y, Dejong RJ. Fibrinogen-bearing protein genes in the snail Biomphalaria glabrata: characterization of two novel genes and expression studies during ontogenesis and trematode infection. Dev Comp Immunol. 2008;32(10):1119–30. doi: 10.1016/j.dci.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang SM, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. 2004;341:255–66. doi: 10.1016/j.gene.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang SM, Loker ES. The FREP gene family in the snail Biomphalaria glabrata: additional members, and evidence consistent with alternative splicing and FREP retrosequences. Dev Comp Immunol. 2003;27(3):175–87. doi: 10.1016/s0145-305x(02)00091-5. [DOI] [PubMed] [Google Scholar]
- 74.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004 Jul 9;305(5681):251–4. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 75.Zhang SM, Zeng Y, Loker ES. Expression profiling and binding properties of fibrinogen-related proteins (FREPs), plasma proteins from the schistosome snail host Biomphalaria glabrata. Innate Immun. 2008;14(3):175–89. doi: 10.1177/1753425908093800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fryer SE, Hull CJ, Bayne CJ. Phagocytosis of yeast by Biomphalaria glabrata: Carbohydrate specificity of hemocyte receptors and a plasma opsonin. Dev Comp Immunol. 1989;13:9–16. doi: 10.1016/0145-305x(89)90011-6. [DOI] [PubMed] [Google Scholar]
- 77.Zhang SM, Zeng Y, Loker ES. Characterization of immune genes from the schistosome host snail Biomphalaria glabrata that encode peptidoglycan recognition proteins and gram-negative bacteria binding protein. Immunogenetics. 2007;59(11):883–98. doi: 10.1007/s00251-007-0245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Renwrantz LR, Richards EH. Recognition of beta-glucuronidase by the calcium-independent phosphomannosyl surface receptor of haemocytes from the gastropod mollusc, Helix pomatia. Dev Comp Immunol. 1992;16(23):251–6. doi: 10.1016/0145-305x(92)90024-7. [DOI] [PubMed] [Google Scholar]
- 79.Stout B, Adema CM, Zhang S-M, Loker ES. The biology of FREPs: diversified lectins with fibrinogen-related domains from the freshwater snail B. glabrata. In: Vasta GR, Ahmed H, editors. Animal Lectins: A Functional View. Cleveland, OH: CRC Press Taylor & Francis; 2008. in press. [Google Scholar]
- 80.Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005 Sep 16;309(5742):1874–8. doi: 10.1126/science.1116887. Epub 2005 Aug 18. [DOI] [PubMed] [Google Scholar]
- 81.Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, Buckley KM, Brockton V, Nair SV, Berney K, Fugmann SD, Anderson MK, Pancer Z, Cameron RA, Smith LC, Rast JP. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006 Dec 1;300(1):349–65. doi: 10.1016/j.ydbio.2006.08.065. Epub 2006 Sep 3. [DOI] [PubMed] [Google Scholar]
- 82.Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto MR, Marino R, Shida K, Ikeda M, Ikeda M, Arai M, Inoue Y, Shimizu T, Satoh N, Rokhsar DS, Du Pasquier L, Kasahara M, Satake M, Nonaka M. Genomic analysis of immunity in a urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics. 2003 Nov;55(8):570–81. doi: 10.1007/s00251-003-0606-5. Epub 2003 Oct 7. [DOI] [PubMed] [Google Scholar]
- 83.Basch PF, Samuelson J. Cell biology of schistosomes. I Ultrastructure and transformations. In: Wyler DJ, editor. Modern Parasite Biology. 106 Vol. 91. New York WH: Freeman & Co.; 1990. [Google Scholar]
- 84.Bayne CJ, Yoshino TP. Determinants of compatibility in mollusc-trematode parasitism. Am Zoologist. 1989;29:399–407. [Google Scholar]
- 85.Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ. Disruption of ERK signalling in Biomphalaria glabrata defence cells by Schistosoma mansoni: Implications for parasite survival in the snail host. Dev Comp Immunol. 2008;32(12):1561–71. doi: 10.1016/j.dci.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 86.Sentman CL, Olsson MY, Kärre K. Missing self recognition by natural killer cells in MHC class I transgenic mice. A ‘receptor calibration’ model for how effector cells adapt to self. Semin Immunol. 1995;7(2):109–19. doi: 10.1006/smim.1995.0015. [DOI] [PubMed] [Google Scholar]
- 87.Lehr T, Beuerlein K, Doenhoff MJ, Grevelding CG, Geyer R. Localization of carbohydrate determinants common to Biomphalaria glabrata as well as to sporocysts and miracidia of Schistosoma mansoni. Parasitology. 2008;135(8):931–42. doi: 10.1017/S0031182008004514. [DOI] [PubMed] [Google Scholar]
- 88.Plows LD, Cook RT, Davies AJ, Walker AJ. Carbohydrates that mimic schistosome surface coat components affect ERK and PKC signalling in Lymnaea stagnalis haemocyteS. Int J Parasitol. 2005;35(3):293–302. doi: 10.1016/j.ijpara.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 89.Humbert E, Coustau C. Refractoriness of host haemocytes to parasite immunosuppressive factors as a putative resistance mechanism in the Biomphalaria glabrata-Echinostoma caproni system. Parasitology. 2001;122(Pt 6):651–60. doi: 10.1017/s003118200100782x. [DOI] [PubMed] [Google Scholar]
- 90.Dikkeboom R, Bayne CJ, van der Knaap WPW, Tijnagel JMGH. Possible role of reactive forms of oxygen in in vitro killing of Schistosoma mansoni sporocysts by hemocytes of Lymnaea stagnalis. Parasitol Res. 1988;75:148–54. doi: 10.1007/BF00932715. [DOI] [PubMed] [Google Scholar]
- 91.Hahn UK, Bender RC, Bayne CJ. Production of reactive oxygen species by hemocytes of Biomphalaria glabrata: carbohydrate-specific stimulation. Dev Comp Immunol. 2000;24:531–41. doi: 10.1016/s0145-305x(00)00017-3. [DOI] [PubMed] [Google Scholar]
- 92.Hahn UK, Bender RC, Bayne CJ. Killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata: role of reactive oxygen species. J Parasitol. 2001;8:292–9. doi: 10.1645/0022-3395(2001)087[0292:KOSMSB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 93.Hahn UK, Bender RC, Bayne CJ. Involvement of nitric oxide in killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata. J Parasitol. 2001;87(4):778–85. doi: 10.1645/0022-3395(2001)087[0778:IONOIK]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 94.Boemhler A, Fryer SE, Bayne CJ. Killing of Schistosoma mansoni sporocysts by Biomphalaria glabrata hemolymph in vitro: alteration of hemocyte behavior after poly-L-lysine treatment of plastic and the kinetics of killing by different host strains. J Parasitol. 1996;82:332–5. [PubMed] [Google Scholar]
- 95.Bender RC, Broderick EJ, Goodall CP, Bayne CJ. Respiratory burst of Biomphalaria glabrata hemocytes: Schistosoma mansoni resistant snails produce more extracellular H2O2 than susceptible snails. J Parasitol. 2005;91:275–9. doi: 10.1645/GE-415R. [DOI] [PubMed] [Google Scholar]
- 96.Goodall CP, Bender RC, Broderick EJ, Bayne CJ. Constitutive differences in Cu/Zn superoxide dismutase mRNA levels and activity in hemocytes of Biomphalaria glabrata (Mollusca) that are either susceptible or resistant to Schistosoma mansoni (Trematoda) Mol Biochem Parasitol. 2004;137:321–8. doi: 10.1016/j.molbiopara.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 97.Goodall CP, Bender RC, Brooks JK, Bayne CJ. Biomphalaria glabrata cytosolic copper-zinc dismutase (SOD1) gene: association of SOD1 alleles with resistance/susceptibility to Schistosoma mansoni. Mol Biochem Parasitol. 2006;147(2):207–10. doi: 10.1016/j.molbiopara.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 98.Bender RC, Goodall CP, Blouin MS, Bayne CJ. Variation in expression of Biomphalaria glabrata SOD1: A potential controlling factor in susceptibility/resistance to Schistosoma mansoni. Dev Comp Immunol. 2007;31(9):874–8. doi: 10.1016/j.dci.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 99.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–3. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 100.Mei H, LoVerde PT. Schistosoma mansoni: the developmental regulation and immunolocalization of antioxidant enzymes. Exp Parasitol. 1997;86(1):69–78. doi: 10.1006/expr.1997.4150. [DOI] [PubMed] [Google Scholar]
- 101.Myers J, Ittiprasert W, Raghavan N, Miller A, Knight M. Differences in cysteine protease activity in Schistosoma mansoni-resistant and -susceptible Biomphalaria glabrata and characterization of the hepatopancreas cathepsin B Full-length cDNA. J Parasitol. 2008;94(3):659–68. doi: 10.1645/GE-1410R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]