Abstract
Melanocytic nevi are thought to be senescent clones of melanocytes that have acquired an oncogenic BRAF mutation. BRAF mutation is considered to be a crucial step in the initiation of melanocyte transformation. However, using immunomagnetic separation or laser-capture microdissection, we examined BRAF mutations in sets of approximately 50 single cells isolated from acquired melanocytic nevi from 13 patients and found a substantial number of nevus cells that contained wild-type BRAF mixed with nevus cells that contained BRAFV600E. Furthermore, we simultaneously amplified BRAF exon 15 and a neighboring single nucleotide polymorphism (SNP), rs7801086, from nevus cell samples obtained from four patients who were heterozygous for this SNP. Subcloning and sequencing of the polymerase chain reaction products showed that both SNP alleles harbored the BRAFV600E mutation, indicating that the same BRAFV600E mutation originated from different cells. The polyclonality of BRAF mutations in acquired melanocytic nevi suggests that mutation of BRAF may not be an initial event in melanocyte transformation.
CONTEXT AND CAVEATS
Prior knowledge
Oncogenic BRAF mutation has been considered to be an early step in the formation of melanocytic nevi, but recently, clonal heterogeneity was found in a few nevi.
Study design
Approximately 50 single cells were separated from each of 13 melanocytic nevi either by using immunomagnetic beads or by laser-capture microdissection and were subjected to single-cell polymerase chain reaction and sequencing to determine BRAF mutations. In another experiment, BRAF and a neighboring single-nucleotide polymorphism were simultaneously amplified from nevi of four patients who were heterozygous for the polymorphism.
Contribution
Although BRAF mutations were always found among nevus cells, cells that contained only wild-type BRAF predominated in nine of 13 nevi. When BRAF was sequenced from both alleles of four patients heterozygous for an adjacent polymorphism, both alleles harbored BRAF mutations.
Implications
These results suggest that BRAF may be mutated as a late event in melanocytic tumorigenesis.
Limitations
A small number of nevi of only two histological types were examined.
From the Editors
v-raf murine sarcoma viral oncogene homolog B1 (BRAF) is a member of the raf gene family that encodes a cytoplasmic serine/threonine kinase that signals in the mitogen-activated protein kinase pathway and mediates cell proliferation and differentiation. Mutations in the BRAF gene, most commonly including BRAFV600E, which encodes an amino acid substitution that leads to RAS-independent autoactivation of the BRAF kinase with subsequent stimulation of the downstream kinases and transcription factors, have been frequently found in melanoma lesions and melanocytic nevi (1,2). This finding has been interpreted to suggest that mutation of BRAF is a crucial step in the initiation of melanocyte transformation (2). Furthermore, two recent studies have shown that sustained BRAFV600E expression in normal melanocytes causes either cell cycle arrest accompanied by the induction of both p16INK4a protein expression and senescence-associated acidic β-galactosidase activity (3) or substantial reduction of cell proliferation accompanied by the induction of nuclear p16INK4a expression (4). Both p16INK4a protein expression and acidic β-galactosidase activity were also demonstrated in melanocytic nevi in situ (3,4). These results suggest that melanocytic nevi are benign tumors that originate from the clonal expansion of a single melanocyte that acquires a BRAFV600E mutation and temporarily proliferates in response to oncogenic BRAF signaling, followed by growth arrest due to oncogene-induced senescence (5).
However, in a recent analysis of BRAF mutations in a large series of melanocytic nevi (6), we observed clonal heterogeneity in terms of BRAF mutations among the cells in a few small congenital nevi (6). Because of these findings and the possibility that they contradict the currently accepted model of melanocytic neoplastic progression (5), we tested the polyclonality of BRAF mutations in acquired melanocytic nevi.
The study was approved by the medical ethics committee of the Shinshu University School of Medicine. All patients gave written informed consent. We obtained acquired melanocytic nevi from 14 patients. After bisection, one-half of the nevus tissue was routinely fixed and embedded in paraffin. The other half was used to isolate single nevus cells. For six nevi (numbers 1–6 in Table 1), we removed the epidermis, minced the tissue, and collected nevus cells using a cocktail of human high molecular weight-melanoma-associated antigen (HMW-MAA)–specific monoclonal antibodies and magnetic beads (Dynabeads CELLection Pan Mouse IgG Kit; Invitrogen Dynal AS, Oslo, Norway), as described previously (7). Separated nevus cells were smeared on film-coated glass slides (Meiwafosis Co Ltd, Osaka, Japan) and subjected to laser-capture microdissection using a PALM-MB microdissection system (PALM Microlaser technologies, Bernried, Germany) to isolate approximately 60–65 cells.
Table 1.
Polyclonality of BRAF mutations in acquired melanocytic nevi as revealed by single-cell polymerase chain reaction (PCR) and sequencing of DNA from approximately 50 cells per nevus*
| Sample No. | Age, y | Sex | Site | Histology (Type) | Method | No. of cells with wild-type BRAF | No. of cells with heterozygous V600E mutation | No. of cells with other BRAF mutations† | No. of cells with homozygous V600E mutation | PCR failure | Total No. of cells (valid PCR) | P‡ |
| N-1 | 34 | F | Nose | Intradermal nevus (Miescher) | IMS | 15 | 34 | 0 | 1 | 9 | 59 (50) | <.001 |
| N-2 | 28 | F | Abdomen | Intradermal nevus (Unna) | IMS | 11 | 38 | 0 | 1 | 9 | 59 (50) | .018 |
| N-3 | 31 | F | Nose | Intradermal nevus (Miescher) | IMS | 26 | 22 | 0 | 2 | 12 | 62 (50) | <.001 |
| N-4 | 44 | M | Face | Intradermal nevus (Miescher) | IMS | 33 | 11 | 2 (V600K)3 (T599I) | 1 | 14 | 64 (50) | <.001 |
| N-5 | 52 | F | Neck | Intradermal nevus (Unna) | IMS | 28 | 21 | 0 | 1 | 13 | 63 (50) | <.001 |
| N-6 | 35 | F | Face | Intradermal nevus (Miescher) | IMS | 30 | 20 | 0 | 2 | 10 | 62 (52) | <.001 |
| N-7 | 13 | F | Abdomen | Intradermal nevus (Unna) | FTS | 29 | 16 | 1 (V600A) | 2 | 15 | 63 (48) | <.001 |
| N-8 | 65 | F | Nose | Compound nevus (Miescher) | FTS | 31 | 14 | 0 | 5 | 9 | 59 (50) | <.001 |
| N-9 | 81 | M | Face | Compound nevus (Miescher) | FTS | 21 | 23 | 1 (V600E/G) | 4 | 16 | 65 (49) | .012 |
| N-10 | 28 | F | Arm | Intradermal nevus (Unna) | FTS | 31 | 16 | 0 | 3 | 15 | 65 (50) | <.001 |
| N-11 | 27 | F | Face | Intradermal nevus (Miescher) | FTS | 13 | 30 | 0 | 7 | 13 | 63 (50) | .510 |
| N-12 | 25 | F | Sole | Junctional nevus (Acral) | FTS | 42 | 3 | 0 | 1 | 16 | 62 (46) | <.001 |
| N-13 | 37 | M | Sole | Compound nevus (Acral) | FTS | 45 | 5 | 0 | 0 | 13 | 63 (50) | <.001 |
| N-14 | 27 | F | Abdomen | Intradermal nevus (Unna) | FTS | — | — | — | — | — | — | — |
| 928mel | — | — | — | Melanoma cell line | IMS | 1 | 47 | 0 | 2 | 13 | 63 (50) | — |
| 928mel | — | — | — | Melanoma cell line | FTS | 7 | 38 | 0 | 5 | 12 | 62 (50) | — |
FTS = Frozen tissue section. Six-μm cryosections were prepared from the nevus tissues or melanoma cell suspension embedded in agarose; IMS = Immunomagnetic cell separation using antibodies against human high molecular weight-melanoma-associated antigen.
V600K, GT1798-99→AA; T599I, C1796→T; V600A, T1799→C; V600E/G, T1799→A + T1799→G.
Two-sided binomial tests were conducted using the 95% confidence limit of each method (0.11 for IMS and 0.27 for FTS) with the null hypothesis that all the wild types were due to allele dropout.
Immunohistochemical staining with monoclonal antibodies showed high levels of expression of HMW-MAA on nevus cells (Supplementary Figure 1, available online). No staining of melanocytes was detected. Although a few keratinocytes within the hair follicles and the basal layer of the epidermis expressed HMW-MAA, as reported previously (8), virtually no cells were captured when we performed immunomagnetic cell separation using normal skin. To further confirm the specificity of the nevus cell isolation, we stained the immunomagnetically separated cells with a rabbit polyclonal anti–S-100 antibody (Dako, Glostrup, Denmark) (1:300 dilution) followed by DAKO Envision System (Dako, Carpinteria, CA), for three samples (sample numbers 3–5), and observed positive staining for almost all of the separated cells.
For the remaining eight nevi (numbers 7–14 in Table 1), one-half of the nevus tissue was snap frozen in optimal cutting temperature compound, cut 6 μm thick, stained with toluidine blue, and subjected to laser-capture microdissection. Approximately 60–65 isolated single nevus cells were collected into lysis buffer containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 4 mg/mL proteinase K (Roche Diagnostics, Basel, Switzerland), and 3% Tween 20 and incubated for 16 hours at 50°C (6). We amplified exon 15 of the gene using heminested polymerase chain reaction (PCR) (Supplementary Table 1, available online). PCR products were purified and sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). To prevent potential contamination, aerosol resistant tips with filters were used and gloves and a mask were worn at all times. Samples without target DNA and/or DNA from single epidermal keratinocytes containing wild-type BRAF (similarly isolated by laser-capture microdissection) were always included as negative controls to monitor PCR contamination.
We successfully isolated single nevus cells from immunomagnetically captured cell smears and from frozen tissue sections (Figure 1, A and B). The success rate of PCR amplification from single cells ranged from 74% to 85%, and we obtained BRAF sequences from approximately 50 single nevus cells for each patient sample. Sequence analyses revealed that all the 13 samples of the acquired melanocytic nevi contained nevus cells that had wild-type BRAF and cells that were heterozygous for BRAFV600E (Table 1). Nevus cells containing wild-type BRAF predominated in nine of 13 nevi. Consistent with the results of our previous study (6), nevus cells harboring BRAF mutations were very rarely detected in the two acral nevi that we examined (sample numbers 12 and 13) compared with nevi from other sites, which suggested a possible role for UV exposure in the acquisition of BRAF mutations in acquired melanocytic nevi. Cells with rare BRAF mutations, such as BRAFV600K, BRAFV600A, BRAFV600E/G, and BRAFT599I, all of which have been previously described in melanoma lesions (http://www.sanger.ac.uk/genetics/CGP/cosmic), were found in three nevi (numbers 4, 7, and 9). Except for one compound heterozygous mutation of BRAFV600E/G that was identified in sample number 9 (Figure 1, B), all of the other variant mutations were heterozygous. The protein products of two of these variant mutations (BRAFT599I and BRAFV600K) were shown to have much lower kinase activity compared with BRAFV600E (9). Thus, cell clones harboring these variant mutations may have weaker growth advantage than those with the BRAFV600E mutation, and may have been overlooked in the previous conventional sequence analyses, because the heterozygous mutation could be reliably discernible only when the samples contained more than 20% of clonal mutant cells (6).
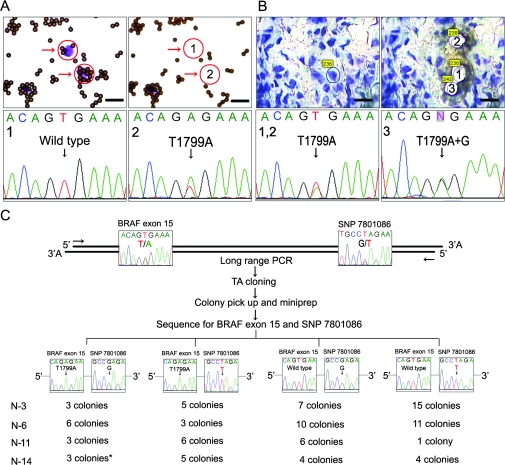
Figure 1.
Polyclonality of v-raf murine sarcoma viral oncogene homolog B1 (BRAF) mutations in acquired melanocytic nevi. A) Selection of single nevus cells after immunomagnetic separation. Single nevus cells (purple dots with arrows) were captured by high molecular weight-melanoma-associated antigen–specific monoclonal antibodies bound to immunomagnetic beads (pink dots). The cells (encircled) were procured by laser-capture microdissection (top; bar = 20 μm). Polymerase chain reaction (PCR) amplification and subsequent sequencing of single nevus cells showed wild-type BRAF and BRAFV600E mutations (bottom). B) Laser-capture microdissection of frozen tissue section of acquired melanocytic nevi followed by direct sequencing of BRAF exon 15 (top; bar = 20 μm). Sequencing revealed two of the contiguous single nevus cells to have the BRAFV600E mutation and one to have a compound heterozygous BRAFV600E (T1799A) and BRAFV600G (T1799G) mutation, showing a heterogeneous pattern of BRAF mutations in proximal cells on a single-cell level (bottom). C) Subcloning and subsequent sequencing of BRAF exon 15 and the single nucleotide polymorphism (SNP) rs7801086. This SNP maps approximately 2 kb telomeric from BRAF exon 15. Four nevi (numbers 3, 6, 11, and 14) were excised from patients who were heterozygous for this SNP. DNA was extracted from hundreds of nevus cells isolated either by using immunomagnetic beads (numbers 3 and 6) or laser-capture microdissection of frozen tissue sections (numbers 11 and 14). A 2859-bp fragment containing both BRAF exon 15 and the SNP rs7801086 was amplified by long-range PCR. Subcloning was carried out using this fragment as an insert. Sixteen to 30 colonies were randomly picked from each patient sample and analyzed for the sequence of both BRAF exon 15 and rs7801086. In all four patient samples, colonies with BRAFV600E as well as wild-type BRAF were accompanied by different SNP alleles, some harboring the G allele and others harboring the T allele. In sample number 14, one colony (*) showed a tandem BRAFV600E/K601E (T1799→A and A1802→G) mutation.
Detection of the homozygous BRAFV600E mutation in a few single cells was considered to be due to the failure of PCR to amplify the wild-type allele, that is, allele dropout, which is a common problem in single-cell PCR (10). To assess the actual frequency of allele dropout, we performed a control experiment using the melanoma cell line 928mel, which is known to harbor the heterozygous BRAFV600E mutation (11). Using the methodology used to obtain single nevus cells, single melanoma cells were isolated by laser-capture microdissection either from immunomagnetically captured cell smears or 6-μm cryosections of cells suspended in agarose and embedded in optimal cutting temperature compound. We found wild-type BRAF sequences and homozygous BRAFV600E mutations, both of which were thought to be due to allele dropout, in three of 50 (6%) single melanoma cells isolated by immunomagnetic beads and in 12 of 50 (24%) of those obtained from 6-μm frozen sections. The much higher frequency of allele dropout in the latter is explained by the nuclear damage during tissue sectioning. This was reflected in the results that showed a lower frequency of homozygous BRAFV600E mutations in samples obtained by immunomagnetic isolation (numbers 1–6) than frozen tissue sections (numbers 8–13). Then, we conducted a two-sided binomial test for each nevus sample using the 95% confidence limits of allele dropout in control melanoma cells as the test proportion, that is, 0.11 for immunomagnetic cell separation and 0.27 for frozen tissue sections. We used SPSS 15.0 software package (SPSS Inc, Tokyo, Japan) for this analysis. At the statistical significance level of .05, the possibility that all of the wild-type sequences were due to drop out of the mutant alleles was denied in all but one nevus (number 11), indicating that most acquired melanocytic nevi were composed of both BRAF-mutated cells and BRAF-wild-type cells.
We then further investigated whether BRAF mutant cells in acquired melanocytic nevi are monoclonal or polyclonal. For this purpose, we examined four acquired melanocytic nevi (numbers 3, 6, 11, and 14) that were excised from patients who were heterozygous for the single nucleotide polymorphism (SNP) rs7801086 (GCCGAGA vs GCCTAGA) (Figure 1, C). This SNP maps to chromosome 7, about 2 kb telomeric from exon 15 of the BRAF gene. We obtained pure nevus cell populations (but not single nevus cells) from bisected nevus tissues either by using immunomagnetic beads with HMW-MAA–specific monoclonal antibodies (for sample numbers 3 and 6) or by laser-capture microdissection of multiple nevus cell nests from 6-μm frozen tissue sections (for sample numbers 11 and 14). After simultaneous amplification of DNA fragments containing both BRAF exon 15 and SNP rs7801086 using the long-range Expand High FidelityPLUS PCR System (Roche Applied Science, Mannheim, Germany), we subcloned the PCR products of separate alleles in bacteria. We then sequenced BRAF exon 15 and SNP rs7801086 from 16 to 30 individual bacterial colonies (for PCR primers, see Supplementary Table 1, available online). Among the clones containing PCR products from each of the four acquired melanocytic nevi, we found both colonies that harbored the BRAFV600E (T1799→A) mutation and the T allele of SNP rs7801086 and colonies that harbored the BRAFV600E (T1799→A) mutation and the G allele of SNP rs7801086. Colonies that contained wild-type BRAF also harbored both G and T alleles. We used a high fidelity Taq polymerase in the PCR and did not find any base substitutions other than those encoding T1799A in exon 15 of the BRAF gene, except in one bacterial subclone from sample number 14 that carried a BRAFV600E/K601E (T1799→A and A1802→G) tandem mutation. These results indicate that even the same type of BRAFV600E mutation could originate from different cells in the same nevus and that multiple BRAF mutations were possible among the cells within a given nevus. Collectively, our data strongly suggest marked polyclonality of BRAF mutations in acquired melanocytic nevi.
This result suggests that acquired melanocytic nevi may be polyclonal lesions, of multicellular origin, that result from random proliferation of cells containing wild-type BRAF as well as cells containing mutant BRAF. The polymorphic X-linked human androgen receptor gene has been used previously to show that acquired melanocytic nevi are of clonal origin (12). However, a recent study questioned the validity of the human androgen receptor gene as a marker of tumor clonality because the clonal patch is relatively large in humans, often greater than 4 mm in diameter in the aorta, and even larger in the colon and breast, and because polyclonality can only be demonstrated at the borders of X-inactivation patches (13).
The reason why melanocytes in acquired melanocytic nevi are so susceptible to mutation is unknown. One possibility is that genetically aberrant clones of melanocytes might already exist in the lesional skin of acquired melanocytic nevi, which would expand and acquire multiple mutations from stimuli such as UV radiation. Alternatively, the BRAF mutation might be a second hit after the clonal proliferation of nevus cells, which is initiated by either an as yet unknown mutation or other mechanisms. Cell proliferation itself may render melanocytes prone to mutation by the leakage of genotoxic species, such as reactive oxygen species (14).
It should be noted that most of the melanocytic nevi we examined were Unna’s nevi and Miescher's nevi. We did not examine Clark's nevi, which are most commonly seen in adult Caucasians and are sometimes seen in association with melanoma (15). Nevertheless, polyclonality of BRAF mutations in the lesions of acquired melanocytic nevi suggests an alternative to the view that BRAF mutation is an initial event in melanocytic neoplasia (5).
Funding
Grant-in-Aid for Cancer Research from the Ministry of Health, Labor, and Welfare of Japan (15–10 to T.S.) and by Public Health Service grant awarded by the National Cancer Institute (R01 CA105500 to S.F.). J.L. is a PhD student supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Supplementary Material
Footnotes
The study sponsor(s) had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
References
- 1.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33(1):19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 3.Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 4.Gray-Schopfer VC, Cheong SC, Chong H, et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br J Cancer. 2006;95(4):496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaloglou C, Vredeveld LC, Mooi WJ, Peeper DS. BRAF(E600) in benign and malignant human tumours. Oncogene. 2008;27(7):877–895. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- 6.Ichii-Nakato N, Takata M, Takayanagi S, et al. High frequency of BRAFV600E mutation in acquired nevi and small congenital nevi, but low frequency of mutation in medium-sized congenital nevi. J Invest Dermatol. 2006;126(9):2111–2118. doi: 10.1038/sj.jid.5700366. [DOI] [PubMed] [Google Scholar]
- 7.Goto Y, Arigami T, Kitago M, et al. Activation of Toll-like receptors 2, 3, and 4 on human melanoma cells induces inflammatory factors. Mol Cancer Ther. 2008;7(11):3642–3653. doi: 10.1158/1535-7163.MCT-08-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campoli MR, Chang CC, Kageshita T, Wang X, McCarthy JB, Ferrone S. Human high molecular weight-melanoma-associated antigen (HMW-MAA): a melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit Rev Immunol. 2004;24(4):267–296. doi: 10.1615/critrevimmunol.v24.i4.40. [DOI] [PubMed] [Google Scholar]
- 9.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 10.Piyamongkol W, Bermudez MG, Harper JC, Wells D. Detailed investigation of factors influencing amplification efficiency and allele drop-out in single cell PCR: implications for preimplantation genetic diagnosis. Mol Hum Reprod. 2003;9(7):411–420. doi: 10.1093/molehr/gag051. [DOI] [PubMed] [Google Scholar]
- 11.Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med. 2006;203(7):1651–1656. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson WA, Lemon M, Elefanty A, Harrison-Smith M, Markham N, Norris D. Human acquired naevi are clonal. Melanoma Res. 1998;8(6):499–503. doi: 10.1097/00008390-199812000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Novelli M, Cossu A, Oukrif D, et al. X-inactivation patch size in human female tissue confounds the assessment of tumor clonality. Proc Natl Acad Sci USA. 2003;100(6):3311–3314. doi: 10.1073/pnas.0437825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miranda M, Amicarelli F, Poma A, et al. Cyto-genotoxic species leakage within human melanoma melanosomes. Molecular-morphological correlations. Biochem Mol Biol Int. 1994;32(5):913–922. [PubMed] [Google Scholar]
- 15.Ackerman AB, Magana-Garcia M. Naming acquired melanocytic nevi. Unna's, Miescher's, Spitz's, Clark's. Am J Dermatopathol. 1990;12(2):193–209. doi: 10.1097/00000372-199004000-00012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.