Abstract
Small regulatory RNAs have recently emerged as key regulators of eukaryotic gene expression. Here we used high-throughput sequencing to determine small RNA populations in the germline and soma of the African clawed frog Xenopus tropicalis. We identified a number of miRNAs that were expressed in the female germline. miRNA expression profiling revealed that miR-202-5p is an oocyte-enriched miRNA. We identified two novel miRNAs that were expressed in the soma. In addition, we sequenced large numbers of Piwi-associated RNAs (piRNAs) and other endogenous small RNAs, likely representing endogenous siRNAs (endo-siRNAs). Of these, only piRNAs were restricted to the germline, suggesting that endo-siRNAs are an abundant class of small RNAs in the vertebrate soma. In the germline, both endogenous small RNAs and piRNAs mapped to many high copy number loci. Furthermore, endogenous small RNAs mapped to the same specific subsets of repetitive elements in both the soma and the germline, suggesting that these RNAs might act to silence repetitive elements in both compartments. Data presented here suggest a conserved role for miRNAs in the vertebrate germline. Furthermore, this study provides a basis for the functional analysis of small regulatory RNAs in an important vertebrate model system.
Short RNAs have recently emerged as abundant regulators of gene expression in many eukaryotes, including plants, animals, and fungi (Sharp 2009). The lin-4 and let-7 miRNAs were the first type of endogenous short regulatory RNAs to be identified in eukaryotes (Lee et al. 1993; Reinhart et al. 2000); since then many functional small RNAs have been identified in organisms as diverse as roundworms, flies, fish, frogs, mammals, flowering plants, mosses, anemones, sponges, and even viruses, using genetics, molecular cloning, and predictions from bioinformatics (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Llave et al. 2002; Reinhart et al. 2002; Lim et al. 2003; Pfeffer et al. 2004; Arazi et al. 2005; Axtell and Bartel 2005; Watanabe et al. 2005; Grimson et al. 2008). In cells, miRNAs are tightly bound by proteins of the Ago clade of the Argonaute superfamily of RNA-binding proteins (Cerutti et al. 2000). miRNAs are thought to inhibit efficient translation of target mRNAs or to control mRNA decay.
Another class of small RNAs, 21–24 nucleotides (nt) endogenous siRNAs, was first discovered in plants in response to viral infection (Hamilton and Baulcombe 1999; Llave et al. 2002). These RNAs are thought to represent endogenous instances of short interfering RNAs (siRNAs), the mediators of RNAi (Fire et al. 1998; Tuschl et al. 1999; Zamore et al. 2000). More recently, endo-siRNAs have also been identified in Caenorhabditis elegans (Ambros and Lee 2004), Drosophila (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008), and mouse oocytes (Tam et al. 2008; Watanabe et al. 2008). endo-siRNAs are enriched in the germline of animals and map to various genomic loci including repetitive elements, pseudogenes, palindromes, and regions where both strands are transcribed. Like miRNAs, endo-siRNAs interact with Argonaute proteins. endo-siRNAs likely have roles in silencing of transposable elements or pseudogenes (Okamura et al. 2008).
A third class of 25–30 nt RNAs has been identified in Drosophila, zebrafish, mice, rats, anemones, and sponges and has been named Piwi-associated RNAs or piRNAs (Grimson et al. 2008; Klattenhoff and Theurkauf 2008). By definition piRNAs interact with proteins of the Piwi clade of the Argonaute superfamily. piRNA populations are complex; there are hundreds of thousands of unique piRNAs in mammals. Piwi and piRNAs are required for transposon silencing: for example, in Drosophila the piRNAs of the flamenco locus control the gypsy retrotransposon (Desset et al. 2003; Brennecke et al. 2007). The piRNAs of C. elegans are unique in that they are 21 nt short RNAs with distinct genomic organization and biogenesis, but a conserved role in transposon silencing (Ruby et al. 2006; Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008).
Previously, small RNAs have also been grouped together based on their genomic location as repeat-associated small RNAs (rasiRNAs) in plants, fungi, Drosophila, and zebrafish (Llave et al. 2002; Reinhart et al. 2002; Aravin et al. 2003; Chen et al. 2005b). These can now be reclassified as endo-siRNAs or piRNAs based on their size, biogenesis, and associated Argonaute superfamily proteins (Okamura et al. 2008; Malone and Hannon 2009). Although endo-siRNA and piRNA pathways are distinct, in animals, endo-siRNAs and piRNAs are 2′O-methylated at the 3′ end (Horwich et al. 2007; Tam et al. 2008; Watanabe et al. 2008). While the function of this modification remains unclear in animals, in plants, 2′O-methylation stabilizes miRNAs and endo-siRNAs (Yang et al. 2006).
Xenopus laevis has been used widely as a model system for the study of oocyte development and maturation, including the regulation of gene expression at the level of translation and RNA localization. Xenopus oogenesis is subdivided into six stages (I–VI) based on features such as diameter, pigmentation color, and the amount of yolk in the cytoplasm. Stage VI oocytes are arrested in first meiotic prophase, and can be matured into eggs, arrested in MII metaphase, by progesterone. While previous work in Xenopus has identified a number of miRNAs through cloning and comparative genomic approaches, little is know about small RNAs population during Xenopus oogenesis. Microarrays, Northern blotting, and in situ hybridization have been used to determine miRNA expression during embryogenesis and adult frog (Watanabe et al. 2005; Hikosaka et al. 2007; Michalak and Malone 2008; Tang and Maxwell 2008; Walker and Harland 2008). However, recent advances in sequencing technology have allowed the more complete assessment of small RNA species in animals, plants, and fungi.
Here we applied Illumina sequencing (formerly known as Solexa sequencing) to determine the expression of small RNAs in the Xenopus tropicalis germline and somatic tissues. This work represents the first example of small RNA high-throughput sequencing in an amphibian. Using this approach we identify abundant populations of miRNAs, piRNAs, and other small RNAs in the germline and soma of X. tropicalis. We hope that these data might set the stage for the biochemical analysis of small RNA pathways in a powerful model system, the Xenopus oocyte.
Results
The Xenopus female germline expresses different classes of small RNAs
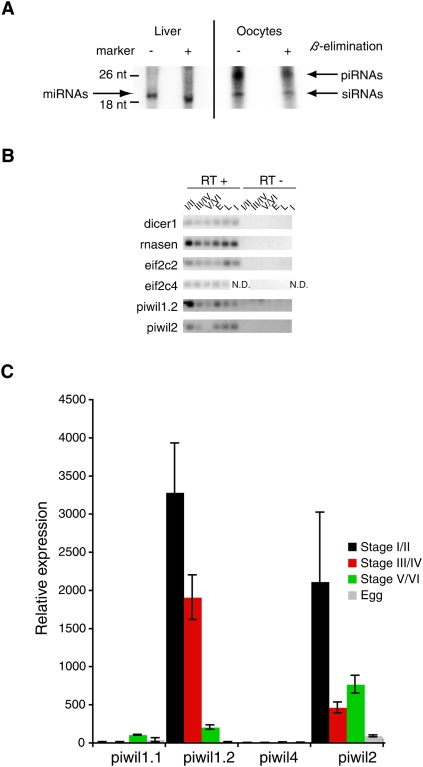
We first isolated total RNA from oocytes of Xenopus tropicalis and Xenopus laevis at different stages of oogenesis. Small RNAs were size-selected and resolved on polyacrylamide gels (Supplemental Fig. S1). We observed a strikingly similar pattern of small RNAs in the different oogenic stages in X. tropicalis and X. laevis, where short ∼22, ∼24, and ∼30 nt RNAs were most abundant. Given the known sizes of microRNAs (miRNAs), endogenous siRNAs (endo-siRNAs), and Piwi-interacting RNAs (piRNAs) in other species we suspected that these bands could represent miRNAs, endo-siRNAs, and piRNAs in X. tropicalis and X. laevis, respectively. To test this hypothesis, we took advantage of the fact that in mouse and Drosophila melanogaster endo-siRNAs and piRNAs are 2′O-methyl-modified at the 3′-most nucleotide, while miRNAs are not (Horwich et al. 2007). We purified small RNA fractions from X. tropicalis stage I and stage II oocytes and adult liver and subjected these to β-elimination, a reaction that removes the 3′-most nucleotide of RNA if this nucleotide is not 2′O-methyl-modified (Horwich et al. 2007). We then 5′ end-labeled control untreated RNA and RNA after β-elimination and separated them on a denaturing polyacrylamide gel (Fig. 1A). The small, ∼22 nt RNA band from adult liver shifted about 2 nt in mobility after β-elimination, suggesting that this RNA was not 2′O-methyl-modified. In contrast, in RNA from stage I and stage II oocytes, two small RNA bands did not shift mobility after β-elimination, which was consistent with 2′O-methylation of the 3′-most nucleotide. Based on these observations we concluded that the most abundant small RNAs in adult liver were miRNAs, whereas the most abundant small RNAs in oocytes likely were endo-siRNAs and piRNAs.
Figure 1.
Expression of small RNAs and core protein components in X. tropicalis. (A) Total RNA was isolated from X. tropicalis adult liver and stage I and stage II oocytes. RNA was size-selected using the miRVana kit. Ten micrograms of this RNA was subjected to β-elimination or not as indicated and analyzed on a denaturing gel after 5′ end-labeling. Arrows indicate RNA that likely represent miRNAs, siRNAs, and piRNAs. (B,C) The expression of core components of small RNA pathways in Xenopus oocytes, eggs, and somatic tissues was assayed using RT-PCR and qRT-PCR. For each experiment equivalent oocyte total RNA was reverse transcribed. E, egg; L, liver; I, intestine; N.D., not done. (B) RT-PCR experiment. Control experiment without the addition of reverse transcriptase. RT-PCR primers are listed in Supplemental material, Armisen_SupData4.xls. (C) qRT-PCR for the four Piwi-related genes described in X. tropicalis.
The Xenopus female germline expresses key small RNA pathway genes
We next tested if genes required for small RNA biogenesis and function are expressed in the germline or the soma of X. tropicalis. We designed specific reverse transcription RNA polymerase chain reaction (RT-PCR) assays to detect the expression of RNase enzymes required for miRNA biogenesis (dicer1, rnasen), Ago family Argonaute proteins (eif2c2, eif2c4), and Piwi family Argonaute proteins. We then assayed their expression during oogenesis, in eggs, adult liver, and adult intestine. As shown in Figure 1B, we detected expression of all four genes thought to be involved in miRNA biogenesis and function (dicer1, rnasen, eif2c2, eif2c4) in all tissues tested. We recently demonstrated that at least two Argonaute proteins of the Piwi family, piwil1.1/piwil1.2 (also known as Xiwi1a/Xiwi1b), and piwil2 (also known as Xili), are present in Xenopus oocytes (Wilczynska et al. 2009). We detected piwil1.2 and piwil2 mRNA in both the germline and the soma; however, no Piwi proteins were detected in the soma (Supplemental Fig. S2). We also confirmed expression of the Piwi family Argonaute proteins using quantitative RT-PCR (qRT-PCR) (Fig. 1C) and by searching X. tropicalis EST libraries (data not shown).
High-throughput sequencing identifies abundant miRNAs, piRNAs, and other small RNAs in X. tropicalis
Next, we prepared RNA from oocytes of different stages of oogenesis (stages I/II, III/IV, and V/VI) and from adult liver and skin for high-throughput sequencing of small RNAs as described previously (Das et al. 2008; Supplemental material). Small RNA libraries were subjected to high-throughput sequencing using the Illumina platform. As shown in Supplemental Table S1, we obtained over six million primary reads for each sample. After stripping off of adaptor sequences, 758,445–3,641,616 of these reads mapped perfectly to at least one locus in the current release of the X. tropicalis genome sequence (JGI v4.1, http://www.jgi.doe.gov/). All reads that did not match perfectly to the available genome sequence were discarded. All sequencing data were submitted to the GEO database at NCBI (www.ncbi.nlm.nih.gov/geo/) and are freely accessible (see Supplemental material for accession numbers). We termed all primary reads that perfectly matched the genome “reads” and after collapsing identical reads termed unique reads “tags.” We then analyzed the size distribution of reads and tags in all five small RNA libraries. As shown in Figure 2, the size distributions of the germline libraries (oocyte stages I/II, III/IV, and V/VI) and the somatic libraries (liver, skin) were more similar within these groups than between the groups. Focusing on the length distributions of reads (Fig. 2A) we found that the most abundant reads in the two somatic libraries were ∼22 nt long, which is consistent with the size distribution expected for miRNAs (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008). Indeed, this peak disappeared in the length distributions of tags (Fig. 2B), which is to be expected for miRNAs, as miRNA genes produce identical reads with high accuracy that collapsed into few tags. Conversely, we found that the most abundant reads in the three germline libraries were 25–30 nt long, which is consistent with the size distribution expected for piRNAs (Fig. 2A). Indeed, the broad 25–30 nt peaks in read length distributions were matched by similar peaks observed for tag length distributions (Fig. 2B). These observations were consistent with the hypothesis that these peaks indeed represented piRNAs as piRNA populations were found to be highly complex in D. melanogaster and mouse oocytes (Okamura and Lai 2008; Malone and Hannon 2009). However, although the size distributions of germline and somatic libraries were clearly distinct, we also observed some similarities. Indeed, germline libraries also contained abundant reads of ∼22 nt RNAs that were reduced in the tag length distributions, indicative of miRNAs. Both somatic and germline libraries contained 20–24 nt complex populations, indicative of endo-siRNAs.
Figure 2.
(A,B) Length distributions of filtered short RNA sequences in oocyte and somatic cell libraries. (A) Counting total reads per library at each length. (B) Counting unique tags per library at each length.
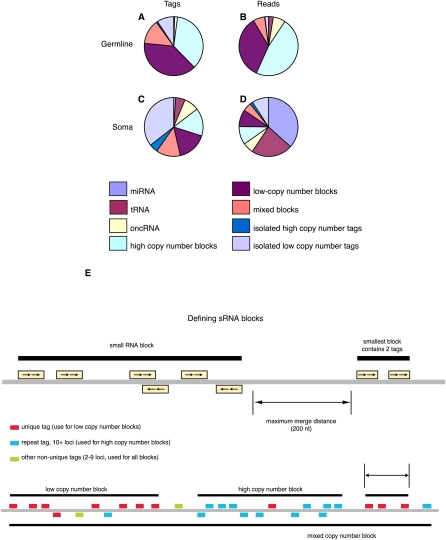
To separate small RNAs into distinct groups we compared all reads to known RNA species using the Rfam database and the miRNA registry (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008; Gardner et al. 2009). In Figure 3A–D we summarize these data by grouping all oocyte libraries together as a germline group and the liver and skin libraries as a somatic group. Small RNAs that did not match any known RNA species were grouped according to local small RNA density in the X. tropicalis genome, i.e., by occurrence in “blocks” of neighboring tags. Blocks were defined by groups of tags of a given type with no gaps between neighbors greater than 200 bases, as used previously in plants (Fig. 3E; Mosher et al. 2008). However, this definition was irrespective of read length and included putative endo-siRNAs or piRNAs. Tag types used were single locus tags, tags with 10+ loci, and all tags, defining low copy number, high copy number, and mixed blocks, respectively. Tags not in blocks were termed isolated. This terminology seemed to us particularly useful to analyze endo-siRNAs and piRNAs, which are highly complex populations and often are found in groups and often map to repeat elements (Okamura et al. 2008). As shown in Figure 3, we found that most reads from somatic libraries were miRNAs, as expected. However, we also found substantial numbers of small RNAs in high, mixed, or low copy number blocks, indicative of endo-siRNAs or piRNAs. We then analyzed the length distribution of these somatic RNAs that occur in blocks and found these to peak at 21 nt (Supplemental Fig. S3). These size distributions were consistent with endo-siRNAs, but not piRNAs and we postulated that the vertebrate Xenopus does contain somatic endo-siRNAs. In contrast, we found that the majority of small RNAs in germline libraries were in high, mixed, or low copy number blocks (Fig. 3). We again analyzed the size distribution of the germline small RNAs found in blocks. As shown in Supplemental Figure S3 we observed distinct length distributions for small RNAs in blocks: Low copy and mixed copy number blocks showed a single peak at 25–30 nt, consistent with the length distributions of piRNAs. In contrast, high copy number blocks contained reads with peak lengths of 21 and 25–30 nt. These data suggested that high copy number blocks were distinct in that they contained abundant endo-siRNAs and piRNAs. In comparison to somatic small RNAs, miRNAs were underrepresented in germline small RNAs.
Figure 3.
(A–D) Distribution of small RNA (sRNA) types in oocytes (A,B) and somatic cells (C,D), by unique tag (A,C) and by read (B,D) count. Filtered tags were identified either by BLAST similarity to known RNA types (miRNA, tRNA, rRNA, other noncoding RNA [oncRNA]), or otherwise by occurrence in “blocks” of neighboring tags, or not. Blocks were defined by groups of tags of a given type with no gaps between neighbors greater than a fixed value (200 bases). Tag types used were single locus tags, tags with 10+ loci, and all tags, defining low copy number, high copy number, and mixed blocks, respectively. Tags not in blocks were termed isolated. (E, top) Schematic view of general method for defining short RNA blocks. (E, bottom) Different families of blocks are made using only unique mapping tags (low copy number), tags with ten or more loci (high copy number), and all tags.
miR-202-5p is a germline-enriched miRNA
Next, we focused our analysis on miRNAs. We were able to match both somatic and germline small RNA libraries to known miRNAs of X. tropicalis using the miRNA registry (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008). The most frequently sequenced miRNAs from germline and somatic libraries are shown in Table 1 and Supplemental Table S2, respectively. A full list of all read matches to known miRNAs is shown in the Supplemental material Armisen_SupData1. Although it is currently not clear how well high-throughput sequencing library read frequency is correlated with small RNA concentration in the sample analyzed, it is likely that these two values did correlate in our data, as we showed for selected miRNAs and piRNAs below. The miRNA with the most abundant reads from the liver library was miR-122, a miRNA that had previously been shown to be highly expressed in liver in vertebrates and mammals (Jopling et al. 2005). The most highly represented miRNA in the skin library was miR-451, previously found enriched in zebrafish erythrocytes (Pase et al. 2009), mouse red blood cells (Rathjen et al. 2006), and mouse lung stem cells (Qian et al. 2008). As expected from our global analysis of small RNA reads from germline libraries in Figure 2, we also identified a number of reads representing known miRNAs in the germline libraries (Table 1; Supplemental Data 1). All three oocyte libraries showed similar miRNA read profiles, although there were differences in the ranking of individual miRNA reads. In addition to known miRNAs we also identified a number of candidate novel miRNAs in X. tropicalis (Supplemental Table S3). To identify miRNA candidates we mapped all tags to the genome, extracted surrounding sequence information, and used RNAfold (Hofacker and Stadler 2006) to predict local secondary structure. From these candidates and based on the rules for miRNA secondary structure (Ambros et al. 2003) and miR and miR* reads in our library, we confidently identified two new miRNAs in X. tropicalis in the soma, xtr-miR-2184 and xtr-miR-2188, which have been submitted to the miRNA registry (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008; Supplemental Table S3).
Table 1.
Most frequently sequenced known miRNAs from oocyte libraries
We next aimed to validate the expression of miRNAs in the germline of X. tropicalis. We selected three miRNAs that were highly represented in the germline small RNA libraries (miR-148a, miR-101, and miR-202-5p) and performed Northern blotting to assess the expression of the miRNAs in oocytes at different stages of oogenesis and in eggs (Fig. 4A–C). As expected from our high-throughput sequencing data, we found that all three miRNAs were expressed in oocytes. Interestingly, we observed distinct expression patterns for miR-202-5p and the other two miRNAs. miR-202-5p was most highly expressed in the early stages of oogenesis and was not detected in eggs. In contrast, miR-148a and miR-101 were strongly expressed in eggs. The expression of a set of miRNAs in the female germline of Xenopus was surprising and to exclude the possibility of any somatic contamination, we compared the expression of all three miRNAs in oocytes with their expression in follicular cells, the only possible source of somatic contamination during oocyte preparation. As shown in Supplemental Figure S4 for miR-148a, we do not detect this miRNA in follicular cells. We obtained the same results for the other two miRNAs (data not shown). We also confirmed expression of all three miRNAs in oocytes of X. laevis (Supplemental Fig. S5). Next, we assayed the expression of miR-148a, miR-101, and miR-202-5p in somatic tissues. While miR-148a and miR-101 were expressed in a number of adult somatic tissues including muscle, heart, brain, and liver, miR-202-5p was not detected in these tissues (Fig. 4D–F). These data were consistent with the high-throughput sequencing data we obtained for liver and skin (Supplemental Table S2; Armisen_SupData1). We concluded that miR-202-5p is a germline-enriched miRNA in Xenopus.
Figure 4.
(A–C) Expression of three miRNAs (miR-101, miR-202-5p, and miR-148a) was assessed during oogenesis and in eggs. Stages I and II, III and IV, and V and VI were pooled. One hundred and fifty oocytes were used for each experiment. (D–F) The expression of the same three miRNAs was assessed in early stage oocytes (stages I and II) and a number of adult somatic tissues using Northern blotting. Ten micrograms of total RNA were used for each lane. EDC was used to crosslink small RNAs to the membrane prior to hybridization. 5S rRNA is shown as a loading control.
Xenopus piRNAs
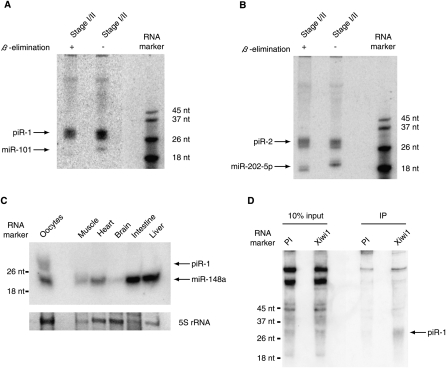
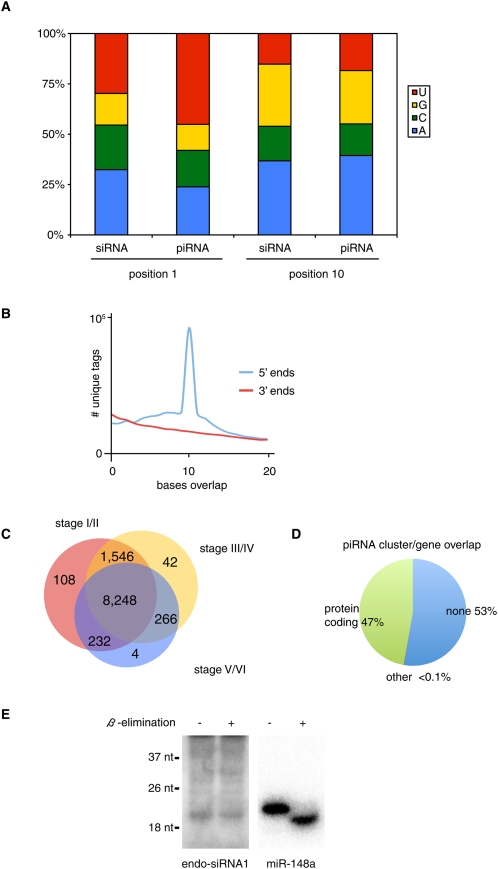
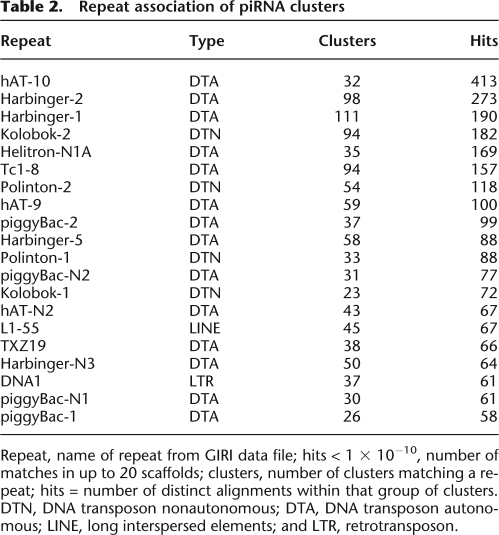
One major difference between the germline and somatic small RNA libraries was the abundance of longer (25–30 nt) RNAs, which were prominent in germline libraries, but were underrepresented in somatic libraries (Fig. 2). These 25–30 nt RNAs likely were piRNAs. First, these RNAs were of the same size as piRNAs from D. melanogaster, zebrafish, and mice (Klattenhoff and Theurkauf 2008). Second, these RNAs were 2′O-methyl-modified at the 3′-most nucleotide (Fig. 1A). Third, we previously demonstrated that one Piwi protein of X. tropicalis, piwil1.1/piwil1.2, associated specifically with ∼30 nt RNAs in Xenopus oocytes (Wilczynska et al. 2009). Next, we validated two of these candidate piRNAs, named piR-1 and piR-2, using Northern blotting. As shown in Figure 5, piR-1 and piR-2 were both expressed in stage I/II oocytes. Furthermore, β-elimination did not affect the mobility of either piRNA, whereas it did affect the mobility of two control miRNAs (Fig. 5A,B; Supplemental Fig. S6), indicating that piR-1 and piR-2 were 2′O-methylated at the 3′-most nucleotide. We also analyzed the expression of piR-1 in different tissues of X. tropicalis (Fig. 5C). We detected piR-1 expression throughout oogenesis, but not in muscle, heart, brain, intestine, or liver (Supplemental Fig. S6). Moreover, we were able to specifically immunoprecipitate piR-1 using an anti-piwil1.1/piwil1.2 antibody (Fig. 5D). Next, we examined our small RNA libraries for other features of piRNAs. As shown in Figure 6A, small RNAs of 25–30 nt (piRNAs) preferentially had uracil as the first base in germline libraries, but not in somatic libraries (data not shown). We also detect a bias for a 10 base overlap of 5′ ends of piRNAs of opposite strands (Fig. 6B), indicative of ping-pong amplification. In addition to their enrichment in the germline, piRNAs also differed from other small RNAs in their genomic location. As shown in Supplemental Figure S3, piRNAs were found in low, mixed, and high copy number blocks in the germline. In contrast, germline endo-siRNAs were mostly enriched in high copy number blocks. piRNAs from the three different oocyte libraries (stages I/II, III/IV, and V/VI) showed largely overlapping piRNA populations (Fig. 6C). Defining piRNA clusters analogous to blocks (see above), but only considering 25–30 nt RNAs, we found that the piRNA clusters of X. tropicalis are highly strand-biased (Supplemental Table S4). Of 19,657 total piRNA clusters analyzed 2338 matched at least one repeat sequence as annotated in Repbase (Table 2; Jurka et al. 2005). Forty-seven percent of piRNA clusters overlapped with protein-coding genes (Fig. 6D).
Figure 5.
(A) piRNA piR-1 (5′-TGAAGACGGACAGAAGATGGGTTAATTATTT-3′) expression was validated using Northern blotting of early stage oocytes (stages I and II). One hundred and fifty oocyte equivalents were used to analyze small RNA expression. A miR-101 probe was used as a control. β-Elimination was performed to assay for 2′O-methyl-modified 3′ nucleotides. EDC was used to crosslink small RNAs to the membrane prior to hybridization. (B) Experiments were performed as in A, but with probes for piR-2 (5′-TGAATTGTAGAACAATGTACAGGTACACCAT-3′) and miR-202-5p. (C) Expression of piR-1 and miR-148a was assessed in a number of adult somatic tissues and early stage oocytes (stages I and II) using Northern blotting. Ten micrograms of total RNA were used for each lane. 5S rRNA is shown as a loading control. (D) Northern blot analysis of immunoprecipitated piRNAs using piwil1.1/piwil1.2 specific antibody (PI: pre-immune).
Figure 6.
(A) Base composition at positions 1 and 10 was analyzed separately for piRNAs and 20–24 nt RNAs (siRNAs). (B) Analysis of 5′ and 3′ base overlap for unique tags. (C) Overlap of piRNA populations cloned from different stages of oogenesis. We considered only unique tags with ≥10 reads. (D) Overlap between piRNA clusters and annotated genes. (E) Identification of endo-siRNA in Xenopus tropicalis. The expression of endo-siRNA1 (5′-ACGGCCGGGGGCATTCGTATT-3′) was validated using Northern blotting after β-elimination, to assay for 2′O-methyl-modified 3′ nucleotides. miR-148a was used as a control for β-elimination as well as a loading control.
Table 2.
Repeat association of piRNA clusters
Repeat, name of repeat from GIRI data file; hits < 1 × 10−10, number of matches in up to 20 scaffolds; clusters, number of clusters matching a repeat; hits = number of distinct alignments within that group of clusters. DTN, DNA transposon nonautonomous; DTA, DNA transposon autonomous; LINE, long interspersed elements; and LTR, retrotransposon.
Other small RNAs of the germline and soma
As we showed in Figure 2 and Supplemental Figure S3, many short endogenous small RNAs map to blocks of varying copy numbers in the germline and the soma. These 20–24 nt RNAs likely represented endo-siRNAs. In support of this hypothesis, we found that these RNAs were resistant to β-elimination as a population (Fig. 1A) and individually, as shown for a single somatic small RNA (Fig. 6E). The 20–24 nt RNAs lack a 5′ U bias, suggesting that these are unlikely to be degradation products of piRNAs (Fig. 6A). Many 20–24 nt RNAs mapping to opposite strands overlap, which is consistent with these RNAs being Dicer products, but there was no apparent peak of 2 nt overhangs (Supplemental Fig. S7). It had previously been shown that many endo-siRNAs and piRNAs map to repetitive elements (Okamura et al. 2008). We therefore asked if small RNAs from somatic and germline libraries mapped together to repetitive elements in the X. tropicalis genome sequence. We analyzed the overlap between instances of all repeats in the genome and small RNA (endo-siRNA and piRNA) loci. We summarized these data for germline and somatic small RNAs in Supplemental Tables S5 and S6, respectively. The complete data are available as Armisen_SupData2 (germline) and Armisen_SupData3 (somatic), respectively. The overall number of small RNAs mapping to repeat elements in the genome was much higher in the germline than the soma, as we expected from our analysis of high copy number blocks (Fig. 3). Nevertheless, the overall pattern of repeat elements that matched small RNAs in the germline and the soma was similar. Seventy-five percent of blocks of all mapped tags found in the somatic libraries overlapped with those found in the oocyte libraries (30% when only considering unique tags). These data suggested that small RNA pathways might have similar functions in the silencing of repeat elements in the germline and the soma. In both cases small RNAs matched to abundant classes of repetitive elements in the X. tropicalis genome, including DNA transposons of the Harbinger, hAT, and piggyBac superfamilies. However, the repeat elements that were the source of the largest number of reads in germline and somatic libraries were DNA transposons of the Polinton family, which are not among the abundant repeat elements in X. tropicalis. Similarly, a LTR retrotransposon of the ERV family was overrepresented in these data (Supplemental Table S5).
Discussion
Here we report the first unbiased analysis of small RNA populations of X. tropicalis using a high-throughput sequencing approach. Historically, the majority of work in frogs has been carried out using X. laevis, but this species is allo-tetraploid, and its smaller diploid cousin, X. tropicalis, has been adopted by the Xenopus community as a more useful genomic and genetic model. The current X. tropicalis genome assembly (JGI v4.1, http://www.jgi.doe.gov/) has an estimated coverage of over 90% in 19,759 scaffolds, and although not yet fully assembled into chromosomes, has become a powerful tool for functional sequence mining and comparative genomics. Furthermore, as high-throughput RNA sequencing, unlike microarray approaches, is independent of the known genome sequence, the data presented here will remain suitable for further analysis as the X. tropicalis assembly improves.
In this study we identify miR-202-5p as a miRNA enriched in Xenopus oocytes, raising the possibility that this miRNA might function as a regulator of gene expression in the female germline of Xenopus. Previous analysis of miRNA expression in Xenopus oocytes found many miRNAs to be absent from oocytes, but found an enrichment of some miRNA precursors (Tang and Maxwell 2008), which raised the possibility that the miRNA pathway might not be functional in the oocyte. Indeed, it was reported that pre-miRNAs injected into stage VI oocytes are inefficiently processed (Lund et al. 2004; Lund and Dahlberg 2006). Our northern data indicate that fully processed miRNAs are present in Xenopus oocytes. Although analysis of embryos derived from dicer1 mutant germline clones suggested that miRNAs are not essential for early development in the zebrafish, miRNAs have been identified in oocytes of Drosophila and mice (Hatfield et al. 2005; Tang et al. 2007; Tam et al. 2008). Moreover, maternal miRNAs are essential for zygotic development in the mouse (Murchison et al. 2007; Tang et al. 2007). miR-202-5p is a vertebrate-specific miRNA and is highly expressed in mouse oocytes, ovary, and testis, but not in other adult tissues (Ro et al. 2007; Tam et al. 2008). Despite the fact that miR-202-5p is the most conserved part of the miR-202 pre-miRNA and that miR-202-5p has been cloned most frequently, it was previously annotated as miR-202* in mirBase (and conversely miR-202-3p as miR-202) and has subsequently been missed in a number of microarray studies. Although miR-202-5p appears to be absent in eggs and a number of adult tissues, including liver, skin, and intestine, our study is not sufficient to demonstrate that miR-202-5p is specific for oocytes. Indeed, previous work identified miR-202-5p as a miRNA highly expressed in Xenopus testis (Michalak and Malone 2008). Interestingly, this study suggested that miR-202-5p expression levels differed in wild-type X. tropicalis testis and their sterile hybrids. Although there is currently no published functional data on this miRNA in any vertebrate system, it is tempting to speculate that miR-202-5p has a conserved role in germline development in vertebrates.
This study identifies abundant piRNAs and putative endo-siRNAs in the germline of X. tropicalis. Interestingly, both classes of small RNAs often map to high copy number blocks or highly repetitive elements within the genome (Fig. 3). Previous work has demonstrated a role for piRNAs in the silencing of repeat elements (Klattenhoff and Theurkauf 2008). In the case of endo-siRNAs, such a role has at least been proposed (Okamura and Lai 2008; Malone and Hannon 2009). Our data suggests that endo-siRNA and piRNA pathways in Xenopus might be closely linked, raising the question of the relationship between these two pathways. In C. elegans, piRNAs act upstream of an endo-siRNA pathway (Das et al. 2008). In contrast, it has been proposed that in Drosophila and mammals endo-siRNAs and piRNAs act redundantly in the female germline (Okamura and Lai 2008; Malone and Hannon 2009). Our data does not distinguish between these two possibilities. However, we have found a strong overlap between endo-siRNAs in the germline and the soma of Xenopus. In both cases, endo-siRNAs mapped to a similar subset of repeat elements (Supplemental Tables S5 and S6). With piRNAs either absent or strongly underrepresented in somatic tissues (Fig. 2), we postulate that in the soma endo-siRNAs might be sufficient to provide protection against transposable elements.
Work in Drosophila has demonstrated a role for piRNAs in the silencing of DNA transposons and retrotransposons (Klattenhoff and Theurkauf 2008). However, we observe a strong underrepresentation of small RNAs that map to retrotransposons (Supplemental Tables S5 and S6) with regard to the abundance of these elements in the genome of X. tropicalis. Indeed, the repeat element with most small RNA read matches in germline and soma is a newly discovered type of DNA transposon called Polintons (Kapitonov and Jurka 2006). Polintons are the most complex DNA transposons discovered so far, encoding at least four distinct proteins. Their unique integration mechanism is thought to lead to the frequent formation of genomic palindromes. Palindromes were recently identified as a source of endo-siRNAs in the soma of Drosophila, and the large number of small RNAs matching Polintons in Xenopus suggest that Polinton-induced palindromes might be the source of these endo-siRNAs. Polintons in Xenopus laevis were also recently described as a source of piRNAs (Kirino et al. 2009).
We find that the majority of non-miRNA small RNAs in the germline and the soma of X. tropicalis map to multiple loci in the genome. These observations are a direct consequence of the association of small RNAs with repeat elements and consistent with previous work in other organisms (Malone and Hannon 2009). Therefore, the precise origin of any of these small RNAs cannot be determined. Tags mapping to multiple loci often have a low read count and the full complexity of these tags might not have been captured in our small RNA libraries. While many blocks of small RNAs show a stark strand bias consistent with an RNAi type mechanism (Supplemental Tables S5 and S6), in cases where strand bias is lacking, this may be due to the superposition of small RNAs from different loci. Furthermore, populations of repeat elements in the genome are highly heterogeneous, consisting of active and inactive elements, elements that are part of protein-coding genes and isolated elements. To understand how small RNAs contribute to the regulation of these elements it will be important to identify the correct source of these RNAs. The analysis of longer RNA transcripts using high-throughput sequencing technology might provide insights here.
A substantial number of reads in our libraries correspond to tRNAs, rRNAs, and other noncoding RNAs (Fig. 3). In addition, a number of reads match the sense strand of abundant mRNAs in the corresponding tissues (data not shown). Similar to previous studies based on high-throughput sequencing of small RNAs, we have disregarded these RNAs as likely RNA degradation products. However, as it is currently impossible to separate initiators, effectors, and products of RNAi and related small RNA pathways, it remains unclear if these other RNAs might not contain important information and deserve attention. For example, 5.8S RNA processing has recently been shown to generate a small RNA and to involve a small RNA pathway gene, eri-1 (Gabel and Ruvkun 2008).
Methods
RNA isolation
Ovarian lobes were removed from Xenopus tropicalis and Xenopus laevis females and collagenase treated for 3 h. Oocytes were staged and washed in modified Barth's solution (8.8 mM NaCl, 1 mM KCl, 330 μM Ca(NO3)2, 410 μM CaCl2, 820 μM MgSO4, 2.4 mM NaHCO3, 10 mM HEPES-NaOH pH 7.4). Total RNA was isolated from X. tropicalis and X. laevis staged oocytes, or from X. tropicalis laid eggs and adult tissues using TRIzol reagent (Invitrogen) and according to the manufacturer's protocol. In all experiments, we pooled the same number of oocytes from different stages. Total RNA extracted was enriched for small RNAs using the miRVana kit (Ambion), resolved in a 15% denaturing polyacrylamide gel and stained using SYBR green (Invitrogen).
β-Elimination, 5′ end-labeling, and Northern blotting
β-Elimination and 5′ end-labeling was performed essentially as described previously (Horwich et al. 2007). Small RNA Northern blotting using DNA probes was performed as described previously (Miska et al. 2004; Pall and Hamilton 2008). See Supplemental material for more detail.
RT-PCR and qRT-PCR
Briefly, total RNA from oocyte equivalents was isolated and treated with RQ1 RNase-Free DNase, phenol/chloroform extracted and ethanol precipitated. Reverse transcription (RT) was performed according to the Superscript II protocol using an oligo-dT primer. Two microliters of RT were used for standard PCR. qRT-PCR was performed using Quantitect SYBR green PCR mix (see also Supplemental material). RT-PCR and qRT-PCR primer design was as described previously (Chen et al. 2005a). Sequences of oligonucleotides used for qRT-PCR are listed in the Supplemental material (Armisen_SupData4.xls).
High-throughput sequencing
For high-throughput sequencing small RNAs were enriched using the miRVana kit, resolved on a 15% denaturing polyacrylamide gel, and the size range corresponding to the bands indicated with an asterisk in Supplemental Figure S1 were isolated (∼18–34 bases). Small RNA libraries were cloned through ligation of 5′ and 3′ adapters as described previously (Das et al. 2008). All libraries were sequenced using an Illumina GA2 instrument (Illumina). Read length was 45 cycles. Detailed information on sequencing data analysis, block analysis, and prediction of miRNA candidates can be found in the Supplemental material. Briefly, raw Illumina reads were processed into unique tags after removing the adapters sequences and blasted against known RNA families: miRNAs, ribosomal RNA, transfer RNAs, and other noncoding RNAs deposited in Rfam, (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008; Gardner et al. 2009). Groups of neighboring tags were clustered together in blocks for further analysis. For prediction of miRNA candidates, tags were tested for potential miRNA precursor folds using RNAfold (Hofacker and Stadler 2006).
Acknowledgments
We thank Jim Smith and David Simpson for providing materials and technical support. We thank Partha Das for technical advice. We thank Leonard Goldstein for discussions on data analysis. We thank Nelson Lau for sharing unpublished observations. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (UK) to N.S. and E.A.M., a Cancer Research UK Program Grant to E.A.M. (C13474), and core funding to the Wellcome Trust/Cancer Research UK Gurdon Institute provided by the Wellcome Trust (UK) and Cancer Research UK.
Footnotes
[Supplemental material is available online at http://www.genome.org. Short read sequence data from this study have been submitted to NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE14952.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093054.109.
References
- Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005a;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes & Dev. 2005b;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. doi: 10.1093/genetics/164.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gabel HW, Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat Struct Mol Biol. 2008;15:531–533. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1023–1024. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Hikosaka A, Takaya K, Jinno M, Kawahara A. Identification and expression-profiling of Xenopus tropicalis miRNAs including plant miRNA-like RNAs at metamorphosis. FEBS Lett. 2007;581:3013–3018. doi: 10.1016/j.febslet.2007.05.054. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros VR. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros VR. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P, Malone JH. Testis-derived microRNA profiles of African clawed frogs (Xenopus) and their sterile hybrids. Genomics. 2008;91:158–164. doi: 10.1016/j.ygeno.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes & Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protocols. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Qian S, Ding JY, Xie R, An JH, Ao XJ, Zhao ZG, Sun JG, Duan YZ, Chen ZT, Zhu B. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem Biophys Res Commun. 2008;377:668–673. doi: 10.1016/j.bbrc.2008.10.052. [DOI] [PubMed] [Google Scholar]
- Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Song R, Park C, Zheng H, Sanders KM, Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GQ, Maxwell ES. Xenopus microRNA genes are predominantly located within introns and are differentially expressed in adult frog tissues via post-transcriptional regulation. Genome Res. 2008;18:104–112. doi: 10.1101/gr.6539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes & Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Harland RM. Expression of microRNAs during embryonic development of Xenopus tropicalis. Gene Expr Patterns. 2008;8:452–456. doi: 10.1016/j.gep.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA. 2009;15:337–345. doi: 10.1261/rna.1422509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]