Abstract
T-cell receptor (TCR) genomic loci undergo somatic V(D)J recombination, plus the addition/subtraction of nontemplated bases at recombination junctions, in order to generate the repertoire of structurally diverse T cells necessary for antigen recognition. TCR beta subunits can be unambiguously identified by their hypervariable CDR3 (Complement Determining Region 3) sequence. This is the site of V(D)J recombination encoding the principal site of antigen contact. The complexity and dynamics of the T-cell repertoire remain unknown because the potential repertoire size has made conventional sequence analysis intractable. Here, we use 5′-RACE, Illumina sequencing, and a novel short read assembly strategy to sample CDR3β diversity in human T lymphocytes from peripheral blood. Assembly of 40.5 million short reads identified 33,664 distinct TCRβ clonotypes and provides precise measurements of CDR3β length diversity, usage of nontemplated bases, sequence convergence, and preferences for TRBV (T-cell receptor beta variable gene) and TRBJ (T-cell receptor beta joining gene) gene usage and pairing. CDR3 length between conserved residues of TRBV and TRBJ ranged from 21 to 81 nucleotides (nt). TRBV gene usage ranged from 0.01% for TRBV17 to 24.6% for TRBV20-1. TRBJ gene usage ranged from 1.6% for TRBJ2-6 to 17.2% for TRBJ2-1. We identified 1573 examples of convergence where the same amino acid translation was specified by distinct CDR3β nucleotide sequences. Direct sequence-based immunoprofiling will likely prove to be a useful tool for understanding repertoire dynamics in response to immune challenge, without a priori knowledge of antigen.
T-cell receptors (TCRs) are dimeric (αβ or γδ), highly variable T lymphocyte membrane proteins that recognize antigenic peptides presented on heterologous cells by the major histocompatibility complex (MHC) (Davis and Bjorkman 1988; Bassing et al. 2002). Recognition specificity for diverse peptide-MHC (pMHC) complexes is provided by the three complementarity-determining regions (CDRs) of the TCR. CDR1 and CDR2 are coded for by germline sequences while CDR3, the highly polymorphic principal recognition site, is created when TCR genomic loci undergo somatic recombination between gene segments during development of T lymphocytes in the thymus (Gellert 1992, 2002; Jung and Alt 2004). For the α locus and the γ locus, recombination occurs between variable (V) and joining (J) segments. For the δ locus and the β locus, there is recombination between V and J segments, but also the inclusion of one of two short diversity (D) segments. The combinatorial diversity of the human β locus is illustrated in Figure 1A. At CDR3 recombination junctions, further complexity is generated through the deletion of germline-encoded bases and the addition of random nontemplated bases. The resulting hypervariable sequences of the CDR3 make possible the recognition of diverse peptide-MHC (pMHC) complexes. During T-cell maturation, all T cells expressing rearranged receptors capable of binding pMHC with high enough affinity to be biologically relevant are retained (positive selection), but only T cells with rearranged receptors that do not interact strongly with self-pMHC complexes ultimately exit the thymus (negative selection). It should be noted that V(D)J recombination is not entirely random, and the prevalence of specific gene segments and combinations of gene segments shows marked variation in the repertoire. Contributions to this bias are introduced even before thymic selection, through variation in the efficiency of recombination of different gene segments (Manfras et al. 1999; Krangel 2003). The peripheral blood thus contains a large repertoire of T lymphocytes with the potential to recognize diverse antigens. Binding of a naïve T cell's TCR to a structurally compatible pMHC on an antigen-presenting cell will, with the appropriate interaction of co-stimulatory molecules, initiate rapid clonal expansion to generate a population of effector cells. This acute response occurs on the order of days, and is followed by a gradual contraction of the expanded pool over the course of several weeks, with differentiation into a small number of long-lived memory cells. Thus, the T-cell repertoire is not static, but rather is constantly molded by immune challenge (for reviews, see Nikolich-Zugich et al. 2004; Harty and Badovinac 2008).
Figure 1.
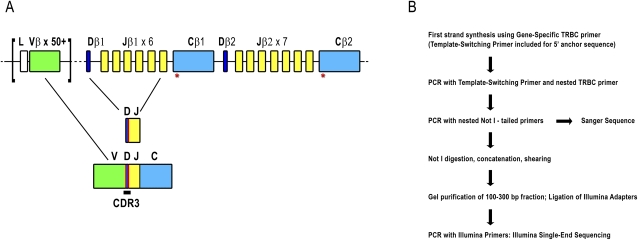
(A) Representation of the TCRβ locus at human chromosome 7q34. The TCRβ locus spans 620 kb and includes over 50 TRBV genes (green) belonging to 30 subgroups. There are two TRBC genes (light blue) each downstream from a TRBD (dark blue) and six or seven TRBJs (yellow). Recombination first occurs between TRBJ and TRBD genes, followed by recombination to a TRBV gene. (Red lines) Addition of nontemplated bases. After transcription, intervening sequences are spliced out so that a TRBC is adjacent to the recombined V-D-J sequence. Gene width and distances are not to scale. (Red asterisks under TRBC) Location of primers used for 5′-RACE and PCR reactions. Refer to Supplemental Figure 1A for primer locations. A detailed locus map can be obtained from IMGT (www.imgt.org/textes/IMGTrepertoire). (B) Flowchart illustrating 5′-RACE and Illumina library construction. For more detail, please refer to Methods and Supplemental Figure 1.
There has been remarkable progress in characterizing the size and dynamics of the T-cell repertoire, but the task remains daunting due to the enormous combinatorial diversity that is theoretically possible (>1015 distinct αβ receptors, or clonotypes [Davis and Bjorkman 1988; Murphy et al. 2007]) and the limited power of existing tools for interrogation. Previously, a method called TCR spectratyping (Pannetier et al. 1993; Gorski et al. 1994) had been used to probe the T-cell repertoire. This approach involves the use of V and J gene segment-specific primers for RT-PCR amplification of the CDR3. In TCR spectratyping, CDR3 amplicons are separated according to size by polyacrylamide gel electrophoresis. Typically, six or so distinct amplicons are observed per primer pair, spaced at 3-nucleotide (nt) intervals in accordance with reading frame. An experimental estimate of repertoire size of ∼106 beta chains in blood has been obtained (Arstila et al. 1999) by exhaustive Sanger sequencing of a single amplicon from a TRBV18-TRBJ1-4 spectratype, then extrapolating the observed diversity according to the relative abundance of this amplicon in the spectratype and the estimated frequency of TRBV18-TRBJ1-4 pairing in the repertoire. Of course, actual TCR diversity will be higher still, due to αβ heterodimerization (Fuschiotti et al. 2007; Ozawa et al. 2008).
Advances in sequencing technology (Holt and Jones 2008; Shendure and Ji 2008) now permit interrogation of complex sequencing targets at unprecedented depth and reasonable cost. Here, we describe a method for deep sampling of the TCR repertoire at sequence-level resolution. Our approach relies on massively parallel Illumina sequencing of CDR3β amplification products and a novel TCR-specific short read assembly strategy (Warren et al. 2009).
Results
Experimental strategy
We used 5′ rapid amplification of cDNA ends (RACE) to obtain CDR3β transcript sequences from a commercially available mRNA sample prepared from normal human peripheral blood leukocytes (PBL) pooled from 550 individuals (Fig. 1B; Supplemental Fig. 1). Peripheral blood from different individuals will include different frequencies of naïve and memory T cells. Because individual memory repertoires are skewed due to historical antigen encounter and the individual's HLA type, our results do not reflect the expected repertoire of any individual, but rather are reflective of average clonotype abundance in a population.
The RACE approach avoids the potential bias associated with the use of the multiple primer sets required to amplify from all TRBV sequences (Boria et al. 2008) and takes advantage of the conserved sequences offered by TRBC1 and TRBC2 (96% nucleotide sequence identity). Reverse transcription to generate cDNA was performed using a primer specific for the TRBC genes (Ozawa et al. 2008) as well as a template-switching primer (Peters et al. 1999; Douek et al. 2002) to provide a 5′ anchor for subsequent PCR. First-round PCR reactions with a nested TRBC primer and the template-switching primer produced a high level of background amplification. A second round of PCR using nested primers was performed to obtain a cleaner product of ∼520 bp. (See Methods for primer sequences and Supplemental Fig. 1A for TRBC primer locations.) The RACE product was then gel-purified and an aliquot was cloned and Sanger sequenced to confirm the presence of CDR3β amplicons. The RACE product was too long to directly sequence the CDR3β region with short-read technology, so it was ligated to produce concatamers that were then sheared by sonication. A 100- to 300-bp size fraction was isolated by PAGE and shotgun-sequenced on the Illumina platform (www.illumina.com). The initial sequencing runs generated 18,829,563 36-nt reads. During the course of this analysis, a protocol to produce longer read lengths became available, so further 21,752,666 50-nt reads were generated and analysis was performed on the pooled set of 40,582,229 reads (Table 1).
Table 1.
Sequencing and assembly statistics
aComplete CDR3β sequences in correct reading frame.
iSSAKE assembly and analysis of reconstructed TCRβ sequences
We have recently described a system for profiling TCR diversity using short sequence reads and the assembly software package we call iSSAKE (immuno-Short Sequence Assembly by K-mer search and 3′ read Extension; Warren et al. 2009). Sequence reads that have homology with a TRBV gene segment but have unmatched bases at their ends (corresponding to the beginning of the recombined CDR3β sequence) are used as seeds to initiate directional, de novo CDR3β assemblies, as described in the Methods section. Briefly, with iSSAKE, reads from the shotgun data set derived from the CDR3β amplicon are optimally aligned to each seed to generate CDR3β sequence contigs. An important feature of iSSAKE is that reads can be reused, which allows the assembly of different CDR3β sequences that end in the same TRBJ segment. However, this also means that read depth is not uniform throughout CDR3β and cannot be used to determine clonotype frequency. Depth is exceptionally high over the TRBJ segment, where only 14 TRBJ possibilities exist. Some seeds are themselves long enough to cover most or all of the CDR3β-encoded bases without being significantly extended by other reads, so read depth in these instances may be low. For the current study, we set the assembly parameters to only extend a contig when two or more reads aligned at each base position (−o 2). Further, only reads with overlaps 15 bases or longer (−m 15) were considered. Read error was mitigated using the iSSAKE internal error-handling algorithm and consensus bases were called when bases from reads agreed 70% of the time or more (−r 0.7). The output comprises contiguous sequences (contigs) that contain the last 15 nt of TRBV, any nontemplated and TRBD bases, and the first 15 recognizable TRBJ segment bases. Upon completion of assembly, contigs are compared and those that have matching sequence, and therefore represent the same beta-chain clonotype, are grouped together. The number of matching contigs for a given clonotype provides the relative frequency of that clonotype in the original sample. From previous simulations we know that contig depth determined in this manner is proportional to clonotype frequency (correlation coefficient >0.999; Warren et al. 2009).
The results of the analysis of the sequence data generated in the present study are outlined in Table 1. From the complete data set of >40.5 M total reads we identified 310,614 assembly seeds. From these seeds, CDR3β sequences were assembled for 35,762 distinct TCR beta-chain clonotypes. The sequence of each clonotype is provided in Supplemental File 1. A large proportion of seeds did not yield additional distinct clonotypes, as there was not adequate sequence coverage of our sample to extend these seeds into TRBJ. Clonotype sequences were screened for open reading frames (ORFs), and 2098 (5.9%) that were found to contain stop codons in frame with TRBJ were removed, leaving 33,664 distinct beta-chain clonotypes. The relatively high proportion of clonotypes containing stop codons does not appear to be due to sequencing error or misassembly. Rather, we find that 96% of these stop codons map to nontemplated bases at the V-D-J junction and likely represent real events captured by our assay. During T-cell maturation, in the event that the first T-cell receptor beta chain (TCRβ) rearrangement in a given cell is nonproductive, rearrangement of the second TCRβ allele is initiated. However, after thymic selection, down-regulation of the nonproductive allele is not absolute (Li and Wilkinson 1998), and these transcripts may account for the premature termination codons we identify in the present study.
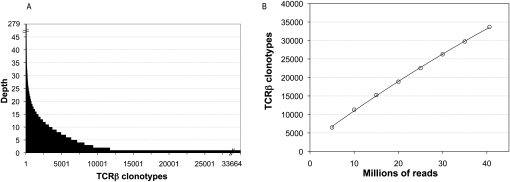
Clonotype abundance varied from one to a maximum of 279 (Fig. 2A). Rare clonotypes detected as single copies represented 65.3% of all clonotypes but only 18.8% of the 117,052 total CDR3β sequence assemblies. Moderately abundant clonotypes detected at a copy number between two and 19 represented 32.6% of clonotypes and 64.1% of assemblies. Finally, there were 720 clonotypes (2.1% of clonotypes) with copy number >20, and this small number of highly abundant clonotypes represented 17.1% of all assemblies. Since the data are generated from RNA pooled from many individuals, we expect that the majority of clonotypes will originate from the more prevalent effector and memory cells of the population sampled. It is possible that the most abundant clonotypes therefore represent highly expanded effector cells from those individuals with a recent antigen encounter. Additionally, some abundant clonotypes may exemplify the phenomenon of public T-cell responses, that is, identical TCR rearrangements from multiple individuals in response to the same antigen (Venturi et al. 2008).
Figure 2.
(A) TCRβ diversity. A total of 33,664 TCRβ clonotypes were identified from complete and in-frame CDR3β sequences assembled by iSSAKE. Clonotypes with a copy number of one (clonotypes identified by a single iSSAKE contig) account for 65.3% of all clonotypes. Clonotypes identified by two to 19 iSSAKE contigs account for 32.6% of all clonotypes, and high-abundance clonotypes (contig depth ≥20) account for 2.1% of the total. (B) Saturation analysis. In duplicate experiments we chose independent sets of 5, 10, 15, 20, 30, and 35 M reads at random from the pool of 40,582,229 total sequence reads in our data set. These subsets of reads were assembled and clonotypes counted as the set of complete, in-frame, nonredundant CDR3β sequences. The number of clonotypes (mean ± SD for the duplicate experiments) is plotted as a function of the number of reads. Error bars are contained within the symbols.
To determine if the depth of sequencing in the current study showed any trend toward saturation, we took random subsamples of sequences at intervals of 5 million reads. These were independently assembled and the number of distinct clonotypes at each point was plotted (Fig. 2B). The relationship is linear (Pearson coefficient = 0.999), indicating we have not begun to approach saturation. This is expected, given the fact that we obtained 33,664 distinct clonotypes from our sequencing target of pooled peripheral blood mononuclear cells (PBMCs), and, as noted above, the repertoire size of just one individual has been estimated previously to be ∼106 beta chains.
TRBV, TRBD, and TRBJ usage
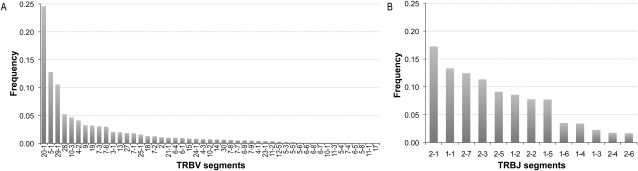
With our experimental approach, only the portion of the TRBV sequence that is present in the seed sequence is available for assignment of a particular TRBV gene to an assembled CDR3β. This fact, together with the often high sequence homology among certain TRBV genes and the replacement of deleted TRBV ends with nontemplated bases makes it impossible to assign a single unique TRBV gene to every clonotype. We were successful, however, in making an unambiguous assignment for 22,704 (67.4%) of the assembled clonotypes (Table 1; Supplemental Fig. 2) to one of 49 distinct TBRV genes. Subsequent analysis is based on this portion of the data set. Usage of this set of 49 TBRV genes ranged from 24.6% for TRBV20-1 to 0.01% for TRBV17 (Fig. 3A).
Figure 3.
TRBV and TRBJ usage. (A) Relative frequency of usage of TRBV segments was for the subset of clonotypes with an unambiguous TRBV gene segment assignment (n = 22,704). (B) Relative frequency of usage of TRBJ segments for the set of all clonotypes (n = 33,664).
Several TRBV genes identified in our assembly are ORFs (open reading frame, International ImMunoGeneTics [IMGT] nomenclature) that either have changes at conserved amino acid positions (TRBV6-7, TRBV17, and TRBV7-1) or noncanonical splice donor sites (TRBV5-3 and TRBV23-1). We also found assemblies containing the pseudogene TRBV21-1, which has a frameshift in the leader sequence. As with the appearance of rearrangements that contain stop codons (see above), it is possible that these assemblies represent the transcription of an initial nonproductive rearrangement. TRBV21-1 and TRBV23-1 have previously been shown to be transcribed (summarized by Folch and Lefranc 2000), and for TRBV23-1, expression has also been demonstrated at the protein level (Leslie et al. 2006). The transcription of TRBV7-1 is unexpected, as it is also deficient in sequences essential for recombination. It is possible that functional alleles of TRBV7-1 exist.
All known, functional TRBJ genes are represented in the sequence assembly and can be assigned unambiguously within the full set of 33,664 potential clonotype sequences. Usage ranged from 17.2% for TRBJ2-1 to 1.6% for TRBJ2-6 (Fig. 3B). The pseudogene TRBJ2-2P was not detected.
TRBD segments sustain substantial base deletion and overall transformation, so that the segments are usually unrecognizable without ambiguity. Clonotypes where TRBD could be identified unambiguously and accurately (with minimum length = 8 nt) represent 5497 out of the 22,704 sequences with accurate TRBV assignment. For these 5597 clonotypes, we calculate that 49.9% are TRBD1 and 50.1% are TRBD2.
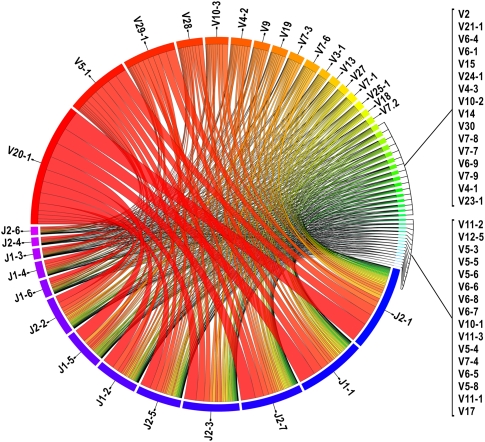
From our set of 49 positively identified TRBV and 13 TRBJ, there are 637 potential pairings. We find that 562 of these TRBV–TRBJ combinations are represented in our data for the 22,704 unique clonotypes (Fig. 4; Supplemental Fig. 3; Supplemental Table 1). The most frequent pairing is of TRBV20-1 to TRBJ2-1, accounting for 4.1% of all pairings, whereas 58 TRBV–TRBJ pairs were identified only once.
Figure 4.
Frequencies of V-J pairing calculated from the subset of clonotypes with an unambiguous TRBV gene segment assignment (n = 22,704). (Blue to purple rectangular bands) TRBJ segments, (red to cyan rectangular bands) TRBV segments. The width of the bands is proportional to the number of times the TRBV and TRBJ genes connected by the band co-occur in CDR3β sequences. TRBV and TRBJ segments are arranged from left to right and right to left, respectively, and ordered by total pairing links they share. (This illustrates the data contained in Supplemental Table 1.) This figure was generated using the Circos software package (Krzywinski et al. 2009).
Sequence diversity of CDR3β
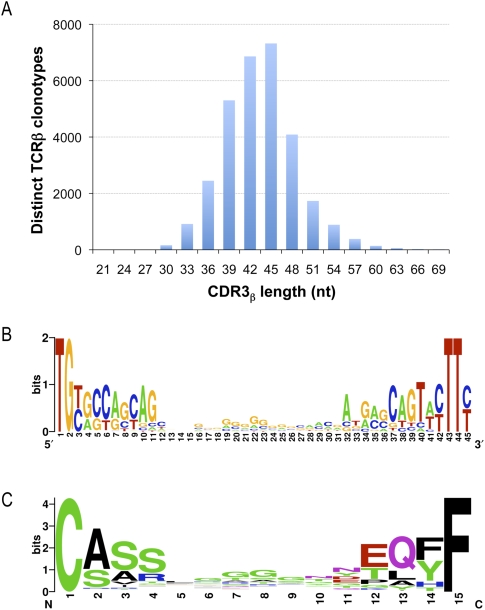
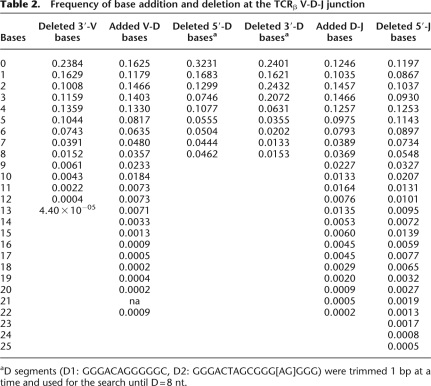
We examined the frequency of base addition and deletion at the V-D-J junction. The boundary of the CDR3β is not absolute. In order to compare sequences, we defined CDR3β coordinates as starting at the codon for the last cysteine of TRBV and ending at the phenylalanine in the conserved TRBJ segment motif FGXG. In our data set this defines a subset of 30,366 CDR3β sequences. The length of CDR3 varies from 21 to 81 nt with a peak at 45 nt (Fig. 5A). While most rearrangements involve the removal and addition of a few residues, the extent of change can be considerable. Nontemplated bases and/or deletions were detected in all D-J junctions examined and in only two instances was there no net change in sequence at the V-D junction. Nontemplated bases at the V-D junction are 62.9% GC and the nontemplated bases at the J-D junction are 54.3% GC. The 7319 CDR3β sequences contained in the 45-nt peak were used to create nucleotide and amino acid sequence logos (Fig. 5B,C). The logos are a graphical representation of a nucleic acid or amino acid multiple sequence alignment (Schneider and Stephens 1990; Crooks et al. 2004). We find no evidence of any overrepresented sequence in CDR3β other than the prevalence of guanines in the center of the nucleic acid logo (Fig. 5B) and glycines in the center of the amino acid logo (Fig. 5C), which simply reflect the sequence and coding potential of the guanine-rich TRBD segments. The conservation apparent at the left and right ends of the logos reflects the contribution of TRBV and TRBJ sequences, respectively. Finally, in our sequence assemblies, there are many examples of independent recombination events that have produced the same CDR3β amino acid sequence. In 659 instances, the same CDR3β nucleotide sequence is detected in association with different TRBV and TRBJ sequences. In addition, we find 1573 examples of rearrangements where the same CDR3β amino acid sequence can be translated from divergent nucleotide sequences.
Figure 5.
CDR3β nucleotide length distribution and sequence composition of the most abundant CDR3β length. To explore CDR3β length variation we used a precise length criterion, defined as all bases between the last cysteine of TRBV and the phenylalanine in the TRBJ segment motif FGXG. Of the 33,664 total clonotypes, 30,366 could be classified in this manner, and the length distribution of this subset was plotted (A). The most frequently observed length was 45 nt. For the subset of clonotypes with 45 nt CDR3β sequences, we created logos for the nucleotide (B) and inferred amino acid (C) composition, using WebLogo (Crooks et al. 2004).
Verification
Validation of the assembly and analysis was obtained from the intersection of iSSAKE-generated data with that obtained by Sanger sequencing of 5′-RACE clones. A total of 288 unique TCR sequences were obtained by Sanger sequencing of the cloned 5′-RACE products, and, of these, 69% were found in our assembled Illumina data. In addition, comparison of the usage frequencies of TRBJ genes from the iSSAKE contigs and the Sanger sequences shows a very similar profile (Pearson coefficient 0.904, data not shown) and provides a validation of the iSSAKE sequence assembly. Comparison of the frequencies for TRBV was limited to those 49 genes that could be unambiguously identified from the Illumina assembly (Pearson coefficient 0.822). Next, to estimate sequence error attributable to PCR and reverse transcription, we used Sanger sequencing to evaluate TCRβ 5′-RACE products amplified and cloned from the cell line DES M26, a melanoma-specific T-cell line that carries a single TCRβ rearrangement. Single-pass sequence reads from 311 independent colonies were quality-trimmed to Q50 (representing the highest quality subset with only one predicted error per 100,000 bases) and assembled, demonstrating an accuracy of 99.91%. This sets a practical upper limit on the contribution of reverse transcriptase- and polymerase-introduced errors in the method as it is described here.
Discussion
By massively parallel sequencing of CDR3β amplicons from pooled leukocytes, we have identified 33,664 distinct human CDR3β sequences. As of May 2009 there were only 3187 unique human TCRβ mRNA sequences in GenBank, not all of which include the CDR3β region. The IMGT database reports 5303 rearranged human TCRβ sequences, 2927 of which are shared with GenBank. Thus, in a single experiment, we have increased the number of known sequences by an order of magnitude. Analysis of the data from the present study has provided information, at unprecedented precision, on many fundamental TCRβ properties such as preferences for nucleotide removal and addition at recombination junctions (Table 2) and the extent of CDR3β length diversity (Fig. 5A). As expected from all previous studies, we see that certain TRBV and TRBJ genes are commonly utilized while others are quite rare (Fig. 3A,B) and the pairing of TRBV and TRBJ is not random (Fig. 4; Supplemental Table 1; Rosenberg et al. 1992; Even et al. 1995; Hall and Lanchbury 1995; Roldan et al. 1995; Manfras et al. 1999). The reasons for bias are not clearly understood but are likely due to a combination of proximity effects and recombination signal sequence compatibilities that influence initial TCR development, plus thymic selection and immune challenge that modify the representation of selected clones in the extant repertoire (Krangel 2003). It must be emphasized that the results presented here for TRBV and TRBJ frequency and pairing are obtained from a biased sample, where the individual repertoires of subjects contributing to the pool have been skewed by antigen encounter and the individual's HLA type. These results cannot be taken to represent the innate TRBV and TRBJ usage and pairing preferences of the V(D)J recombination process during T-cell development. Further, it is possible that preferential PCR amplification of certain TRBV and TRBJ sequences over others has skewed the usage frequencies reported here. We have attempted to mitigate this by using 5′-RACE, rather than TRBV-specific primers, in order to reduce bias from differential primer annealing or variable amplicon lengths. The uniformity of the CDR3 length distribution presented in Figure 5 suggests that length bias has not been an issue. However, future studies with independent replicates from multiple individuals may be informative regarding other, unanticipated sources of bias.
Table 2.
Frequency of base addition and deletion at the TCRβ V-D-J junction
aD segments (D1: GGGACAGGGGGC, D2: GGGACTAGCGGG[AG]GGG) were trimmed 1 bp at a time and used for the search until D = 8 nt.
To differentiate the bias in TRBV and TRBJ representation that is incurred during V(D)J recombination and thymic selection from bias that is cased by antigen encounter in the periphery, it should be possible to sort and independently profile the naïve (CD25−, CD44−, CD45RA+, CD62L+) and memory/effector (CD25+, CD44+, CD45RO+, CD62L−) peripheral T-cell compartments. However, biases that are strictly due to recombination can only be delineated by profiling rearranged but pre-selected double-negative (CD4−, CD8−) cells from the thymic cortex. Studies of this nature would be most tractable in mouse.
Individual TCR diversity has been estimated as ∼106 beta chains (Arstila et al. 1999). This estimate relied on the calculation that TRBV18 to TRBJ1-4 pairing would occur at a frequency of 0.00024 in the repertoire. Our findings do not conflict with this assumption. We see pairing of TRBV18 and TRBJ1-4 in 4 of 22,704 unique clonotypes (which represents a frequency of 0.00018). However, at the present time we cannot provide insight into individual repertoire size because our sample is derived from blood pooled from multiple individuals. In fact, there may not be a “typical” individual peripheral blood T-cell repertoire, given that repertoires are skewed by many factors, including HLA type, historical antigen encounters, and current responses to acute infection.
There is considerable utility in our method of TCR sequence profiling (or its adaptation to immunoglobulin sequencing) even in the absence of heroic sequencing depth, since clonotypes that respond to a given immune challenge (effector and subsequently memory cells) will be present in the peripheral blood in higher copy number. There are many types of immune challenge, for example, acute and latent infections, autoimmunity, organ transplantation, and vaccination against infectious agents and malignancies. In these scenarios, and without a priori knowledge of the antigen, sequencing prospective samples to moderate depth should reveal responsive clonotypes. Additional applications may include the evaluation of immune reconstitution following, for example, bone marrow transplant or the initiation of highly active antiretroviral therapy (HAART) for HIV infection. It should also be possible to more readily identify the public T-cell clonotypes associated with infectious agents or tumor neoantigens and correlate these with effective immune responses.
Methods
5′-RACE
Peripheral leukocyte polyA+ RNA isolated from 165 L of peripheral blood pooled from 380 males (ages 18–40) and 170 females (ages 18–40) was obtained from a commercial supplier (Clontech #636170). First-strand cDNA was synthesized using a published TRBC primer (5′>CACGTGGTCGGGGWAGAAGC<3′) (Ozawa et al. 2008). A target-switching oligo (5′>AAGCAGTGGTAACAACGCAGAGTACGCGGG<3′) (Peters et al. 1999) was added to provide a 5′ template for RACE (reaction conditions: 100 ng of RNA, oligonucleotides 1 μM each, 2 mM DTT, 1 mM each dNTP, 25 mM Tris-HCl pH 8.3, 37.5 mM KCl, 1.5 mM MgCl2, and 400 units of Superscript II [Invitrogen] in a 20-μL volume. Extension was 90 min at 42°C followed by inactivation for 7 min at 72°C.). A control reaction with no enzyme was included to ensure that subsequent PCR products were the result of amplification from a reverse-transcribed template. PCR was performed using Platinum Pfx (Invitrogen) and 0.5 μL of first-strand reactions with the target-switching oligonucleotide (above) and a nested TRBC primer (5′> TGGTGCGGCCGCTCTCTGCTTCTGATGGCTCAAAC<3′) tailed with a NotI restriction site (reaction conditions: 1 unit of enzyme, 2× Pfx amplification buffer, 1 mM MgSO4, oligonucleotides 0.3 μM each, and 0.3 mM each dNTP in a 50-μL volume; 2 min denaturation at 94°C was followed by 30 cycles of 30 sec at 94°C, 30 sec at 55°C, and 45 sec at 68°C, plus a final extension for 5 min at 68°C). In order to obtain a cleaner product, PCR was performed on 0.1 μL of the first-round reaction with a nested target-switching oligonucleotide (5′>AGTTGCGGCCGCTAACAACGCAGAGTACGCGGG<3′), and an equimolar combination of two primers (5′>CACAGCGGCCGCGGGTGGGAACACCTTGTTCAGGT<3′), and (5′>CACAGCGGCCGCGGGTGGGAACACGTTTTTCAGGT<3′) specific for TRBC1 and TRBC2, respectively (reaction conditions: 1 unit of enzyme, 2× Pfx amplification buffer, 1 mM MgSO4, 0.3 μM oligonucleotides, and 0.3 mM each dNTP in a 50-μL volume; 2 min denaturation at 94°C was followed by 20 cycles of 30 sec at 94°C and 75 sec at 68°C, plus a final extension for 5 min at 68°C.). The nested PCR reaction was loaded on a 12% polyacrylamide gel and the band centered at 520 bp was visualized using SYBR Green (Lonza), excised, and processed for sequencing (below).
Preparation of 5′-RACE products for Illumina sequencing
Eight nested PCR reactions were pooled and purified using 20 μL of QIAEX II matrix (Qiagen). The eluate was digested with NotI (reaction conditions: 4.5 μg of DNA, 50 units of NotI [New England Biolabs], 1× NEB3 Buffer in 150 μL volume, 22 h at 37°C), and the band centered at 520 bp was purified from a 12% polyacrylamide gel. The fragment was then concatenated by ligation (reaction conditions: 500 ng of DNA, 1× NEB T4 DNA ligase buffer, 200 units of T4 DNA ligase [New England Biolabs] in a 5-μL volume) and stored at 4°C. Prior to sonication, the ligation product was cleaned with QAIEX II and eluted in a 20-μL volume. After 20 min sonication, the sample was loaded on a 8% polyacrylamide gel, and the fraction from 100–300 bp was excised, purified, and blunted (reaction conditions: 1× NEB Blunting Buffer, 100 μM dNTPs, 1 μL of Blunting Enzyme Mix [New England Biolabs E1201S] in a 25-μL volume) for 30 min at 21°C. The product was purified by phenol/chloroform extraction and ethanol precipitation prior to A-tailing (reaction conditions: 5 units of Klenow Fragment [3′→5′ exo–] [New England Biolabs], 1× reaction buffer, 200 μM dATP in a 50-μL volume, 30 min at 37°C). The product was purified by phenol/chloroform extraction and ethanol precipitation in preparation for ligation to Illumina TS adapters (www.illumina.com) (reaction conditions: 1× NEB T4 DNA ligase buffer, 1200 units of T4 DNA ligase [New England Biolabs], 1 μL of TS adapters in a 30-μL volume, 15 min at 21°C). The product was purified using a QiaQuick column (Qiagen) and eluted in a volume of 30 μL. Ten milliliters was then amplified by PCR using Illumina primers 1.1 and 2.2 (reaction conditions: 1 unit of enzyme, 2× Pfx amplification buffer, 1 mM MgSO4, 0.3 μM oligonucleotides, and 0.3 mM each dNTP in a 25-μL volume. Two minute denaturation at 94°C was followed by 15 cycles of 30 sec at 94°C, 30 sec at 65°C, and 30 sec at 68°C, plus a final extension of 5 min at 68°C.). The PCR product was purified using a MinElute column (Qiagen) with a final volume of 13 μL.
Illumina sequencing and analysis
We generated 18.8 million 36-nt reads and 21.7 million 50-nt single ends reads with the Illumina GAII analyzer, using sequencing chemistry version 2 (Illumina FC-204-2036) and cleavage reagent version 2 (Illumina #1005159). The combined set of 40,582,229 short reads were assembled using iSSAKE (with parameters: −m 15 −o 2 −r 0.7), as previously described (Warren et al. 2009). Briefly, reads aligning to the end of Ensembl TRBV gene predictions (Flicek et al. 2008) and having consecutively three or more unmatched bases in the adjacent CDR3β were used to seed iSSAKE de novo assemblies of CDR3β. Only contigs with a depth of at least two reads at each position were retained for analysis. However, we did not set a requirement of double coverage for the seed sequences themselves. Given that a seed sequence could be long enough to span the complete CDR3β, and there is no requirement for redundant coverage of seed sequences, 21,973 of the 33,664 clonotypes in our data set are represented by a single sequence read. This could, in principle, artifactually inflate the diversity of the repertoire, but in reality there is probably very little influence given that previous benchmarking of our assembly method using simulated, error-prone short sequence reads from 1 million computationally modeled TCRβ sequences showed 93% sensitivity and 99.96% accuracy for CDR3β clonotypes present at >3 p.p.m. (Warren et al. 2009).
The iSSAKE contigs (TCRβ reconstructions) were searched for the presence of 15 consecutive TRBJ segment bases. For any TRBJ segment, any 15-letter word from base position 1 to 25 characterizes uniquely that segment and allows the identification of the precise TRBJ segment boundary as well as the number of TRBJ bases deleted. TRBV segments, TRBV segment boundaries, and the exact number of deleted TRBV bases were inferred by tracing back the seed alignments that yielded the contigs and singlets under scrutiny. Sequence clonotypes were identified by extracting the contiguous bases spanning the last 15 bases of TRBV to the first recognizable 15 TRBJ segment bases, inclusively. During this automated process, we tracked clonotypes originating from seeds that aligned equally well to more than one TRBV segment, checked the sequence frame and peptide translation of the TCRβ reconstructions, and extracted the CDR3β, if applicable. The mined data was written to file and organized into a MySQL relational database for further analysis.
Fine-resolution analysis of N-diversity mechanisms at the V-D-J junction was made possible by searching for TRBD1 (12 nt) and TRBD2 (16 nt) bases between the TRBV and TRBJ boundaries using the longest to shortest TRBD word sizes. To favor accuracy over yield (some bases deletion/addition yield no recognizable TRBD bases), we chose to search until a minimum word of 8 nt for both TRBD segments. Unambiguous detection of TRBD bases allows precise identification of TRBD segment boundaries, the characterization of nontemplated bases at the V-D and D-J junction, and the frequency calculation of TRBD deleted bases for all clonotypes.
Sanger sequencing and analysis
Purified fragment was inserted into the vector pCR-4 using the recommended conditions for Invitrogen's Zero Blunt TOPO PCR Cloning Kit for Sequencing and One Shot MAX Efficiency DH5α-T1R competent cells. M13FP and M13RP were used to prime Sanger Sequencing reactions. We generated paired reads from 384 clones.
Low-quality bases were trimmed and vector sequences screened using Cross_match (www.phrap.org). A total of 736 quality and vector-trimmed paired-end reads remained and were assembled using CAP3 (Huang and Madan 1999). Five hundred eighty-two reads (331 clones) contained the complete V(D)J sequence and collapsed into 220 unique contigs (including single-read contigs, or singlets). The resulting contigs and singlets were analyzed for the frequency of predicted Vβ and 14 Jβ gene segments. Briefly, we aligned contigs and singlets against two separate databases of Ensembl (Flicek et al. 2008) human gene predictions for 54 Vβ and 14 Jβ gene segments using WU-BLAST (default parameters with −b 3000 −v 3000). For each of the database sequences we tallied the Vβ and/or Jβ gene alignments having the highest sequence identity to the contig and singlet sequences. Sequence alignments were analyzed using custom scripts, noting both the exact position of each 3′-Vβ and/or 5′-Jβ segment onto the mRNA, and a report of Vβ frequency and Jβ frequency was generated. Mapping of both the Vβ and Jβ segment positions and ensuring that the translation frame was preserved and consistent with peptide predictions permitted the extraction of variable CDR3β bases between the two segments (not shown).
Acknowledgments
This work was funded by Genome Canada and Genome British Columbia. R.A.H. is a Michael Smith Foundation for Health Research scholar. We thank Martin Krzywinski for generating Figure 4 using the Circos software package (Krzywinski et al. 2009). The melanoma-specific T-cell clone DES M26 was kindly provided by Cassian Yee, Fred Hutchinson Cancer Research Centre, Seattle, WA.
Footnotes
[Supplemental material is available online at http://www.genome.org. The TCRβ cDNA sequence and quality-score files have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA008633.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092924.109.
References
- Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl.):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Boria I, Cotella D, Dianzani I, Santoro C, Sblattero D. Primer sets for cloning the human repertoire of T cell receptor variable regions. BMC Immunol. 2008;9:50. doi: 10.1186/1471-2172-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai K, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Even J, Lim A, Puisieux I, Ferradini L, Dietrich PY, Toubert A, Hercend T, Triebel F, Pannetier C, Kourilsky P. T-cell repertoires in healthy and diseased human tissues analysed by T-cell receptor beta-chain CDR3 size determination: Evidence for oligoclonal expansions in tumours and inflammatory diseases. Res Immunol. 1995;146:65–80. doi: 10.1016/0923-2494(96)80240-9. [DOI] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch G, Lefranc MP. The human T cell receptor beta variable (TRBV) genes. Exp Clin Immunogenet. 2000;17:42–54. doi: 10.1159/000019123. [DOI] [PubMed] [Google Scholar]
- Fuschiotti P, Pasqual N, Hierle V, Borel E, London J, Marche PN, Jouvin-Marche E. Analysis of the TCR α-chain rearrangement profile in human T lymphocytes. Mol Immunol. 2007;44:3380–3388. doi: 10.1016/j.molimm.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Gellert M. Molecular analysis of V(D)J recombination. Annu Rev Genet. 1992;26:425–446. doi: 10.1146/annurev.ge.26.120192.002233. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Gorski J, Yassai M, Zhu X, Kissela B, Keever C, Flomenberg N. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR 3 size spectratyping: Correlation with immune status. J Immunol. 1994;152:5109–5119. [PubMed] [Google Scholar]
- Hall MA, Lanchbury JS. Healthy human T-cell receptor β-chain repertoire quantitative analysis and evidence for Jβ-related effects on CDR3 structure and diversity. Hum Immunol. 1995;43:207–218. doi: 10.1016/0198-8859(95)00013-t. [DOI] [PubMed] [Google Scholar]
- Harty JT, Badovinac VP. Shaping and reshaping CD8 T cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- Holt RA, Jones SJ. The new paradigm of flow cell sequencing. Genome Res. 2008;18:839–846. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Alt FW. Unraveling V(D)J recombination: Insights into gene regulation. Cell. 2004;116:299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- Krangel MS. Gene segment selection in V(D)J recombination: Accessibility and beyond. Nat Immunol. 2003;4:624–630. doi: 10.1038/ni0703-624. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: An information aesthetic for comparative genomics. Genome Res. 2009 doi: 10.1101/gr.092759.109. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, Crawford H, Honeyborne I, Asher TE, Luzzi G, et al. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J Immunol. 2006;177:4699–4708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- Manfras BJ, Terjung D, Boehm BO. Non-productive human TCR β chain genes represent V-D-J diversity before selection upon function: Insight into biased usage of TCRBD and TCRBJ genes and diversity of CDR3 region length. Hum Immunol. 1999;60:1090–1100. doi: 10.1016/s0198-8859(99)00099-3. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Travers P, Walport M. Janeway's immunobiology. Garland Science; London, UK: 2007. [Google Scholar]
- Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Tajiri K, Kishi H, Muraguchi A. Comprehensive analysis of the functional TCR repertoire at the single-cell level. Biochem Biophys Res Commun. 2008;367:820–825. doi: 10.1016/j.bbrc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DG, Kassam AB, Yonas H, O'Hare EH, Ferrell RE, Brufsky AM. Comprehensive transcript analysis in small quantities of mRNA by SAGE-lite. Nucleic Acids Res. 1999;27:e39. doi: 10.1093/nar/27.24.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan EQ, Sottini A, Bettinardi A, Albertini A, Imberti L, Primi D. Different TCRBV genes generate biased patterns of VDJ diversity in human T cells. Immunogenetics. 1995;41:91–100. doi: 10.1007/BF00182318. [DOI] [PubMed] [Google Scholar]
- Rosenberg WM, Moss PA, Bell JI. Variation in human T cell receptor V beta and J beta repertoire: Analysis using anchor polymerase chain reaction. Eur J Immunol. 1992;22:541–549. doi: 10.1002/eji.1830220237. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- Warren RL, Nelson BH, Holt RA. Profiling model T cell metagenomes with short reads. Bioinformatics. 2009;25:458–464. doi: 10.1093/bioinformatics/btp010. [DOI] [PubMed] [Google Scholar]