Abstract
Although the emergence of high-throughput sequencing technologies has enabled whole-genome sequencing from extinct organisms, little progress has been made in accelerating targeted sequencing from highly degraded DNA. Here, we present a novel and highly sensitive method for targeted sequencing of ancient and degraded DNA, which couples multiplex PCR directly with sample barcoding and high-throughput sequencing. Using this approach, we obtained a 96% complete mitochondrial genome data set from 31 cave bear (Ursus spelaeus) samples using only two 454 Life Sciences (Roche) GS FLX runs. In contrast to previous studies relying only on short sequence fragments, the overlapping portion of our data comprises almost 10 kb of replicated mitochondrial genome sequence, allowing for the unambiguous differentiation of three major cave bear clades. Our method opens up the opportunity to simultaneously generate many kilobases of overlapping sequence data from large sets of difficult samples, such as museum specimens, medical collections, or forensic samples. Embedded in our approach, we present a new protocol for the construction of barcoded sequencing libraries, which is compatible with all current high-throughput technologies and can be performed entirely in plate setup.
Genetic studies with fossil remains and museum specimens allow direct assessment of genotypes and genetic diversity from past species and populations, thereby providing a unique opportunity to observe evolution in “real time.” However, the DNA isolated from such material, commonly referred to as “ancient DNA,” is typically present in low amounts, heavily fragmented, chemically modified, and contaminated with environmental DNA (Willerslev and Cooper 2005), imposing great difficulties on the retrieval of large amounts of sequence data. The emergence of high-throughput sequencing technologies has brought a technical solution to some of these problems, especially fragmentation. Being ideally suited for sequencing millions of short molecules, these technologies have given a great boost to the field of paleogenomics (Poinar et al. 2006), enabling the recovery of genome sequences from extinct organisms, such as mammoth and Neanderthal (Green et al. 2006; Miller et al. 2008).
Despite successes in whole-genome sequencing, population studies with ancient DNA are still confined to comparatively short sequences, rarely exceeding a few hundred base pairs per sample. This is due to the fact that target regions must still be reconstructed through PCR amplification of short overlapping fragments. Barcoding methods were developed for sequencing many different PCR products in parallel on a high-throughput platform (Binladen et al. 2007; Meyer et al. 2007), but the amplifications are still performed in separate reactions, imposing serious limitations on the scope of sequence data generation. In the absence of alternatives, whole-extract shotgun sequencing has been proposed as an approach for generating targeted sequence data from ancient DNA. In a proof-of-principle study (Gilbert et al. 2007), this approach was used to reconstruct 15 mammoth mitochondrial genome sequences from hair, a rare type of tissue in the fossil record. However, approximately one full 454 Life Sciences (Roche) Genome Sequencer (GS) FLX run was required per sample, despite exceptional DNA preservation, little environmental contamination, and an elevated ratio of mitochondrial to nuclear DNA. More typically for ancient DNA, the recent recovery of a single Neanderthal mitochondrial genome from a bone sample required a sequencing depth only reached as part of a whole-genome sequencing effort (Green et al. 2008).
Although there is a broad choice of target enrichment methods for modern high-quality DNA, none of them has yet been successfully applied to ancient DNA. Typical ancient DNA molecules are very short, which presents a potential handicap for circularization-based amplification assays (Dahl et al. 2005, 2007; Porreca et al. 2007; Krishnakumar et al. 2008). Selective hybridization of targets to probes on microarrays (Albert et al. 2007; Hodges et al. 2007) or in solution (Gnirke et al. 2009) may be confounded by the complex composition of metagenomic samples. In addition, all these methods require enzymatic manipulations in the presence of the template DNA prior to amplification, e.g., through restriction digestion, ligation, or library preparation, inevitably causing a loss of material in purification steps or through reaction inefficiencies. In contrast, it has been shown that multiplex PCR, i.e., a PCR reaction containing more than one primer pair, can effectively pre-amplify targets directly from low copy number extracts, providing full single-molecule sensitivity (Dear and Cook 1993). Multiplex PCR has been implemented for a variety of applications, including, for example, single nucleotide polymorphism and short tandem repeat typing, linkage mapping, and also the amplification of ancient DNA (Krause et al. 2006). However, multiplex PCR amplification is commonly believed to produce overwhelming amounts of primer dimers and other spurious side products without extensive optimizations. For this reason, ancient DNA studies currently use multiplex PCR only as a first step, followed by singleplex PCR second steps (Römpler et al. 2006). This approach saves precious extract, but not reagents, time, or work.
Particularly for highly degraded and ancient DNA, it would be desirable to couple multiplex PCR directly with high-throughput sequencing. Previous studies with modern high-quality DNA have shown that this is in principle possible (Fredriksson et al. 2007; Varley and Mitra 2008; Goossens et al. 2009), but the proposed methods require cost- or time-consuming measures to avoid sequencing PCR artifacts. In one study, all primers were computationally checked for self-dimer and cross-dimer interactions, and the concentration of each primer was optimized in singleplex PCRs (Goossens et al. 2009). Despite these efforts, only small sets of primers could eventually be used in each multiplex amplification reaction (below 10 pairs). Two other studies achieved a higher level of multiplexing by introducing a second round of target selection after amplification, either by selective circularization (Fredriksson et al. 2007) or by target-specific adapter ligation (Varley and Mitra 2008). However, this requires more reaction steps and the synthesis of additional long oligos for each target. Furthermore, these methods require modified multiplex PCR primers (Varley and Mitra 2008), produce concatemeric products (Fredriksson et al. 2007), or are not easily compatible with sample barcoding (Fredriksson et al. 2007; Goossens et al. 2009).
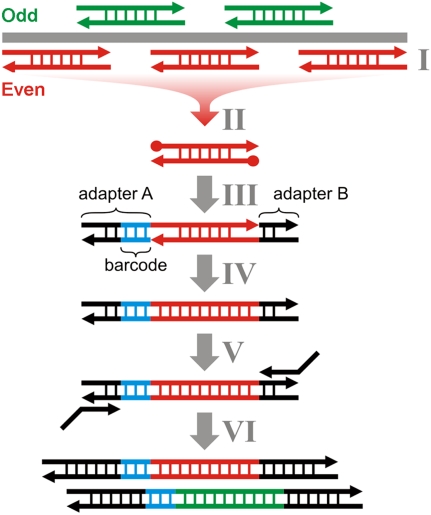
Here, we present direct multiplex PCR sequencing (DMPS), a novel method for targeted high-throughput sequencing of ancient and other degraded DNA, which combines standard multiplex PCR with sample barcoding and high-throughput sequencing in a straightforward way. Our method consists of four major steps (Fig. 1): (1) Many different targets are simultaneously amplified in one multiplex PCR reaction with reduced cycle number. (2) Spurious amplification products are eliminated by size-selective purification using solid phase reversible immobilization (SPRI) technology (DeAngelis et al. 1995). (3) Multiplex PCR products from different samples are barcoded and converted into sequencing libraries. To efficiently implement this process, we developed a new barcoding protocol for 96-well plate setup, which combines elements of the 454 library preparation protocol and one of the previously published barcoding methods (Meyer et al. 2008b). We took special care to reduce the formation of adapter artifacts, in order to allow barcoding of small amounts of input material. (4) The barcoded libraries are pooled and sequenced in parallel on the GS FLX platform from 454 Life Sciences (Roche).
Figure 1.
Schematic description of DMPS. Multiplex PCRs with nonoverlapping primer sets (“Odd” and “Even”) are performed in plate setup (I). After removing primer dimers and other spurious amplification products by size selective purification with SPRI beads, the double-stranded PCR products are blunt end repaired (II). Two different truncated double-stranded 454-adapters (A and B), one of which is barcoded (blue), are randomly ligated to either end of the double-stranded molecules by single-stranded ligation (III). Nicks are closed using a strand-displacing polymerase (IV), and the adapters are extended to full length in an amplification step with 5′-tailed primers (V). Library molecules carrying the same adapter on both ends are excluded from amplification, because they form hairpin structures that prevent primer annealing. Regardless, such molecules would not interfere with sequencing. Double-stranded barcoded sequencing libraries from multiple PCR products are pooled in equimolar ratios (VI) and sequenced on a high-throughput platform.
We demonstrate the efficiency of this method by sequencing—in replicates—the complete mitochondrial genomes of 31 cave bears (Ursus spelaeus), a species that went extinct 25,000 yr ago. Cave bears display a large amount of genetic diversity and possibly comprise more than one species, but until now their intraspecific relationships could not be resolved due to a lack of sequence data (Hofreiter et al. 2002; Orlando et al. 2002; Knapp et al. 2009). Therefore, they are well suited for showing the robustness of the approach as well as the power of the resulting data to resolve complex within-species relationships.
Results
Characterization of the method
Two nonoverlapping primer sets (termed “odd” and “even”), each containing 64 primer pairs, were designed to amplify the entire mitochondrial genome of cave bears in 128 fragments, ranging in length between 150 and 180 bp. No efforts were made to verify the compatibility of all primers in the multiplex mixes. Fossil bone samples of Pleistocene cave bear were obtained from a broad geographic range across Eurasia. DNA extracts from each bone were screened for the presence of one 175-bp target by singleplex PCR. Among 110 screened extracts, we identified 56 extracts with potentially sufficient DNA preservation for this study.
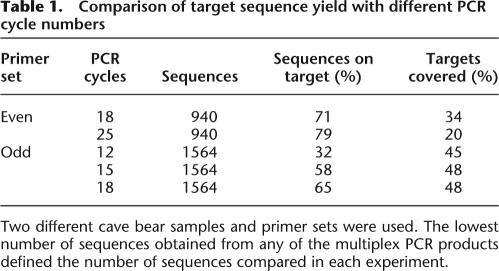
Using two of these extracts, we performed initial multiplex PCR and sequencing experiments to study the effect of PCR cycle number on target sequence yield. At the lowest number of cycles, we observed the highest fraction of sequence reads that did not represent target sequences (Table 1). Interestingly, most of these sequences did not start with a PCR primer sequence, indicating that carryover of template DNA into the library is the biggest source of non-target sequences at low cycle number. Falsely primed amplification products were largely absent, despite the complexity of the template DNA and the primer mixes, suggesting that most spurious amplification products were removed together with the primer dimers in the purification step removing the <100 bp fraction. The protocol should in theory exclude the denatured template DNA from sequencing, but it is likely that unspecific reannealing produces end-repaired template DNA molecules, which—after a low number of PCR cycles and starting from low copy numbers—outnumber the desired double-stranded amplification products. At a high cycle number most sequences matched one of the targets, but the overall number of covered targets did not increase due to a more pronounced overrepresentation of the best amplifying targets.
Table 1.
Comparison of target sequence yield with different PCR cycle numbers
Two different cave bear samples and primer sets were used. The lowest number of sequences obtained from any of the multiplex PCR products defined the number of sequences compared in each experiment.
Mitochondrial genome sequencing
Next we performed 20-cycle multiplex PCRs on all 56 preselected cave bear DNA extracts, using both primer sets in duplicates. Replicated amplification and sequencing is necessary to overcome sequence errors derived from miscoding damage, which occurs at an appreciable frequency in ancient DNA (Hofreiter et al. 2001; Briggs et al. 2007; Brotherton et al. 2007). Given the large number of samples from different ages and locations, we did not expect a uniform level of target enrichment for all DNA extracts. Thus, we first constructed sequencing libraries with only one replicate of each multiplex PCR product. Two small GS FLX plate regions produced sufficient data to determine the success of target enrichment in each reaction. The percentage of target sequences varied over a wide range (1%–100%; see Supplemental Table S1), indicating that some DNA extracts contained higher amounts of background DNA than others. Variation was particularly pronounced in the “even” primer set, probably due to a lower overall amplification efficiency of this primer set. As shown above, this problem could probably be compensated for by increasing the number of multiplex PCR cycles (e.g., from 20 to 30). However, in this study we selected for further sequencing only those samples that produced target percentages >40% in both multiplex primer sets. This subset of 31 samples included all geographical regions of our initial sampling (Supplemental Fig. S1; Supplemental Table S2).
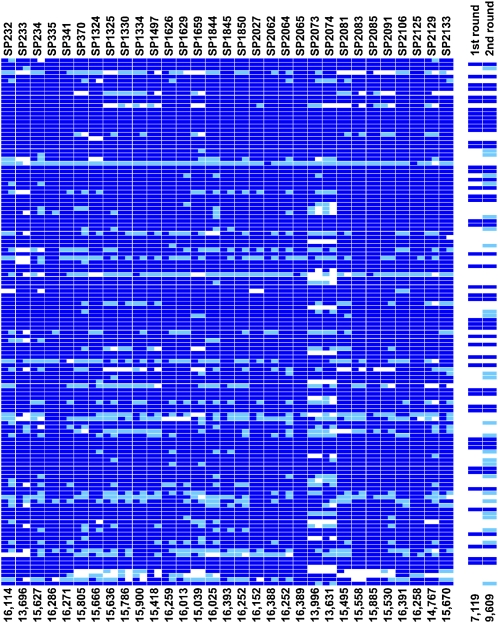
The vast majority of sequences obtained from the run (97.9%) started with a correct barcode. On average, 4610 sequences were obtained from each multiplex PCR product with a homogeneous distribution of sequence reads per barcode (Supplemental Fig. S2). On average, 112 of the 128 mitochondrial targets (87%) were covered by sequence reads (ranging from 72% to 98%, or 92 to 126 targets per specimen and replicate). Filtering for parts of the mitochondrial genome that were covered in replicate for all samples, we obtained 7.1 kb of overlapping sequence data for the complete population (Fig. 2). To increase this number further, we performed a second round of multiplex amplification followed by sequencing on half a GS FLX run. This time we created individual primer mixes for each sample, including only primer pairs that had failed to produce sequences in one or both replicates of the first round. We increased the cycle number to 25 to compensate for the presumably lower amplification efficiency of the remaining targets. After the second round of sequencing, the average coverage of the mitochondrial targets increased to 95.7%. This translated into a final population data set of 9.6 kb of overlapping replicated sequence, which required not more than two GS FLX sequencing plates, including a few small plate regions that were required for titration.
Figure 2.
Target coverage in two rounds of cave bear mtDNA sequencing. (Dark blue) Mitochondrial targets covered in the first round of sequencing, (aqua) targets covered after the second round, (white) uncovered targets. There are two columns per sample, one for each replicate. The amount of replicated sequence data obtained from each sample after two rounds of sequencing is shown at the bottom. Approximately 7 kb and 10 kb of the same mitochondrial sequence were covered in all samples after the first and second rounds of sequencing, respectively.
Phylogenetic reconstructions
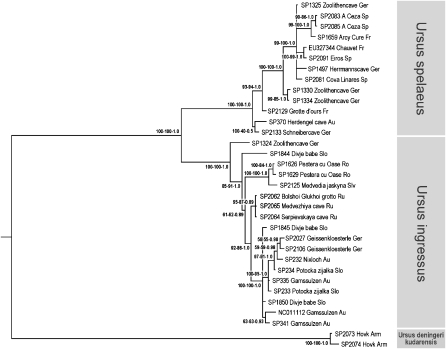
Phylogenetic relationships of cave bears were reconstructed using standard algorithms including neighbor-joining, maximum likelihood, and a Markov chain Monte Carlo based Bayesian approach (see Supplemental material). Independent of the method used, we recovered a robust phylogeny with strong statistical support on almost all branches (Fig. 3). Our data differentiate three major mitochondrial clades of cave bears, supporting a previous classification based on morphological analyses, which proposed Ursus spelaeus, Ursus ingressus, and Ursus deningeri kudarensis as different subspecies or even species (see Knapp et al. 2009 and references therein).
Figure 3.
Phylogentic relationships among cave bears. Numbers on branches represent neighbor-joining and maximum-likelihood bootstrap support values and Bayesian posterior probability. The tree is based on 9.6 kb of aligned mtDNA sequence and is rooted with brown bear as the outgroup (branch not shown). Abbreviations behind sample names indicate country of origin (see Supplemental Table S2).
DMPS with modern DNA and other primer mixes
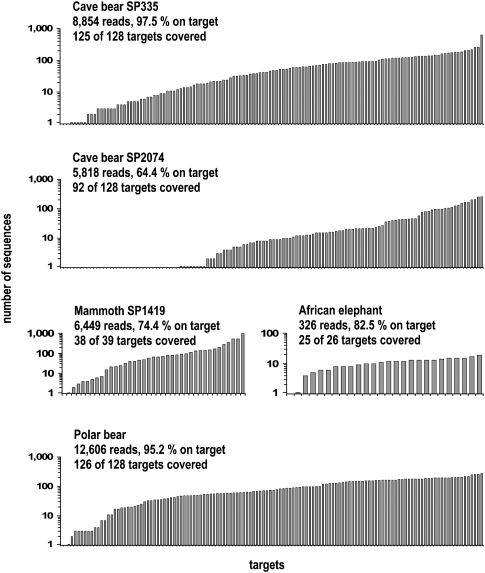
Complexity-related problems with multiplex PCR amplification have been widely discussed in the scientific literature. While some researchers have suggested the addition of certain reagents (Shigemori et al. 2005) or the use of software tools (Nicodeme and Steyaert 1997) in order to increase the compatibility of primers in a reaction, others had seemingly little trouble amplifying hundreds of targets without prior optimizations (Eichinger et al. 2005). To evaluate the general applicability of our approach, we conducted further experiments with both modern DNA and permafrost-derived DNA as different types of template and two additional multiplex PCR primer mixes. First, we used the cave bear primer mixes to amplify and sequence the mitochondrial genome of a polar bear, using high-quality DNA extracted from modern tissue. In the absence of environmental background DNA, we reduced the cycle number to 15. At a similar sequencing depth compared with the cave bear experiments, we obtained sequences for 126 of the 128 targets (Fig. 4). Second, using a mix of 39 primer pairs from a previous study (Rohland et al. 2007), we sequenced half a mitochondrial genome of mammoth from a permafrost sample, covering 38 targets in a single round of sequencing. Third, we tested a primer mix for 27 chromosomal targets of elephants and could recover 26 targets despite very shallow sequencing. These results show that our method works with a variety of different primer mixes and different types of samples without requiring extensive optimizations.
Figure 4.
DMPS applied to different samples and targets. Coverage plots resulting from a single round of amplification and sequencing are shown for mitochondrial or nuclear targets of cave bear, mammoth, polar bear, and African elephant. Only two example plots are shown for cave bears (with high and low target coverage), each representing the sequences obtained from two multiplex PCRs (odd and even).
Discussion
By generating ancient DNA population data on an unprecedented scale, we demonstrate the efficiency of direct multiplex PCR sequencing for targeted high-throughput sequencing of highly degraded DNA. Most of our samples were obtained from middle-European cave sites, exhibiting a state of DNA preservation typical for the kind of samples available for ancient DNA studies outside of permafrost regions and also common with many museum specimens or forensic samples. Already in a single round of amplification and sequencing—and investing only one GS FLX run—we obtained overlapping and replicated sequence from almost half of the mitochondrial genomes from all 31 cave bears. Although this amount of data is in principle sufficient to resolve the phylogenetic relationships among cave bears, we showed that most gaps can be closed with a second round of multiplex PCRs, resulting in a 96% complete data set. Recent studies on Eurasian cave bears suggested the existence of at least three distinct and reproductively isolated subpopulations, Ursus spelaeus, Ursus ingressus, and Ursus deningeri kudarensis (Hofreiter et al. 2004; Rabeder et al. 2004; Knapp et al. 2009). Our large data set for the first time confirms these clades as being reciprocally monophyletic, adding further support for their classification as independent species.
We did not encounter problems with spurious amplification products in any of the primer mixes or with any of the samples tested, suggesting either that they occur to a lower extent than commonly believed, or that they can be efficiently removed by size selection after amplification. Thus, we present a very robust and cost-efficient method for quickly retrieving many kilobases of overlapping sequence data from large sets of highly degraded samples, targeting both mitochondrial and nuclear sequences. Not only will our method facilitate genetic research on the huge numbers of specimens stored in museum collections worldwide, it can also be applied to formalin-fixed tissue and other difficult samples available for medical genetics or forensic research. With the single-molecule sensitivity of multiplex PCR, amplification and sequencing of pathogen DNA from infected tissue may represent another useful application. DMPS is easy to implement and easily scalable for large numbers of samples.
When designing the method, we were faced with the challenge of developing a barcoding protocol suitable for the low amounts of input material generated in low cycle number PCRs. The protocol we present works on all types of double-stranded DNA samples and can be performed in plate setup. Being characterized by simple setup, a high proportion of barcoded sequence reads, and a homogeneous distribution of reads among samples, it represents a good alternative to a number of previously published methods (Meyer et al. 2007; Craig et al. 2008). We did not attempt to avoid a library amplification step, which has been shown to be problematic under some instances (Kozarewa et al. 2009), because we started from already amplified input material. If necessary, amplification could be easily omitted by extending the truncated adapter oligos to their full sequences. By changing the adapter sequences, the method could be easily transformed to other sequencing platforms with higher throughput, such as Illumina's Genome Analyzer.
Methods
DNA extraction and primer design
DNA from cave bear and mammoth bones was isolated in an ancient DNA clean lab using an extraction method published previously (Rohland and Hofreiter 2007). Several blank controls were carried out with each set of DNA extractions. Conserved regions for primer design were identified through a multiple sequence alignment of mitochondrial genomes downloaded from GenBank (brown bear, NC_003427 [Delisle and Strobeck 2002], American black bear, NC_003428 [Delisle and Strobeck 2002], and an already existing mitochondrial genome of cave bear, NC_011112 [Krause et al. 2008]). A repetitive region of roughly 350 bp was excluded, because it could not be broken up and carries no phylogenetic information. All primer pairs were designed either manually or using the software tool primer3 (Rozen and Skaletsky 2000). Primers for the amplification of the mammoth mitochondrial genome were available in-house from a previous study (Rohland et al. 2007). Primers for 27 chromosomal targets of the African elephant were designed using primer3 and the African elephant genome as reference, which is provided freely by the Broad Institute (http://www.broadinstitute.org/ftp/pub/assemblies/mammals/elephant/loxAfr3). All primers exhibit annealing temperatures between 55°C and 62°C.
Multiplex PCR and the construction of barcoded sequencing libraries
Descriptions of the initial PCR screening, multiplex amplification, size selection, the construction of barcoded sequencing libraries, and all oligo sequences are available in the Supplemental material. Sixteen blank controls were carried throughout the DNA extraction, amplification, library preparation, and library amplification processes in the first round, and four were used in the second round of cave bear mitochondrial genome sequencing, thus controlling for contamination in all steps. In total, 70 different barcoded adapters were used throughout the experiments. Barcode sequences of 7 bp length were taken from a previous publication (Meyer et al. 2008b). To minimize the number of sequencing libraries, only two different barcodes per sample were used in each round of sequencing (attaching the same barcode to the first replicates of the odd and even multiplex reactions, and another to the second replicates). The barcoded sequencing libraries were quantified using a quantitative PCR assay described elsewhere (Meyer et al. 2008a). Subsequently, the libraries were pooled in an equimolar ratio, thereby creating final sequencing libraries. The blank controls of the first round were pooled into a separate library and sequenced on a small 454 plate region, because the number of barcodes was limiting. In the second round they were sequenced together with the cave bear samples. All multiplex PCRs that were not part of the cave bear mitochondrial genome sequencing were sequenced together with other samples, using the same barcoding protocol.
Contamination control and consensus calling
Following the standard 454 sequencing routine, barcodes were identified and trimmed using the software tool untag (http://bioinf.eva.mpg.de/pts/). Sequencing errors or cross-contamination among barcoding adapters may lead to an erroneous assignment of sequences to samples. To evaluate this effect, we compared the number of reads carrying unused barcode sequences to the overall number of sequences using a previously published formula (Meyer et al. 2008b). Our estimate of the false-assignment frequency is 0.02%.
Using the software runMapper v2.0.00.20, all sequences were mapped against the reference sequence of the cave bear mitochondrial genome. After trimming primer sequences from all reads, multiple sequence alignments were constructed for each target and replicate. Consensus sequences were called according to the criteria described in the Supplemental material. Consensus sequences from replicates of the same sample were merged and a final super-consensus was called. C/T and G/A mismatches were called as C and G, respectively, because they are known to be derived from deamination of cytosine to uracil, the only type of miscoding ancient DNA damage occurring at appreciable frequency (Briggs et al. 2007; Brotherton et al. 2007). In the entire data set, we observed only three positions (designated “N”) where consensus sequences of replicates differed by types of substitutions different from C/T and G/A. This could be due to sequencing errors, rare miscoding damage of another type, or the amplification of nuclear mitochondrial insertions. However, given their low incidence, they are unlikely to affect the same position twice in independent replicates.
In 26,036 reads obtained from the blank controls, only three reads matched one of the cave bear targets (0.01%). There is no straightforward way to transform this number into a contamination estimate, because the extraction blanks contain much less DNA than the cave bear extracts, requiring a higher level of amplification prior to sequencing. In the absence of other contaminants in the chemicals, even a single contaminating cave bear molecule could in theory produce all sequences obtained from an extraction blank. Thus, we accept 0.01% as an upper estimate of the level of contamination, but expect the real number to be several orders of magnitude smaller. Given that all sequences were replicated and the low estimate of contamination, we consider the effects of contamination and false-assignment negligible.
Additional methods
Phylogenetic tree-building methods are described in Supplemental material.
Acknowledgments
We thank Barbara Hoeffner and Ayinuer Aximu for running the 454 sequencer; Gennady Baryshnikov, Hervé Bocherens, Aurora Grandal d'Anglade, Brigitte Hilpert, Tatjana Kutznetsova, Susanne Münzel, Ron Pinhasi, Gernot Rabeder, Alfred Roca, Wilfried Rosendahl, Erik Trinkaus, and the Leipzig Zoo for providing samples; Knut Finstermeier for help with the figures; and Adrian Briggs for comments on the manuscript. We thank Ed Green, Patricia Heyn, Nadin Rohland, and Svante Pääbo for helpful discussions, and the Max Planck Society for financial support.
Footnotes
[Supplemental material is available online at http://www.genome.org. All cave bear mitochondrial genome sequences from this study have been submitted to GenBank under accession nos. FN390842–FN390872.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.095760.109.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J, Cooper A. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 2007;35:5717–5728. doi: 10.1093/nar/gkm588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis MM, Wang DG, Hawkins TL. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 1995;23:4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear PH, Cook PR. Happy mapping: Linkage mapping using a physical analogue of meiosis. Nucleic Acids Res. 1993;21:13–20. doi: 10.1093/nar/21.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisle I, Strobeck C. Conserved primers for rapid sequencing of the complete mitochondrial genome from carnivores, applied to three species of bears. Mol Biol Evol. 2002;19:357–361. doi: 10.1093/oxfordjournals.molbev.a004090. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S, Baner J, Dahl F, Chu A, Ji H, Welch K, Davis RW. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 2007;35:e47. doi: 10.1093/nar/gkm078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MT, Tomsho LP, Rendulic S, Packard M, Drautz DI, Sher A, Tikhonov A, Dalen L, Kuznetsova T, Kosintsev P, et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. doi: 10.1126/science.1146971. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens D, Moens LN, Nelis E, Lenaerts AS, Glassee W, Kalbe A, Frey B, Kopal G, De Jonghe P, De Rijk P, et al. Simultaneous mutation and copy number variation (CNV) detection by multiplex PCR-based GS-FLX sequencing. Hum Mutat. 2009;30:472–476. doi: 10.1002/humu.20873. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PL, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, Haeseler Av A, Pääbo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Capelli C, Krings M, Waits L, Conard N, Münzel S, Rabeder G, Nagel D, Paunovic M, Jambresic G, et al. Ancient DNA analyses reveal high mitochondrial DNA sequence diversity and parallel morphological evolution of late pleistocene cave bears. Mol Biol Evol. 2002;19:1244–1250. doi: 10.1093/oxfordjournals.molbev.a004185. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Rabeder G, Jaenicke-Despres V, Withalm G, Nagel D, Paunovic M, Jambresic G, Pääbo S. Evidence for reproductive isolation between cave bear populations. Curr Biol. 2004;14:40–43. doi: 10.1016/j.cub.2003.12.035. [DOI] [PubMed] [Google Scholar]
- Knapp M, Rohland N, Weinstock J, Baryshnikov G, Sher A, Nagel D, Rabeder G, Pinhasi R, Schmidt HA, Hofreiter M. First DNA sequences from Asian cave bear fossils reveal deep divergences and complex phylogeographic patterns. Mol Ecol. 2009;18:1225–1238. doi: 10.1111/j.1365-294X.2009.04088.x. [DOI] [PubMed] [Google Scholar]
- Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, Barnes I, Lister AM, Ebersberger I, Pääbo S, Hofreiter M. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- Krause J, Unger T, Nocon A, Malaspinas AS, Kolokotronis SO, Stiller M, Soibelzon L, Spriggs H, Dear PH, Briggs AW, et al. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene–Pliocene boundary. BMC Evol Biol. 2008;8:220. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar S, Zheng J, Wilhelmy J, Faham M, Mindrinos M, Davis R. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Myles S, Prüfer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Briggs AW, Maricic T, Höber B, Höffner B, Krause J, Weihmann A, Pääbo S, Hofreiter M. From micrograms to picograms: Quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 2008a;36:e5. doi: 10.1093/nar/gkm1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat Protoc. 2008b;3:267–278. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]
- Miller W, Drautz DI, Ratan A, Pusey B, Qi J, Lesk AM, Tomsho LP, Packard MD, Zhao F, Sher A, et al. Sequencing the nuclear genome of the extinct woolly mammoth. Nature. 2008;456:387–390. doi: 10.1038/nature07446. [DOI] [PubMed] [Google Scholar]
- Nicodeme P, Steyaert JM. Selecting optimal oligonucleotide primers for multiplex PCR. Proc Int Conf Intell Syst Mol Biol. 1997;5:210–213. [PubMed] [Google Scholar]
- Orlando L, Bonjean D, Bocherens H, Thenot A, Argant A, Otte M, Hänni C. Ancient DNA and the population genetics of cave bears (Ursus spelaeus) through space and time. Mol Biol Evol. 2002;19:1920–1933. doi: 10.1093/oxfordjournals.molbev.a004016. [DOI] [PubMed] [Google Scholar]
- Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, Auch A, et al. Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- Rabeder G, Hofreiter M, Withalm G. The systematic position of the Cave Bear from Potočka zijalka (Slovenia) Mitt Komm Quartärforsch Österr Akad Wiss. 2004;13:197–200. [Google Scholar]
- Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- Rohland N, Malaspinas AS, Pollack JL, Slatkin M, Matheus P, Hofreiter M. Proboscidean mitogenomics: Chronology and mode of elephant evolution using mastodon as outgroup. PLoS Biol. 2007;5:e207. doi: 10.1371/journal.pbio.0050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römpler H, Dear PH, Krause J, Meyer M, Rohland N, Schöneberg T, Spriggs H, Stiller M, Hofreiter M. Multiplex amplification of ancient DNA. Nat Protoc. 2006;1:720–728. doi: 10.1038/nprot.2006.84. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Shigemori Y, Mikawa T, Shibata T, Oishi M. Multiplex PCR: Use of heat-stable Thermus thermophilus RecA protein to minimize non-specific PCR products. Nucleic Acids Res. 2005;33:e126. doi: 10.1093/nar/gni111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Mitra RD. Nested Patch PCR enables highly multiplexed mutation discovery in candidate genes. Genome Res. 2008;18:1844–1850. doi: 10.1101/gr.078204.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev E, Cooper A. Ancient DNA. Proc Biol Sci. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]