Abstract
The genomes of higher organisms are packaged in nucleosomes with functional histone modifications. Until now, genome-wide nucleosome and histone modification studies have focused on transcription start sites (TSSs) where nucleosomes in RNA polymerase II (RNAPII) occupied genes are well positioned and have histone modifications that are characteristic of expression status. Using public data, we here show that there is a higher nucleosome-positioning signal in internal human exons and that this positioning is independent of expression. We observed a similarly strong nucleosome-positioning signal in internal exons of Caenorhabditis elegans. Among the 38 histone modifications analyzed in man, H3K36me3, H3K79me1, H2BK5me1, H3K27me1, H3K27me2, and H3K27me3 had evidently higher signals in internal exons than in the following introns and were clearly related to exon expression. These observations are suggestive of roles in splicing. Thus, exons are not only characterized by their coding capacity, but also by their nucleosome organization, which seems evolutionarily conserved since it is present in both primates and nematodes.
Genomic DNA in eukaryotic organisms is packaged in nucleosomes by wrapping the DNA molecules around a histone octamer, resulting in a nucleoprotein structure known as chromatin. It was originally thought that chromatin only had a structural role, but it was subsequently found to be involved in regulation of several biological processes, such as transcription, replication, DNA repair, and recombination (Groth et al. 2007; Li et al. 2007). The functional importance of chromatin in transcription is particularly well established; it is involved in all major transcription steps, i.e., pre-initiation, initiation, and elongation (Li et al. 2007). The genomes of Homo sapiens and other higher organisms contain transcribed units encoding proteins and functional RNAs, as well as larger regions of unknown function. Most protein-coding genes are organized into exons and introns. The primary transcripts are spliced to leave only the exons in the mature mRNAs. Until recently it was believed that splicing occurred mainly post-transcriptionally and that it was independent of chromatin. However, recent data suggest that in metazoans many exons are spliced co-transcriptionally. This suggests that there is a link between transcription, splicing, and chromatin (Neugebauer 2002; Kornblihtt 2007; Allemand et al. 2008; Pandit et al. 2008).
New technologies, in particular next-generation sequencing, have made it possible to obtain genome-wide data for nucleosome positions (Yuan et al. 2005; Schones et al. 2008; Valouev et al. 2008) and several epigenetic histone modifications (Barski et al. 2007; Mikkelsen et al. 2007; Wang et al. 2008). These data make it possible to investigate how nucleosomes and histone modifications are distributed along protein-coding genes and their relationship to other genomic features. These genomic studies have shown that protein-coding genes have well-positioned nucleosomes around their first exon and a nucleosome-free region (NFR) just upstream of the transcription start site (TSS) (Schones et al. 2008). Both nucleosome positioning and depletion have been shown to be dependent on the presence of RNA polymerase II (RNAPII) (Schones et al. 2008). Moreover, nucleosomes around first exons carry specific epigenetic marks related to the transcriptional status of genes. Examples are H3K4me3, H3K79me3, and H3K9ac for active genes, and H3K27me3, H3K9me2, and H3K9me3 for inactive genes (Barski et al. 2007; Mikkelsen et al. 2007; Wang et al. 2008).
Most of these studies have been focused on genomic regions around the TSSs, which undoubtedly harbor very important information, mainly related to transcriptional regulation. However, knowledge of how nucleosomes and histone modifications are distributed relative to other genomic features could result in valuable and novel insights into other biological processes, such as DNA replication and mRNA splicing (Allemand et al. 2008; Bres et al. 2008; Gottipati and Helleday 2009). It has been previously suggested that intron/exon junctions contain DNA sequences that promote nucleosome positioning (Beckmann and Trifonov 1991; Baldi et al. 1996; Kogan and Trifonov 2005). Positioned nucleosomes at these junctions were proposed to protect splice sites at exon starts from mutations (Kogan and Trifonov 2005).
In this letter, we use publicly available data from genome-wide studies to examine: (1) if nucleosomes are positioned at internal exons; (2) if certain histone modifications are preferentially found at internal exons compared to first exons; and (3) if exon positioning or histone modification is related to the transcriptional level.
Results and Discussion
Nucleosomes are well positioned at internal exons independent of transcription level
In order to investigate intragenic nucleosome positioning we analyzed publicly available sequencing data from human CD4+ T-cells (Schones et al. 2008) and Caenorhabditis elegans cells (Valouev et al. 2008). We constructed footprints of these nucleosome signals centered on the start sites of all exons (first, internal, and last) (Hubbard et al. 2007; see Methods for details). In both organisms, we found the previously reported pattern around the TSS as well as nucleosome depletion in the very end of the gene (Fig. 1A). In addition, we found one well-positioned nucleosome at internal exons with a signal clearly higher than that at the TSS. The peaks were centered at +94 (human) and +101 (C. elegans) base pairs (bp) from the internal exon start. Therefore, this positions the average 5′ end of a nucleosome around position +20 and +27 of the exon in H. sapiens and C. elegans, respectively.
Figure 1.
Nucleosomes are well positioned at internal exons. (A) Footprints of normalized nucleosome signal in human T-cells and C. elegans in a ±1-kb window. Signals were normalized for the total number of sequenced bases and genome size. The windows are centered, from top to bottom, on transcription start sites, intron/exon junctions of internal exons, intron/exon junctions of last exons, and 3′ ends of genes. (B) Partial footprints (left) of nucleosome signal in human (black) and C. elegans (green) at internal exon starts and ends split into six groups according to exon length (bp intervals given in brackets). In the pie charts (right), the percentage of the total number of exons in each exon size category is shown for human (black) and C. elegans (green). Included exons have flanking introns that are at least 100-bp long. (C) Footprints of human nucleosome signals for the same exon categories as in A but divided according to gene expression.
We found no clear indication of well-positioned nucleosomes in flanking introns (Supplemental Fig. S1). Moreover, the nucleosome signals were significantly higher at protein-coding exons than in pseudogene exons (Wilcoxon rank sum test P-value = 10−15) (Supplemental Fig. S2). Internal exons shorter than 50 bp comprise less than 5% of the exons in these genomes. We grouped the internal exons by length, and we found there was little or no signal above background for those shorter than 50 bp (Fig. 1B). For the longer exons, the nucleosome signal was higher within exons than in the vicinity in the flanking introns. The difference in nucleosome signal at exon boundaries clearly showed a preference for positioning in exons over introns. It has previously been suggested that introns may display a higher level of chromatin compaction than exons, due to the presence of repeated sequences (Allemand et al. 2008). Our data show that the hypothetically tightly packed nucleosomes in intronic regions may vary in location, a phenomenon sometimes referred to as fuzzy positioning (Jiang and Pugh 2009). In contrast, the nucleosomes at internal exons of protein-coding genes are well positioned. This pattern resembles that seen around the TSS, where poorly expressed genes have higher nucleosome content, although well-positioned nucleosomes are only found around the TSS of active genes. In conclusion, nucleosomes are well positioned at protein-coding exon starts and, for the longer ones (over 500 bp), also at the ends.
Furthermore, using public gene-expression data also from human CD4+ T-cells (Su et al. 2004), we observed that the nucleosome-positioning signal was present at the internal exons regardless of transcription level, in contrast to the patterns at the TSS and last exons (Fig. 1C). The pattern we observed around the TSS (Fig. 1C) is basically in agreement with that reported previously (Schones et al. 2008). The NFR immediately upstream of the TSS is clearly visible in active genes, i.e., in the high- and medium-expression groups, but it is absent in poorly expressed genes, i.e., in the low-expression group. Moreover, well-positioned nucleosomes are observed flanking the NFR, with more clear phasing patterns downstream from the TSS and among highly expressed genes. On the other hand, the nucleosome-positioning pattern at internal exons does not show any NFR and is minimally affected by gene-expression levels. Hence, this positioning seems almost independent of transcriptional activity, suggesting that it can be directed by exon length and/or sequence signals (Baldi et al. 1996), but not by RNAPII. Previous sequence analyses indicate that some periodic sequences are more common within exons than in flanking introns (Baldi et al. 1996; Kogan and Trifonov 2005). Especially relevant was the observation of dinucleotide periodicities indicative of nucleosome positioning within internal exons. On the basis of these data, it was proposed that nucleosomes are positioned at splice sites in order to protect them from mutation (Kogan and Trifonov 2005). However, our analysis demonstrates that nucleosomes are positioned at internal exons, whereas splice sites are frequently located in linker regions. Interestingly, recent reports have found that base-pair substitutions occur less frequently in linker regions than in nucleosomal DNA (Warnecke et al. 2008; Washietl et al. 2008; Sasaki et al. 2009;), probably because DNA-repair proteins have easier access to the nucleosome-free regions. In any case, exonic-nucleosome positioning seems to be under strong evolutionary constraint, as evidenced by the strikingly similar patterns for it in such distant organisms as H. sapiens and C. elegans.
H3K36me3 is enriched at internal exons of highly expressed genes
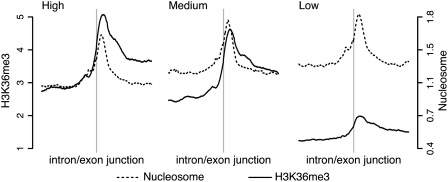
Since the nucleosome-positioning pattern at internal exons was only marginally affected by transcriptional activity, we hypothesized that the nucleosomes could be modified in a transcription-dependent manner, rather than remodeled. For humans, genome-wide data for 38 different histone methylations (Barski et al. 2007) and acetylations (Wang et al. 2008) are publicly available. These data are generated from the same cell type, i.e., CD4+ T-cells, as the nucleosome data (Schones et al. 2008). Human gene-expression measurements for this cell type are also available (Su et al. 2004), which allowed us to investigate if certain histone modifications are enriched at internal exons and how this is related to transcriptional levels (Supplemental Figs. S9–S46, bottom rows, and further described below). For histone modifications enriched at internal exons we would expect a preferential enrichment at intragenic regions without a preference toward 5′ ends of genes. However, H3K36me3 seems to be the only one with that distribution among these histone modifications (Barski et al. 2007). Barski et al. (2007) also report that H3K36me3 levels positively correlate with gene-expression levels. Interestingly, we noticed a conspicuously higher signal for H3K36me3 within internal exons in highly expressed genes than in genes with low expression (Fig. 2), while nucleosome levels remained relatively constant (Figs. 1C, 2). The slightly lower nucleosome level in exons of highly expressed genes may be due to total nucleosome displacement upon RNAPII passage (Kulaeva et al. 2007). Importantly, we found very similar H3K36me3 patterns in data from mouse embryonic stem cells (Mikkelsen et al. 2007), which shows that our findings can be extended to other organisms and cell types (Supplemental Fig. S3).
Figure 2.
H3K36me3 signal is high at internal exons of highly expressed genes. Footprints of normalized (see Fig. 1) H3K36me3 (solid line) and nucleosome (dashed line) signal in human T-cells in a ±1-kb window centered at intron/exon junctions of internal exons in genes with high, medium, and low expression.
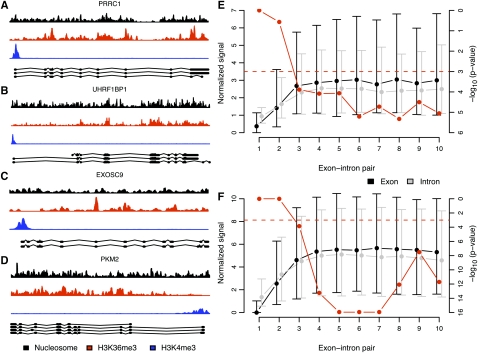
H3K36me3 has been suggested to accumulate over the whole transcribed region, to peak at the 3′ end of genes, and to dip at the very end (Bannister et al. 2005; Barski et al. 2007; Schones and Zhao 2008). We found that, in both mouse and human, the accumulation over the gene body was not simply a result of the progressive increase in H3K36me3 signals toward 3′ ends of transcribed genes. Rather, the intragenic H3K36me3 signal was correlated with the exon distribution within genes since exons are more often located toward 3′ ends (Pearson's r = 0.85 and 0.86 in human and mouse, respectively) (Supplemental Fig. S4). This colocalization is exemplified by the PRRC1, UHRF1BP1, EXOSC9, and PKM2 genes in Figure 3, A–D. We would like to note, however, that in these and other genes, there are H3K36me3 peaks outside internal exons. Sometimes they colocalized with expressed sequence tags (ESTs) (data not shown) and could therefore coincide with uncharacterized exons, or with uncharacterized new sense or anti-sense transcripts. Additionally, it cannot be ruled out that H3K36me3 may have a different function at intronic sequences.
Figure 3.
H3K36me3 signal is overrepresented at internal exons with respect to succeeding introns. (A–D) Example of nucleosome (black), H3K36me3 (red), and H3K4me3 (blue) signal in the human PRRC1 (A), UHRF1BP1 (B), EXOSC9 (C), and PKM2 (D) genes. Ensembl transcripts and corresponding exons are shown below the H3K4me3 signals. Most H3K36me3 signal peaks coincide with the location of exons. (E,F) Median (dots) and interquartile ranges (vertical lines) of average normalized (see Fig. 1) H3K36me3 signal in highly expressed genes in exons (black) and the corresponding succeeding introns (gray) (left vertical axes) in human T-cells (E) and mouse embryonic stem cells (F). The exons are grouped with corresponding succeeding introns in exon–intron pairs. In each exon–intron pair, say, 3, we assure that no exon of lower rank, i.e., 1 or 2, occurs in any annotated Ensembl transcript (see Methods for details). We depict in red (right vertical axes) paired Wilcoxon signed rank test P-values on the alternative hypothesis that signal in exons is higher than in corresponding succeeding introns. The dashed red line indicates a P-value of 0.001.
Finally, we compared the H3K36me3 signal in exons to the signal in the succeeding introns in highly expressed genes. In both human and mouse we found that from exon 3 and onward, the H3K36me3 signal was significantly higher in exons than in the following introns (Fig. 3E,F). This was not so for genes with low expression (Supplemental Fig. S5). Furthermore, exons generally had higher GC content than introns, but we found no bias toward either higher H3K36me3 or nucleosome signals in human internal exons than in the flanking introns of the high-expressed genes caused by content (Supplemental Fig. S6). Thus, we can conclude that there is a strong H3K36me3 signal at the nucleosomes positioned in the internal exons of genes with high expression, compared with those with low expression.
Histone modification marks as quantitative measures of exon expression
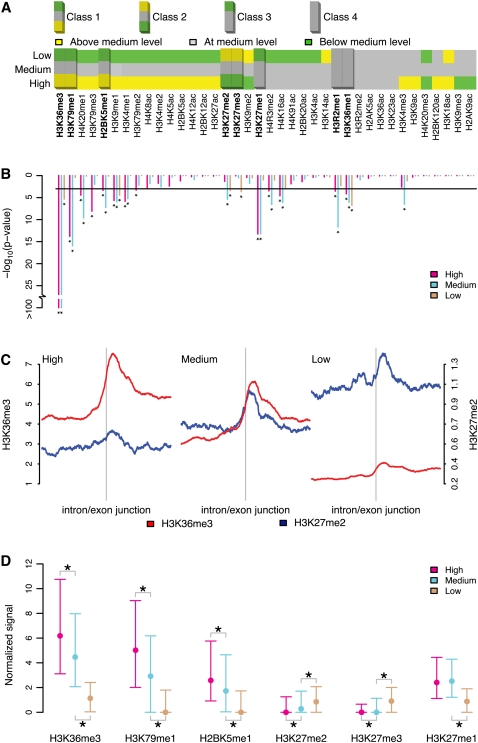
After finding that H3K36me3 is enriched in internal exons of highly expressed genes, we decided to systematically evaluate whether other histone modifications were also enriched at internal exons. Furthermore, we wanted to investigate if this enrichment was related to exon- rather than gene-expression levels. In order to answer these questions, we used expression data for individual exons (Oberdoerffer et al. 2008) from the same human cell type, i.e., CD4+ T-cells, for which the location of 38 histone modifications has been determined (Supplemental Figs S9–S46, top rows). We split the histone modifications into groups of distinct patterns using three criteria (see Methods for details). First, using exon-expression data, we categorized the internal exons of the highly expressed genes as high-, medium-, or low-expressed exons. To identify histone modifications affected by exon expression, for each modification, we compared the exonic signals of high- and low-expressed exons to the exonic signals of medium-expressed exons. This strategy allowed us to classify histone modifications as positively, negatively, or unrelated to changes in exon-expression level (Fig. 4A). Second, only histone modifications with internal exon signals significantly higher than in their corresponding succeeding introns (paired Wilcoxon signed rank test P-value < 0.001) were considered relevant (Fig. 4B). Third, by inspecting all histone marks manually, those with a clear preference for the TSS were disregarded as irrelevant.
Figure 4.
Some histone mark signals are higher in internal exons than in introns in a transcription-dependent manner. (A) For highly expressed genes, the average histone mark signal in exon-expression groups (low and high) (see Fig. 1) was compared to the respective medium-expressed exons to determine whether the signal was above (yellow), below (green) or at the same level (gray). The classes were determined by calculating the fold change (log2) of the average signals in the high-expression and low-expression categories to the average signal in the medium-expression one and then further discretized to above (>0.25), below (<–0.25) or at medium (between –0.25 and 0.25) level. (B) For the alternative hypothesis, that the signal in exons (in each exon-expression group) is higher than in corresponding succeeding introns, paired Wilcoxon signed rank test P-values (-log10) are depicted below each histone mark. Asterisks indicate significant (<0.001, horizontal black line) P-values. Highly relevant histone marks showing (1) any of the major identified trends in A; (2) significantly higher signal in exons than introns (B); and (3) lack of preferential accumulation at TSS-proximal regions (manual inspection) are highlighted (A). (C) Footprints of H3K36me3 (red) and H3K27me3 (blue) signals (±1-kb window) in human T-cells centered on intron/exon junctions of internal exons in highly expressed genes in the three exon-expression groups: high, medium, and low. (D) Median values and interquartile ranges of the exon average signals in the exon-expression groups high, medium, and low in Class 1 (H3K36me3, H3K79me1, and H2BK5me1), Class 2 (H3K27me2 and H3K27me3), and Class 3 (H3K27me1). Significant differences in distributions were tested for the high/medium- and low/medium-expression groups. An asterisk below or above the interquartile ranges indicates significantly (Wilcoxon rank sum test P-value < 10−5) lower or greater signal distribution compared to medium-expressed exons.
As a consequence, we indentified four major classes of histone modifications. The first class (Class 1) consisted of histone modifications with a progressively decreasing exonic signal from the high- to low-expressed exons. This group contained H3K36me3, H3K79me1, and H2BK5me1 (Supplemental Figs. S15, S25, S30). Interestingly enough, for the second class (Class 2), which comprised the marks H3K27me2 and H3K27me3 previously associated with gene silencing, we saw an opposite trend at both the exon- and gene-expression levels (Supplemental Figs. S21, S22). It was recently reported that the H3K27 demethylase KDM6A (also known as UTX) extensively colocalizes with elongating RNAPII and is found within actively transcribed genes (Smith et al. 2008). Since KDM6A demethylates both H3K27me3 and H3K27me2 (Lee et al. 2007), its association with elongating polymerases could partially explain the patterns we observed for these two repressive marks. The Class 1 and 2 histone marks are represented by the profiles of H3K36me3 and H3K27me2 in Figure 4C. The third class (Class 3) included the active mark H3K27me1 (Supplemental Fig. S20), which showed a similar signal in high- and medium-expressed exons, but a lower signal in those with low expression. A fourth class (Class 4) of histone modifications contained H3R2me1 and H3K36me1 and was characterized by a constant signal over all three levels of exon expression (Supplemental Figs. S24, S37).
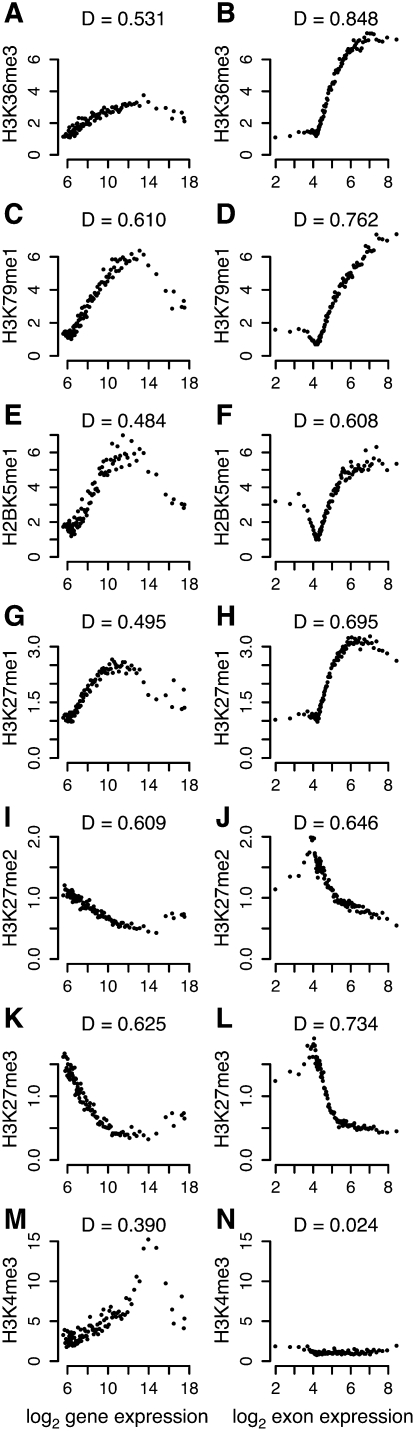
Figure 4D shows the signal distributions of histone modifications in Classes 1–3 at high-, medium-, and low-expressed exons in genes with high expression. These exon expression groups show significant distribution differences (Wilcoxon rank sum test P-value < 10−5). The relationships of these histone modifications to gene-expression level has been previously reported (Wang et al. 2008). We wanted to assess the relationship between each histone modification and expression. Therefore, we coupled the exon expression of all measured internal exons with each corresponding average histone modification signal. Likewise, the gene expression of all measured genes was coupled to each corresponding gene-body average. For each of the identified histone modification with exonic preference, we evaluated the dependencies between expression and histone modification for both relationships (Fig. 5, see Methods for details). In particular, we noticed that H3K36me3 (Fig. 5B) and H3K79me1 (Fig. 5D) were highly dependent on exon-expression level (Hoeffding's D of 0.848 and 0.762, respectively). For all histone modifications in Classes 1–3, we found that the internal exonic signal was more dependent on the exon expression level than what the gene-body average was for the level of gene expression. In contrast, H3K4me3 (Fig. 5M,N), associated with TSS enrichment and RNAPII binding (Barski et al. 2007), was related to the gene-expression level but not to that of exon expression.
Figure 5.
Histone modifications are highly dependent on exon-expression level. Histone modification signals over gene bodies (A,C,E,G,I,K,M) and internal exons (B,D,F,H,J,L,N) related to gene- and exon-expression bins, respectively, for H3K36me3 (A,B), H3K79me1 (C,D), H2BK5me1 (E,F), H3K27me1 (G,H), H3K27me2 (I,J), H3K27me3 (K,L), and H3K4me3 (M,N). The Hoeffding's D statistic (indicated above each plot) measures the dependency of histone modification signal on expression level (see Methods for details).
Footprints provide a means to extract data trends showing averages over a large number of regions with respect to a genomic feature. To further examine the differences in the exonic histone-modification signal in exon-expression groups at individual exons, we generated heat maps of nucleosome, H3K36me3, and H3K79me1 signals over windows centered on all internal exons of high-expressed genes (Fig. 6). We found a clear pattern of nucleosomes positioned downstream from the intron/exon junctions regardless of exon-expression level except for the exons with the highest expression (Fig. 6A). For H3K36me3 (Fig. 6B), we found a clear tendency toward a higher signal at exons with high expression and a lower signal at exons with low expression, in agreement with our previous results (Fig. 4). Even for H3K79me1 (Fig. 6C) this tendency was also present, although less pronounced.
Figure 6.
H3K36me3 and H3K79me1 are found at exonic nucleosomes and have a higher signal at highly expressed exons. Heat maps of nucleosome (A), H3K36me3 (B), and H3K79me1 (C) signal patterns at individual human internal exons. Rows in the heat maps correspond to 2-kb windows centered on intron/exon junctions (I/E junctions) of internal exons in highly expressed genes. Only internal exons with lengths between 100 and 300 bp and with flanking introns with lengths of at least 100 bp are shown. Each window (row in the heat map) was split into subwindows of 100 bp and the average signal calculated. The exons (rows in the heat map) are ordered according to exon expression. The gray tones were assigned using the signal quantiles of considered windows for H3K36me3, H3K79me1, and nucleosome separately. The groups of high-, medium-, and low-expressed exons (Fig. 4) are indicated with black boxes.
These observations raise the question of whether these patterns are present in all measured cells or in a fraction of cells. Higher-sequencing depth of samples or direct measurements from single-molecule sequencing technology (Clarke et al. 2009) are necessary before this can be determined. In summary, our data suggest that histone modifications, besides being functionally important at the TSS, seem to have important roles at the exonic level as well.
Histone modifications may facilitate exon inclusion during co-transcriptional splicing
We have demonstrated that H3K36me3 is the mark with the most significant exon-to-intron differences, at both the gene- and exon-expression levels. The decreasing pattern of the H3K36me3 exonic signal accompanied by the decreasing exon-expression level (Fig. 4C) and its dependence on exon expression (Fig. 5B) suggest a potential role of H3K36me3 in co-transcriptional splicing (Allemand et al. 2008; Pandit et al. 2008). By comparing the expression of each exon to the expression of its corresponding gene, we obtained a relative measurement of the inclusion/exclusion of a given exon into a mature transcript. We observed that the majority of excluded exons, i.e., the alternatively spliced ones, showed very low H3K36me3 levels, while the included exons showed a tendency toward higher H3K36me3 levels (Supplemental Fig. S7). This suggests that H3K36me3 can facilitate exon inclusion. Effects of exon length on splicing have been reported (Hertel 2008), and we have observed low levels of H3K36me3 in the exons shorter than 50 bp. In contrast, there was no such discrepancy for various intron lengths (Supplemental Fig. S8). However, short exon size does not seem to explain the absence of H3K36me3 in excluded exons (data not shown). Instead, exons shorter than 50 bp do not seem to be contained within well-positioned nucleosomes (Fig. 1B); this could make it difficult to add the H3K36me3 mark. Due to their small size, these exons may lack the minimal length of a yet-unknown sequence signal that prevents nucleosome positioning. Alternatively, the absence of clear nucleosome positioning or H3K36me3 in these short exons could partly be due to the small number of exons, compared to other exon size ranges, used to generate the signal footprints. It is noteworthy that a number of highly transcribed exons showed low H3K36me3 levels (Supplemental Fig. S7). This may reflect the fact that splicing is a combinatorial process (Hertel 2008), where other factors can play important roles, e.g., the strength of splice sites. Recent reviews have emphasized the combinatorial nature of splicing, which could be crucial in ensuring the fidelity of the process, especially for metazoan organisms with larger genes and introns (Berget 1995; Kornblihtt et al. 2004; Kornblihtt 2006; Allemand et al. 2008; Hertel 2008). Traditionally, splicing has been proposed to occur by intron-definition mechanisms, with splice-site recognition playing a major role (Berget 1995). However, this seems to apply only when the exons are flanked by introns shorter than 200 bp. In humans and other metazoans, many exons are surrounded by longer introns, and so alternative exon-definition splicing models have been proposed (Berget 1995). It is tempting to speculate that H3K36me3 and other histone modifications such as H3K79me1 may make exons more visible to the splicing machinery, especially when splice sites are weak.
The connection between H3K36me3 and splicing, although speculative, finds further support in the literature. First, while we were submitting this manuscript, Kolasinska-Zwierz et al. (2009) suggested that differences in H3K36me3 signal between exons and introns are evolutionarily conserved, since they also occur in C. elegans. Kolasinska-Zwierz et al. (2009) also reported accumulation of H3K36me3 at internal human and mouse exons, although they noted some preference toward 3′ ends of exons. However, in their analysis, Kolasinska-Zwierz et al. (2009) only considered internal exons with lengths between 350 and 450 bp, which is clearly more than the average exon size in these organisms. We likewise noted accumulation of H3K36me3 at the 3′ end of internal exons, especially within those longer than 200 bp (Supplemental Fig. S8). Importantly, our observations of nucleosome positioning at internal exons in C. elegans suggest that these well-positioned nucleosomes are marked with H3K36me3 in a transcription-dependent manner. Furthermore, after classifying exons as constitutively or alternatively spliced, on the basis of information from genes annotated with multiple transcripts, Kolasinska-Zwierz et al. (2009) found that the putative alternatively spliced exons had lower H3K36me3 levels. When we analyzed exon-expression, H3K36me3, and nucleosome data generated from the same human-cell type, we reached a conclusion consistent with theirs. Our analysis reinforces the link between this histone mark and splicing.
Second, it is intriguing that H3K36me3 has different functions in high and low eukaryotes (Carrozza et al. 2005; Edmunds et al. 2008), which could be due to their different intron–exon structures (Deutsch and Long 1999) and splicing mechanisms, i.e., co-transcriptional or post-transcriptional (Pandit et al. 2008). Moreover, SETD2, a histone H3 methyltransferase responsible for tri-methylation of lysine 36 in vertebrates (Edmunds et al. 2008), interacts with phosphorylated RNAPII that, in turn, interacts with various components of the spliceosome (Morris and Greenleaf 2000; Lin et al. 2004). SETD2 is a part of a complex with the elongation factor SUPT6H (also known as SPT6) and its partner IWS1. All three interact with elongating RNAPII (Yoh et al. 2007, 2008). Interestingly, a mutation or depletion in SUPT6H and IWS1 results in mRNA processing defects, including splicing and mRNA export. The transcripts become longer and accumulate in the nucleus (Yoh et al. 2007). Similarly, SETD2 depletion leads to nuclear retention of transcripts, but effects on transcript length, i.e., splicing, have yet to be tested (Yoh et al. 2008). Since defects in splicing can lead to nuclear-transcript retention (Sommer and Nehrbass 2005), it would not be unexpected if SETD2 also participates in splicing. Very recently, SETD2 was shown to interact in a complex with the heterogeneous nuclear ribonucleoprotein L (HNRNPL) (Yuan et al. 2009). This hnRNP protein is known to participate in various RNA processes, such as exon inclusion during alternative splicing and polyadenylation (Hung et al. 2008). Despite these reports, the role of H3K36me3 in splicing remains speculative at this stage.
In addition to H3K36me3, H3K79me1 seems to be related to splicing. First, the exon-expression level is highly dependent on the H3K79me1 exonic signal (Fig. 5D). Second, the Tudor domain of TP53BP1 is known to interact with mono- and di-methylated forms of H3K79 (Huyen et al. 2004). Interestingly, TP53BP1 has been shown to interact with U2 snRNA, along with several other proteins and RNA (Pryde et al. 2005). This interaction suggests a link between H3K79me1 and splicing, but further experiments are needed.
Guided by our findings and the previously mentioned reports, we believe it is worth considering models that link chromatin and splicing. Therefore, we hypothesize that most internal exons can be made visible to the splicing machinery through well-positioned nucleosomes carrying H3K36me3 and/or H3K79me1. This hypothesis concurs with models of co-transcriptional splicing in which pre-mRNA splice-site recognition is believed to occur while the RNAPII machinery is still engaged in the process of transcription (Beyer and Osheim 1988; Hertel 2008). In these models, splice-site recognition/alternative splicing is guided not only by DNA sequence, but also by other components such as additional spliceosome proteins, the phosphorylation status of RNAPII carboxy-terminal domain (CTD), and epigenetic signals (de la Mata et al. 2003; Batsche et al. 2006; Allemand et al. 2008), for instance the histone modifications H3K36me3 and H3K79me1.
Conclusions
Here we present the novel finding that there are well-positioned nucleosomes at most internal exons in such evolutionarily distant organisms as H. sapiens and C. elegans. We further demonstrate that the following histone modifications—H3K36me3, H3K79me1, H2BK5me1, H3K27me1, H3K27me2, and H3K27me3—function as quantitative measures of exon expression. Moreover, we hypothesize that H3K36me3 and H3K79me1 are involved in pre-mRNA splicing. Recent reports indicate lower substitution rates in linker regions than in nucleosomal DNA (Warnecke et al. 2008; Washietl et al. 2008; Sasaki et al. 2009) as well as higher rates of insertions and deletions longer than 1 bp in linker regions (Sasaki et al. 2009). Our results show that exons are functional units, defined not only by their coding capacity,but also by the way they are packaged in nucleosomes. This may have an impact on their stability and evolution.
Methods
Annotations
Nucleosome and histone modification data for H. sapiens resting CD4+ T-cells (Barski et al. 2007; Schones et al. 2008; Wang et al. 2008) were publicly available in hg18 (March 2006) coordinates. The exon-expression (GSE11384) (Oberdoerffer et al. 2008) and gene-expression data (Su et al. 2004) for human CD4+ T-cells were annotated to Ensembl (Hubbard et al. 2007) (database release 49, NCBI 36) respectively, using hg18 annotation files downloaded from the Affymetrix web page. The C. elegans data (Valouev et al. 2008) were publicly available in ce6 (May 2008) coordinates, and we used the corresponding Ensembl database, release 50 (WS190), as the source of annotations. The Mus musculus embryonic stem cell data (Mikkelsen et al. 2007) were publicly available in mm8 (Feb. 2006) coordinates. Gene annotations for the array platform used for the mouse expression experiment (GSE8024) (Mikkelsen et al. 2007) were extracted from the Ensembl database, release 46 (NCBI 36).
For the human, nematode, and mouse data, we extracted exon, transcript, and gene annotations from the Ensembl database (Hubbard et al. 2007) using the respective releases given above. TSSs were defined as the start of Ensembl genes. All annotated genes were considered unless otherwise stated. To compare the difference of nucleosome content in protein-coding exons to that of pseudogenes (Supplemental Fig. S2), we extracted all exons from the Ensembl database with annotated biotype “protein_coding” and “pseudogene.” An internal exon was defined to be any exon except the first or last one and positioned at least 2 kb from any listed transcript start or end. Using this definition, some exons may not have been truly internal, due to the existence of nonannotated, intragenic alternative TSSs. These are often underestimated in current annotations and are one possible explanation for the observed preference toward 5′ end internal exons in several histone modifications. We believe our definition offers a good compromise between capturing truly internal exons and excessive removal of them.
For the exon–intron comparison in Figure 3, E and F, we first collected all exons annotated as the first exon in any transcript associated with the genes under consideration. These were then compared to the corresponding succeeding introns. To guarantee no overlap with subsequent exons, we proceeded with all exons annotated as exon 2 in any transcript but excluded the ones associated with exon 1, and so on. This strategy guarantees that no annotated alternative TSS occurs in the exon 2 or subsequent groups.
The term “well positioned” is used to refer to a nucleosome well-positioned with respect to a genomic feature, such as intron/exon junctions, as opposed to all cells in the cell population having a nucleosome at the same genomic coordinate.
Signal assembly
All ChIP-seq data originate from the uniquely aligned fragments given by the respective principal investigator (Barski et al. 2007; Mikkelsen et al. 2007; Schones et al. 2008; Valouev et al. 2008; Wang et al. 2008). The information regarding the original lengths of the shredded/digested DNA fragments sequenced was found in the publication corresponding to each data set. We used the original data to construct an overlap signal by extending each aligned fragment to this length and counting the number of overlaps at each individual base pair with the SICTIN tools (S Enroth and J Komorowski, in prep.). The signals were then normalized by dividing each overlap with the normalizing factor nfi = ri * li / L, where ri denotes the total number of uniquely aligned reads for signal i (i.e., histone modification signal or nucleosome signal), li the original lengths of the shredded/digested DNA fragments sequenced, and L the total mappable genome length of the considered species. In this way, each normalized signal represents the fraction over the expected number of overlaps per base pair, assuming that all positions in the mappable parts of the genome are equally probable.
Characterization of genes and exons according to expression
We categorized the Ensembl genes into three groups according to gene-expression measurements from the human CD4+ T-cells (Su et al. 2004) and the mouse embryonic stem cells (GSE8024) (Mikkelsen et al. 2007), respectively. High-expressed genes were defined with an expression level above one standard deviation over the mean. The groups of medium- and low-expressed genes were determined by ensuring the same number of genes in each group. For these two categories, the lowest- and midmost-expressed genes were chosen, respectively. For the exon-expression-related analyses, we categorized the internal exons of highly expressed genes into high,- medium- and low-expressed exons using the above procedure according to the overall exon-expression distribution.
For all exons in highly expressed human genes, we calculated the fold change (log2) between the expression of each exon and the average exon expression of the corresponding gene. These log2 values were then interpreted as indicators of exon-exclusion (negative) or exon-inclusion (around zero or positive) events.
GC-content bias
To test whether the GC-content bias in exons with respect to introns could explain the higher levels of nucleosome and H3K36me3 signal in the human exons, we calculated the GC content of all internal exons and flanking introns in the highly expressed genes. The exons and introns were then grouped according to GC-content intervals and the distributions of average exonic/intronic signal plotted separately for each GC-content interval (Supplemental Fig. S6).
Functional characterization of histone marks
For each histone modification and individual exon in the human genome (Ensembl) of highly expressed genes, the average signal was calculated and normalized as described above. We then categorized these signals according to the expression level of each exon into high-, medium-, and low-expressed exons as we did with the gene-expression data. We calculated the fold change (log2) of the average signals in the high-expression and low-expression categories with respect to the average signal in the medium-expression category. We further categorized each fold change as above (>0.25), below (<–0.25), or unchanged. This procedure yielded a three-digit vector for each modification, corresponding to a high-, medium-, and low-exon-expression level, where each digit in the vector indicated either no change (0), an increase (1), or a decrease (–1) compared to the average signal of medium-expressed exons. We further imposed the selection criteria that histone modifications should have a signal significantly higher in exons than in succeeding introns (paired Wilcoxon signed rank test P-value < 0.001) and without any preference for TSS-proximal regions (by manual inspection). This gave the following classes: Class 1 (1,0,–1), Class 2 (–1,0,1), Class 3 (1,0,0), Class 4 (0,0,0), and inconclusive (any other combination). To further illustrate the differences in signal between the exon-expression groups, we plotted the interquartile ranges of signals in Classes 1–3. Significance of differences between the high-/medium-expression groups and the low-/medium-expression groups was then tested (Wilcoxon rank sum test P-value < 10−5).
Dependency of histone modifications on expression levels
To measure the dependency of histone modification signal on expression level, we binned the exon- and gene-expression data separately into 100 bins of equal size according to expression level. For each exon-expression bin, the average internal exonic histone modification signal of associated exons was calculated. Similarly, for each gene-expression bin, the average gene-body histone modification signal was calculated. For each of the identified histone modifications with exonic preference, we evaluated the relationship between expression and histone modification using the distribution-free Hoeffding's D measure of dependence (Hollander and Wolfe 1999). This statistic measures the average square deviation of sample pairs from independence. Using the implementation of Hoeffding's D in the R (R Development Core Team 2008) package Hmisc (Harrell 2008), the statistic ranges between –0.5 and 1.0. The higher the value of D, the more dependent the variables are on each other. Correlation measures such as the Pearson and Spearman correlation coefficients are not appropriate in this case, because they measure linearity and monotonicity, respectively, while Hoeffding's D may identify a broad range of dependencies.
Heat maps
To examine the H3K36me3, H3K79me1, and nucleosome signal patterns at individual human exons, we generated respective heat maps of 2-kb windows centered on a subset of all internal exons in the highly expressed genes. We considered only internal exons with lengths between 100 and 300 bp and with flanking introns of at least 100 bp. Each window (row in the heat map) was then split into subwindows of 100 bp and the average signal calculated. The windows were then ordered according to exon expression. The gray tones used in the heat maps were determined using the signal quantiles of considered windows for H3K36me3, H3K79me1, and nucleosome separately.
All analyses were performed using the R language (R Development Core Team 2008), with standard methods and packages.
Acknowledgments
R.A., S.E., A.R.-I., and J.K. were partially supported by the Knut and Alice Wallenberg Foundation and by the Swedish Foundation for Strategic Research. A.R.-I. was partially supported by the Olof and Signe Wallenius Foundation. C.W. was supported by the Swedish Research Council for Medicine, Science, and Technology and by the Swedish Cancer Research Foundation. We wish to thank Henric Winell for helpful discussion regarding dependency measures and Terese Bergfors for correcting our language.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092353.109.
References
- Allemand E, Batsche E, Muchardt C. Splicing, transcription, and chromatin: A ménage à trois. Curr Opin Genet Dev. 2008;18:145–151. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Baldi P, Brunak S, Chauvin Y, Krogh A. Naturally occurring nucleosome positioning signals in human exons and introns. J Mol Biol. 1996;263:503–510. doi: 10.1006/jmbi.1996.0592. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Beckmann JS, Trifonov EN. Splice junctions follow a 205-base ladder. Proc Natl Acad Sci. 1991;88:2380–2383. doi: 10.1073/pnas.88.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Deutsch M, Long M. Intron–exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: Link between transcription, replication, and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Harrell FE.Hmisc: Harrell Miscellaneous. 2008. http://cran.r-project.org/web/packages/Hmisc/index.html.
- Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA. Nonparametric statistical methods. 2nd ed. Wiley; New York: 1999. [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: A combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: Advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan S, Trifonov EN. Gene splice sites correlate with nucleosome positions. Gene. 2005;352:57–62. doi: 10.1016/j.gene.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR. Chromatin, transcript elongation, and alternative splicing. Nat Struct Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Studitsky VM. Transcription through chromatin by RNA polymerase II: Histone displacement and exchange. Mutat Res. 2007;618:116–129. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lin KT, Lu RM, Tarn WY. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol Cell Biol. 2004;24:9176–9185. doi: 10.1128/MCB.24.20.9176-9185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie AM, Melton DW, Jullien D, Adachi Y. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- Sasaki S, Mello CC, Shimada A, Nakatani Y, Hashimoto S, Ogawa M, Matsushima K, Gu SG, Kasahara M, Ahsan B, et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol. 2008;28:1041–1046. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer P, Nehrbass U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr Opin Cell Biol. 2005;17:294–301. doi: 10.1016/j.ceb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Batada NN, Hurst LD. The impact of the nucleosome code on protein-coding sequence evolution in yeast. PLoS Genet. 2008;4:e1000250. doi: 10.1371/journal.pgen.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Machne R, Goldman N. Evolutionary footprints of nucleosome positions in yeast. Trends Genet. 2008;24:583–587. doi: 10.1016/j.tig.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes & Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes & Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Yuan W, Xie J, Long C, Erdjument-Bromage H, Ding X, Zheng Y, Tempst P, Chen S, Zhu B, Reinberg D. Heterogeneous nuclear Rribonucleoprotein L is a subunit of human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. J Biol Chem. 2009;284:15701–15707. doi: 10.1074/jbc.M808431200. [DOI] [PMC free article] [PubMed] [Google Scholar]