Abstract
The 11.3-Mb genome of the yeast Lachancea (Saccharomyces) kluyveri displays an intriguing compositional heterogeneity: a region of ∼1 Mb, covering almost the whole left arm of chromosome C (C-left), has an average GC content of 52.9%, which is significantly higher than the 40.4% global GC content of the rest of the genome. This region contains the MAT locus, which remains normal in composition. The excess of GC base pairs affects both coding and noncoding sequences, and thus is not due to selective pressure acting on protein sequences. It leads to a strong codon usage bias and alters the amino acid composition of the 457 proteins encoded on C-left that do not show obvious bias for functional categories, or the presence of paralogs or orthologs of essential genes of Saccharomyces cerevisiae. They share significant synteny conservation with other species of the Saccharomycetaceae, and phylogenetic analysis indicates that C-left originates from a Lachancea species. In contrast, there is a complete absence of transposable elements in C-left, whereas 18 elements per megabase are distributed across the rest of the genome. Comparative hybridization of synchronized cells using high-density genome arrays reveals that C-left is replicated later during S phase than the rest of the genome. Two possible primary causes of this major compositional heterogeneity are discussed: an ancient hybridization of two related species with very distinct GC composition, or an intrinsic mechanism, possibly associated with the loss of the silent cassettes from C-left that progressively increased the GC content and generated the delayed replication of this chromosomal arm.
Lachancea kluyveri is a budding yeast first isolated from the intestinal canal of Drosophila in the Yosemite region of California, described as Saccharomyces kluyveri in 1956 (Phaff et al. 1956), and reclassified in the Lachancea genus (Kurtzman 2003), an assignment confirmed by additional phylogenetic studies (Wu et al. 2008). The Lachancea clade includes L. thermotolerans and L. waltii (formerly classified in the Kluyveromyces) and L. cidri, L. fermentati (formerly classified in the Zygosaccharomyces), and L. meyersii (Naumova et al. 2007). L. kluyveri has been isolated from the Drosophila species in North America, from soil in Europe, and from various tree species in India and North America.
In contrast to S. cerevisiae, L. kluyveri ferments sugars only in the absence of oxygen, and its efficient use of glucose makes it a valuable organism for industrial protein production under aerobic glucose-limited conditions (Møller et al. 2004). Unlike most Saccharomycetaceae, L. kluyveri can use pyrimidines and their degradation products as its sole nitrogen source (Gojkovic et al. 2003).
The type strain of the species (CBS 3082) is diploid, and like all Lachancea species has eight chromosomes (Weinstock and Strathern 1993; Neuvéglise et al. 2000; Naumova et al. 2007). There have been two preliminary surveys of its genome (Neuvéglise et al. 2000; Cliften et al. 2001, 2003), and it has now been fully sequenced and annotated (The Génolevures Consortium 2009). The 11.3-Mb nuclear genome contains 5321 predicted protein-encoding genes and 257 tRNA genes; it is ∼900 kb larger than that of L. thermotolerans with about 300 more genes. The genome contains few transposable elements, with only one family of degenerate class II elements and two families of long-terminal repeat (LTR) retrotransposons. The major family, the Ty1/copia element Tsk1, contains potential active elements that might have been acquired by horizontal transfer (Neuvéglise et al. 2002).
The most intriguing feature of L. kluyveri is the high GC content (52.9%) of the 1-Mb left arm of chromosome C (abbreviated here as “C-left”). The rest of the genome is a homogeneous 40.4% GC (The Génolevures Consortium 2009). The genomes of two related Lachancea, L. thermotolerans (The Génolevures Consortium 2009) and L. waltii (Kellis et al. 2004), do not show large-scale compositional heterogeneity. This is also true for all other yeasts, where GC content variations are limited to local differences between intergenic regions (lower GC content), protein-coding sequences (higher GC content), and genes for noncoding RNAs (highest GC content). For filamentous fungi, large-scale compositional heterogeneity has been reported for the phytopathogen Leptosphaeria maculans, whose genome displays an isochore-like structure (Gout et al. 2006; Fudal et al. 2007). Some of its chromosomes consist of mosaics of GC-rich fragments (51% GC) and AT-rich segments (35% GC) of up to 453 kb that are almost devoid of genes. These AT-rich regions include a few avirulence genes embedded in stretches of highly degenerated LTR retrotransposons due to extensive repeat-induced point mutations. The green algae species Ostreococcus also exhibit significant GC content heterogeneity (Derelle et al. 2006; Palenik et al. 2007). In these species, two chromosomes have a much lower GC content than the rest of the genome, reaching 13% in O. lucimarinus. In these chromosomes, the bias in gene function, intron-containing genes, and transposable element content is associated with a dramatic increase of intrachromosomal rearrangement. Whereas one chromosome is suspected to have been acquired by horizontal transfer, the other one is proposed to be a sex-determining chromosome. The genomes of mammals and other vertebrates have long been known to be mosaics of isochores of GC-rich and GC-poor regions longer than 300 kb (Macaya et al. 1976), correlated with strong biases in gene density or repeated sequences, and with some biases in gene expression, replication timing, and recombination (Bernardi 2007). In the above examples, the GC-content heterogeneity is explained by specific chromosomal organization or genetic content. Here, we describe the unusual characteristics of L. kluyveri C-left, and we present two alternative hypotheses to account for its origin: a unique chromosomal arm exchange versus a progressive GC-content elevation related to delayed DNA replication.
Results
A considerable GC richness is clustered in a 1-Mb chromosomal arm of the L. kluyveri genome
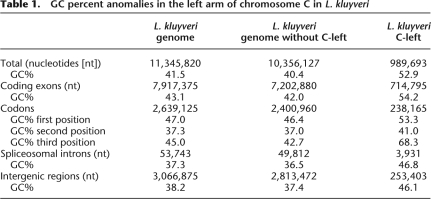
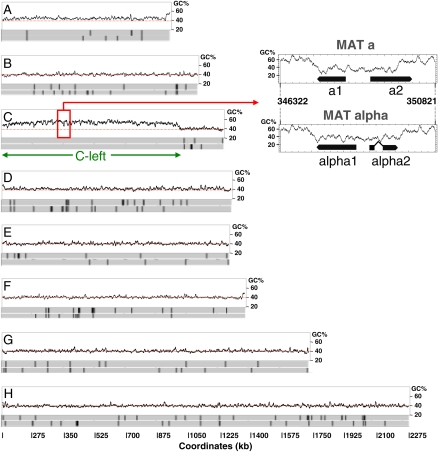
The overall GC content of the 11.3-Mb genome of L. kluyveri, 41.5% (The Génolevures Consortium 2009), is in the midrange of values for hemiascomycetes (36% in Debaryomyces hansenii [Dujon et al. 2004] to 52% in Eremothecium [Ashbya] gossypii [Dietrich et al. 2004]), but a ∼1-Mb region on the left arm of chromosome C stands out with its 52.9% GC content; i.e., 12.5% higher than the rest of the genome (Table 1). This region extends from the left telomere to ∼20 kb before the centromere (positions 1–989,693 of the DNA sequence). We refer to this region as “C-left” (Fig. 1). The remaining part of the chromosome has a GC content similar to the rest of the genome (40.4%).
Table 1.
GC percent anomalies in the left arm of chromosome C in L. kluyveri
Figure 1.
GC content along L. kluyveri chromosomes. (A–H) Shown is the composition of each chromosome (5-kb sliding windows with 2-kb steps) deduced from its DNA sequence (The Génolevures Consortium 2009). (Two solid gray rectangles) Each of the two DNA strands, (underlining) the part of chromosome C corresponding to C-left. The rDNA locus on chromosome H is not represented. (Red rectangle on chromosome C) Position of the MAT locus (enlarged on the right). This locus initially identified by Butler et al. (2004) carries genes that determine either cell type a (MATa1 and MATa2, SAKL0C03674g and SAKL0C03696g, respectively) or alpha (MATalpha1 and MATalpha2, see Methods). It is located between SAKL0C03652g (ortholog of the SLA2 gene of S. cerevisiae) and SAKL0C03718g (ortholog of DIC1 gene of S. cerevisiae) genes. (Vertical black lines) Positions of the Tsk1 retrotransposons (solo LTR, degenerate, and full-length elements) on each chromosomal strand. Note the absence of such elements along C-left.
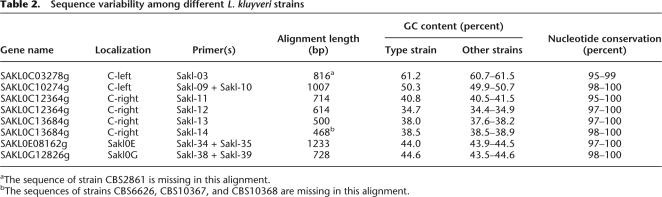
To test whether this heterogeneity is representative of the species L. kluyveri and not simply a peculiarity of the sequenced strain, we determined the nucleotide sequence of a few genes (two on C-left and four in the rest of the genome; Table 2) in 11 strains from various geographical and biological origins (Supplemental Table S1). The data revealed high nucleotide sequence conservation with the type strain CBS 3082 and GC percent very similar to that of the reference genes. We therefore suspect that C-left is GC rich in all other strains with a comparable extension on the chromosomal map (the sequenced gene SAKL0C10274g is close to the right extremity of the GC-rich region in CBS 3082).
Table 2.
Sequence variability among different L. kluyveri strains
aThe sequence of strain CBS2861 is missing in this alignment.
bThe sequences of strains CBS6626, CBS10367, and CBS10368 are missing in this alignment.
The increase in GC content affects the different types of genetic elements on C-left. Coding exons display an average GC content of 54.2%, significantly higher compared with the 42.0% for the rest of the genome (Table 1). The highest GC enrichment is in the third position of codons (68.3% vs. 42.7%), and it greatly exceeds that found in any other hemiascomycetes, which is up to 59% in Yarrowia lipolytica and 61% in E. gossypii. Spliceosomal introns and intergenic regions on C-left also have a much higher GC content than similar elements in the rest of the genome (46.8% vs. 36.5% and 46.1% vs. 37.4%, respectively; Table 1).
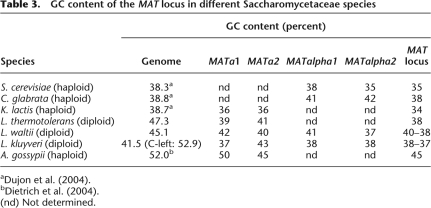
Several small GC-poor wells, however, are dispersed along C-left. Interestingly, one of these encompasses the mating-type locus, located at coordinate ∼350 kb (MAT, Fig. 1), which has a GC percent comparable to that of the rest of the genome, ∼40% vs. 40.4% (Tables 1–3). Note that the GC percent of the MAT loci is lower than the rest of the genome for six of the seven yeast genomes in which it was identified (Table 3). The only exception is Candida glabrata, which has no known sexual cycle.
Table 3.
GC content of the MAT locus in different Saccharomycetaceae species
Another peculiarity of C-left is the excess of microsatellites, i.e., tandem repeats of short DNA sequences, scattered throughout eukaryote and prokaryote genomes (for review, see Richard et al. 2008) and known to present a mutation rate higher than that of unique DNA sequences (for review, see Fan and Chu 2007). We screened for the presence of di-, tri-, and tetranucleotide repeats in the L. kluyveri genome and found a significant enrichment in C-left compared with the rest of the genome (up to twice; Supplemental Table S2). Di- and tetranucleotide repeats are also longer in C-left. Microsatellites are globally poor in GC content (33.1%, 44.2%, and 30.4% GC for di-, tri-, and tetranucleotides, respectively; Supplemental Table S2), but di- and trinucleotide repeats are richer in GC on C-left (45.9% and 54.3%, respectively). Most trinucleotide repeats are located within coding sequences (CDS) in C-left as in the rest of the genome (Supplemental Table S2) and encode preferentially the amino acids Gln, Asp, Ser, Glu, and Ala (data not shown), compatible with the observed specific protein length increase (see below).
High GC content leads to strong biases in codon usage and protein composition
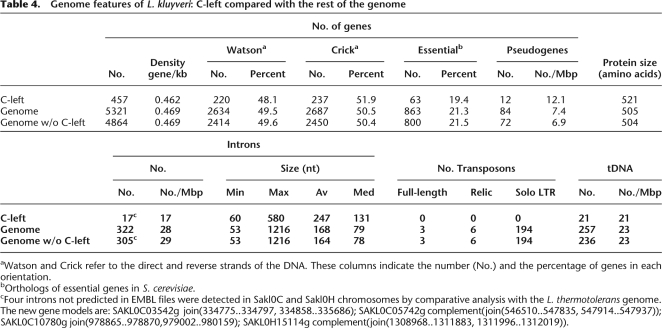
There is no bias in the density of the 457 protein-encoding genes on C-left, nor in their orientation, nor in the number of orthologs of S. cerevisiae essential genes. However, proteins encoded on C-left are, on average, longer than those encoded in the rest of the genome (521 amino acids vs. 504 amino acids; Table 4), and the additional amino acids are biased toward GC-rich codons. This is clearly revealed by comparison between C-left encoding proteins and L. thermotolerans orthologous proteins, which average 22 amino acids shorter (see below the amino acid alignments between L. kluyveri and L. thermotolerans orthologs). This protein length increase is due in part to the overrepresentation of intragenic trinucleotide repeats in C-left and to a 3′ extension of the coding sequences due to A/T toward G/C mutations in the stop codons. Also, C-left genes have slightly fewer introns than other genes (only 17 introns detected in C-left for 28 expected, ddl = 1; χ2 = 5.87 > 3.84; Table 4), and introns in C-left present a structure slightly fewer from that of introns in the rest of the genome (Supplemental Fig. S1).
Table 4.
Genome features of L. kluyveri: C-left compared with the rest of the genome
aWatson and Crick refer to the direct and reverse strands of the DNA. These columns indicate the number (No.) and the percentage of genes in each orientation.
bOrthologs of essential genes in S. cerevisiae.
cFour introns not predicted in EMBL files were detected in Sakl0C and Sakl0H chromosomes by comparative analysis with the L. thermotolerans genome. The new gene models are: SAKL0C03542g join(334775..334797, 334858..335686); SAKL0C05742g complement(join(546510..547835, 547914..547937)); SAKL0C10780g join(978865..978870,979002..980159); SAKL0H15114g complement(join(1308968..1311883, 1311996..1312019)).
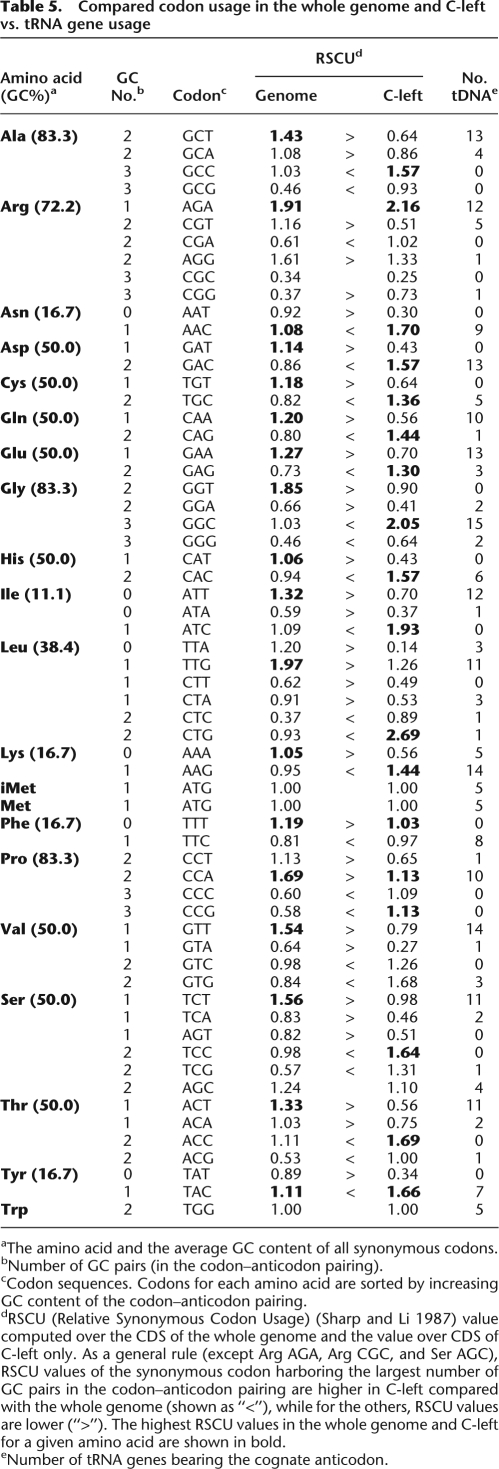
As expected, codon usage is influenced by the GC content (Table 5). The most GC-rich synonymous codons are preferentially used in C-left, the only exceptions being Arg AGA, Arg CGC, Ser AGC, and Phe TTT. For example, of the six leucine codons, those with the fewest GC base pairs (TTA, TTG, CTT, and CTA) are underrepresented while the two most GC-rich leucine codons, CTC and CTG, are overrepresented on C-left relative to the whole genome. Remarkably, in C-left the most GC-rich synonymous codons are preferentially used even if fewer tRNA genes, and therefore fewer molecules, are available to read them: The prevalent Leu codon is the rare CTG (RSCU 2.69; see Table 5, in bold), which is read by the minor tRNA-Leu (CAG) that is encoded by a single gene.
Table 5.
Compared codon usage in the whole genome and C-left vs. tRNA gene usage
aThe amino acid and the average GC content of all synonymous codons.
bNumber of GC pairs (in the codon–anticodon pairing).
cCodon sequences. Codons for each amino acid are sorted by increasing GC content of the codon–anticodon pairing.
dRSCU (Relative Synonymous Codon Usage) (Sharp and Li 1987) value computed over the CDS of the whole genome and the value over CDS of C-left only. As a general rule (except Arg AGA, Arg CGC, and Ser AGC), RSCU values of the synonymous codon harboring the largest number of GC pairs in the codon–anticodon pairing are higher in C-left compared with the whole genome (shown as “<”), while for the others, RSCU values are lower (“>”). The highest RSCU values in the whole genome and C-left for a given amino acid are shown in bold.
eNumber of tRNA genes bearing the cognate anticodon.
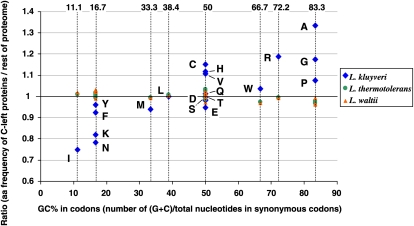
The high GC-content pressure is so strong in C-left that the amino acid composition of proteins is biased toward residues encoded by GC-rich codons. Figure 2 displays ratios of the frequencies of amino acids of C-left proteins versus the rest of the proteome, plotted as a function of the average GC composition of all synonymous codons of each amino acid (blue dots). There is a general tendency in C-left to use residues encoded by GC-rich codons, while residues encoded by GC-poor codons are underrepresented. To exclude the possibility of a compositional bias due to a selective pressure exerted on proteins themselves, we reproduced this plot, replacing the L. kluyveri proteins of C-left with their orthologs from either L. thermotolerans or L. waltii (green and orange dots). In these two cases, no bias toward particular amino acids is apparent (Fig. 2).
Figure 2.
Biased amino acid composition of proteins encoded by C-left. The ratio between the amino acid frequency of proteins encoded by C-left of L. kluyveri and of those encoded by the rest of the genome is plotted along the average nucleotide composition of all synonymous codons for the corresponding amino acid (exact values are on the top of the graph). Amino acids are identified by single-letter code next to blue diamonds. Residues located >1.0 are overrepresented in C-left compared with the rest of the genome; residues <1.0 are underrepresented. Extreme cases are I (isoleucine), which is used 25% less (ratio = 0.75) in C-left than in the rest of the genome, and A (alanine), which is used 33% more (ratio = 1.33). The same analysis was performed for the proteins of L. thermotolerans (green dots) and L. waltii (orange triangles), which are considered orthologous to the C-left proteins of L. kluyveri.
Furthermore, within a set of exchangeable residues, such as the basic amino acids, Arg, and Lys, Arg is more frequently used in C-left than in the whole genome, and the reverse is true for Lys, suggesting that Lys has frequently been exchanged for Arg in C-left-encoded proteins. The same deviation holds for the aliphatic amino acids Val and Ile. Alignments between L. kluyveri C-left proteins and their L. thermotolerans ortholog proteins show that most valine residues are gained in L. kluyveri proteins at the expense of isoleucines (644 amino acids, Supplemental Table S3). Similarly, shifts between lysine residues and arginine involve 383 amino acids (Supplemental Table S3). The amino acid bias in C-left is also due in part to the protein length increase. The majority of the amino acids added in C-left proteins compared with L. thermotolerans proteins are Gly and Ala, which are encoded by 83.3% GC-rich codons, then Thr, Ser, Asp, and Glu, which are encoded by codons that are 50% GC rich; amino acids encoded by AT-rich codons (Ile, Tyr, Phe) are underrepresented among these extra amino acids (Supplemental Table S3).
C-left is totally devoid of transposable elements or their traces
A total of 203 full-length copies or remnants (of which 194 are solo LTRs) of the class I retrotransposon Tsk1 are dispersed throughout the genome (especially near tRNA genes). As no bias in the distribution of tRNA genes is apparent in C-left (Table 4), we would expect to find the same density of Tsk1 elements (i.e., about 19 elements). Instead, C-left is totally devoid of Tsk1 traces (Fig. 1; Table 4).
The phylogeny of Ty1-like elements of Saccharomycetaceae genomes reveals that Tsk1 is very close to Ty1 and Ty2, suggesting that it was acquired through horizontal transfer from a species closely related to the Saccharomyces sensu stricto yeasts (Neuvéglise et al. 2002). Ty1-like transposable elements are also found in L. thermotolerans and L. waltii (The Génolevures Consortium 2009), but their sequences are closer to Tse1 from Saccharomyces exiguus or Tkp1 from Vanderwaltozyma polyspora than to Ty1 or Ty2 from S. cerevisiae. This suggests that the acquisition of Tsk1 occurred after the divergence of L. kluyveri from the L. thermotolerans–L. waltii branch of the phylogeny (Fig. 3A). The Tsk1 element is widespread in L. kluyveri strains as confirmed by successful PCR amplifications with Tsk1 specific primers (data not shown).
Figure 3.
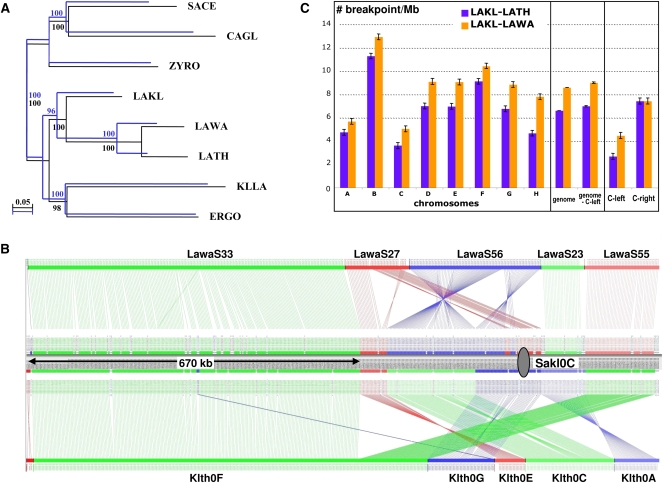
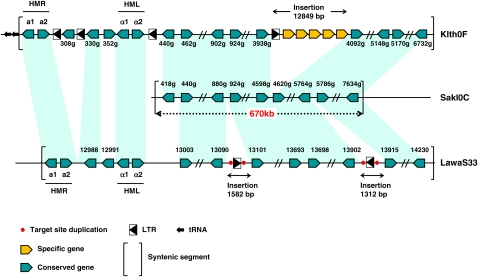
Phylogeny and synteny analyses of L. kluyveri. (A) Two phylogenetic trees were constructed from alignments of universally conserved proteins encoded by C-left (blue; 19 families, 4631 residues) or in the rest of the genome (black; 17 families, 6688 residues) using PhyML, a maximum likelihood method. Species belonging to the Lachancea clade, L. thermotolerans (LATH) and L. waltii (LAWA), were used, as well as more distantly related species such as Kluyveromyces lactis (KLLA), E. gossypii (ERGO), Zygosaccharomyces rouxii (ZYRO), Candida glabrata (CAGL), and S. cerevisiae (SACE). (B) Conservation of synteny was calculated with the AutoGRAPH algorithm (Derrien et al. 2007) from reciprocal best hits between proteins encoded by C-left of L. kluyveri and the proteomes of L. waltii and L. thermotolerans. Each vertical line links orthologs between chromosome C of L. kluyveri and the genomes of L. waltii (top) and L. thermotolerans (bottom). (Gray oval) Centromere of chromosome C. (C) Density of synteny breakpoints per megabase (ordinate), between the genome/chromosomes of L. kluyveri and L. thermotolerans (blue bars) and L. waltii (orange bars). The error bars represent the 99% confidence intervals.
Sequences of the Tsk1 LTR in the sequenced strain range from 71% to 100% sequence identity when aligned, with a majority of sequences diverging from <10% (Supplemental Fig. S2). Solo LTRs are expected to diverge at the neutral rate. The high level of sequence identity (up to 100%) between these sequences suggests that Tsk1 is probably still active.
Phylogeny and synteny conservation associated with a paucity of chromosomal rearrangements support a Lachancea origin of C-left
We tested the possibility that C-left and the rest of the L. kluyveri genome originated from distinct species by comparing phylogenetic trees derived either from proteins encoded by C-left or from proteins encoded by the rest of the genome. Orthologous genes of species belonging to the Lachancea genus (L. thermotolerans and L. waltii), and of more distantly related species such as Kluyveromyces lactis, E. gossypii, Zygosacchromyces rouxii, Candida glabrata, and S. cerevisiae, were used to construct two phylogenetic trees, one from universally conserved proteins encoded by C-left (19 families, 4631 residues), the other one from proteins encoded by genes located in the rest of the genome (17 families, 6688 residues). The resulting tree topologies are identical (Fig. 3A), suggesting that C-left originates from the Lachancea clade.
As the phylogeny of both parts of the L. kluyveri genome is conserved, we investigated synteny conservation among the available genomes of the Lachancea clade. A high level of synteny conservation was found between the two related species, L. thermotolerans and L. waltii (42 conserved regions with an average size of 233 kb, not shown). The two comparisons between the genomes of L. kluyveri/L. thermotolerans and L. kluyveri/L. waltii revealed 91 and 116 regions of conserved synteny, respectively, showing similar size distributions (Supplemental Fig. S3), and an average size of 118 kb for the former comparison and 89 kb for the latter. It is striking that the largest uninterrupted regions of conserved synteny in both comparisons (∼670 kb) correspond to the distal part of C-left (Fig. 3B) when these regions have an unbiased GC composition in L. thermotolerans and L. waltii. Synteny conservation demonstrates that C-left shares a common ancestral origin with the genomes of L. thermotolerans and L. waltii, the most rearranged part of the L. kluyveri genome being the rest of the genome. This observation further rules out the possibility that C-left originated from a non-Lachancea species but does not rule out the possibility that L. kluyveri is a hybrid.
To test whether the apparent lack of genome rearrangements in C-left represents a true deficit compared with other parts of the genome, we measured the global level of genome reorganization, i.e., the density of synteny breakpoints per megabase (bkpt/Mb) for each chromosome. Genome-wide it is 7.3 and 9.5 bkpt/Mb, on average, for the L. kluyveri/L. thermotolerans and the L. kluyveri/L. waltii comparisons, respectively (Fig. 3C). In both comparisons, the most stable chromosomes are chromosomes A and C, but C-left is even more stable (3.0 and 5.0 bkpt/Mb in L. kluyveri/L. thermotolerans and L. kluyveri/L. waltii comparisons, respectively, Fig. 3C).
The silent mating-type cassettes (HML and HMR) are absent from the genome of L. kluyveri (confirmed by Southern blot hybridizations, not shown) but are present in L. thermotolerans and L. waltii (Butler et al. 2004; Fabre et al. 2005; Muller et al. 2007). All three species also lack the HO gene. Interestingly, HML and HMR of L. thermotolerans and L. waltii directly flank the telomere-proximal breakpoint of the 670-kb syntenic region in the genome of L. kluyveri (Fig. 4). A solo LTR lies at this breakpoint in L. thermotolerans. Orthologs of the three L. thermotolerans genes located between HMR and HML are present on different L. kluyveri chromosomes, suggesting successive rearrangements of this subtelomeric region. Since the presence of the two silent cassettes is found in all Saccharomycetaceae species (Butler et al. 2004), the absence of these cassettes in L. kluyveri represents a gene loss specific to the corresponding phylogenetic branch. This gene loss renders L. kluyveri obligatorily heterothallic.
Figure 4.
Breakpoints within the biggest synteny block, between L. kluyveri, L. thermotolerans, and L. waltii. This block corresponds to a 670-kb telomere-proximal region of C-left in L. kluyveri. Reciprocal best hit orthologs are linked with hatched light green parallelepipeds. (Double oblique lines) Synteny is maintained in the regions between two orthologs. Elements at synteny breakpoints such as solo LTR (black triangles) or species-specific genes (orange arrows) are represented. No LTR was found in C-left, whereas some are present in both L. thermotolerans and L. waltii regions, in particular at the breakpoint flanking the MAT locus in L. thermotolerans. While the breakpoint lies at SAKL00418g in L. kluyveri, the synteny blocks between L. thermotolerans and L. waltii extends until the HMR and HML silent cassettes, which are lost in L. kluyveri. Gene names have been abbreviated; for example, 440g on Klth0F corresponds to KLTH0F00440g.
The replication timing of C-left is delayed compared with the rest of the genome
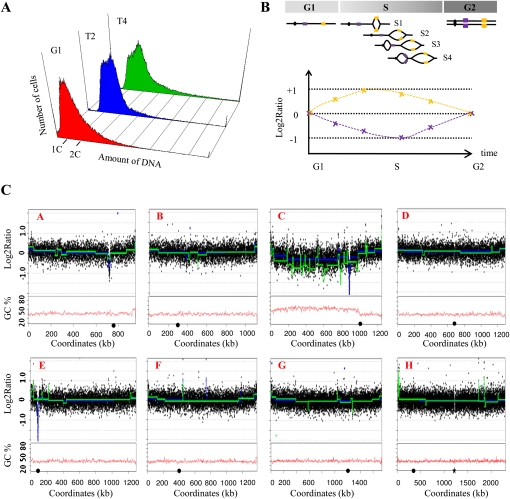
We investigated whether the strong compositional bias of C-left could be attributed to differences in DNA metabolism and, in particular, replication. We used a DNA microarray-based approach (Yabuki et al. 2002) using synchronized cell cultures to compare the timing of replication of C-left with other parts of the genome. Cells were arrested by treatment with α-factor. Flow cytometry of synchronized cells shows that 80% of the cells were arrested in G1 phase, as previously obtained with S. cerevisiae or L. kluyveri α-factor treatments (Hisatomi et al. 1988). Cells harvested after α-factor removal, at T2 and T4, are at the end of the S phase (Fig. 5A). DNA from cells harvested in G1 and either of the two points of the S phase (T2 and T4) were cohybridized to a genome-tiling oligonucleotide microarray, and the log2 of the ratios of the intensities (T2 or T4, divided by G1) were plotted as a function of the chromosomal coordinates. Regions of the genome that replicate early in S phase (yellow, Fig. 5B) present a stronger signal (ratio tends to +1) than the regions that replicate late (violet, Fig. 5B; ratio tends to −1). Seven of the eight chromosomes of L. kluyveri as well as the right arm of chromosome C are essentially fully replicated, exhibiting a relative ratio of zero between G1 and T2 or between G1 and T4 (Fig. 5C). In contrast, C-left presents a very different profile, with a global curve that tends to −1 and 11 sharp peaks whose ratio tends to zero. These peaks are likely to represent active replicons centered on replication origins, indicating that this chromosomal arm is the only part of the genome that is still in an active process of replication. The region showing a replication delay coincides perfectly with the compositional bias (Fig. 5C). Thus, C-left replicates later than the rest of the genome. Few peaks are also visible on chromosomes E, F, and H. Their intensity ratios tend to +1; therefore, they could correspond to the earliest and/or strongest replication origins that fired at the beginning of the following S phase (remember that the cultures contain ∼20% unsynchronized cells). Three very deep wells, two corresponding to centromeres of chromosomes A and E and one on C-left are also visible in T2; the interpretation of these localized decreases is not obvious.
Figure 5.
Replication timing in L. kluyveri. (A) Flow cytometry. Samples of synchronized cells were collected in G1 arrest (red) and in two points (T2 in blue and T4 in green) of the S phase spaced by 5 min. (1C, 2C) DNA content. (B) Schematic representation of the replication of a theoretical chromosome. (Yellow boxes) Early replicated regions, (violet boxes) late replicated regions. In G1, the yellow and violet regions are present in one copy. During the S phase (S1–S4), the copy number of the different cassettes increases to reach two copies in G2. The log2 ratio of the DNA content for the early (yellow) and late (violet) regions are represented according to the time from the G1 to the G2 phases. In G1, the log2 ratio is zero. At the beginning of the S phase, the regions replicated early (yellow box) are present in two copies, while the other regions are just in one copy. Thus, the log2 ratio of the yellow box tends to one, while the log2 ration of the violet box decreases. Later in the S phase, the different parts of the genome replicate until all are present in two copies, and finally in G2 the relative ratio of the intensity tends again to zero. (C) Replication profiles of the chromosomes and their GC content. Microarrays were cohybridized with the DNA from cells arrested in G1 and DNA from cells in T2 or T4 (see Methods). The ratio of the intensities of the two fluorochromes is computed and the log2 ratios are plotted according to the physical position of their corresponding sequences on the different chromosomes (A–H in red, rDNA locus ignored). The curves in blue (T2 vs. G1) and in green (T4 vs. G1) are calculated using a segmentation method for array CGH data analysis (Picard et al. 2007). The peaks underline overrepresented regions corresponding most likely to replication origins. Note that the C-left region is clearly underrepresented, suggesting that the replication of C-left is delayed compared with the rest of the genome, which is almost entirely replicated. (Black circles) Position of the centromeres, (black star) position of the rDNA.
Discussion
GC-content heterogeneities, or “isochores” in vertebrates, have been to date associated with significant changes in genome content. For the simplest case of unicellular eukaryotes, two other cases of compositional heterogeneity have been previously reported. Leptosphaeria maculans contains isochore-like structures that consist of stretches of degenerate LTR retrotransposons with a near absence of protein-coding genes (Gout et al. 2006; Fudal et al. 2007). In the Ostreococcus species, GC-poor chromosomes harbor genes with multiple spliceosomal introns that are smaller than those in the rest of the genome, accumulate transposable elements, and have been subjected to so many rearrangements that synteny was lost (Derelle et al. 2006; Palenik et al. 2007). In contrast, in L. kluyveri, the global gene density, transcriptional orientations, proportion of orthologs to S. cerevisiae essential genes, and proportion of genes in families are comparable between C-left and the rest of the genome. The only differences are modest: the complete absence of any traces of LTR retrotransposons, an overrepresentation of microsatellites, and a moderate increase in CDS length. Thus, the compositional heterogeneity of L. kluyveri is unique so far and provides a unique opportunity to study GC-content heterogeneity without the confounding influence of significant genome content change. Intriguingly, this heterogeneity is not without functional consequence, since we established a significant delay in replication for C-left compared with the rest of the genome. This may provide important clues as to the origin of this heterogeneity.
Yet, two mutually exclusive hypotheses about its origin must be admitted: L. kluyveri could be a hybrid between a GC-rich (∼53%) and a GC-poor (∼40%) ancestor, or the compositional heterogeneity of C-left could result from an intrinsic mechanism that progressively affected its GC content. The latter would have important consequences for understanding isochores in eukaryotes.
In the “hybrid” hypothesis, each ancestor would have transmitted contiguous, though unequal, parts of its genome to the present L. kluyveri genome. These putative ancestors must have differed from each other by at least 12% in their GC composition and therefore by an even higher level of sequence divergence. However, we must rule out that they were phylogenetically distant, since similar phylogenies are obtained for C-left genes and genes in the rest of the genome. Considering the high synteny conservation of C-left with L. thermotolerans and L. waltii, the GC-rich ancestor could belong to the Lachancea clade. Candidate species, close to L. thermotolerans and L. waltii, are L. cidri, L. fermentati, or L. meyersii (Naumova et al. 2007), whose genome sequence is presently not available. As the compositional heterogeneity seems to exist in all other strains of L. kluyveri tested, it follows that the present L. kluyveri genome would have been formed from the hypothetical hybrid ancestor prior to the separation of the various geographical isolates, whose nucleotide divergence reaches up to 5% (i.e., a relatively long separation). The class I transposon Tsk1 is also present in all L. kluyveri strains tested; its complete absence from C-left suggests that its ancestor was devoid of this element while it was brought in by the GC-poor ancestor. This hypothesis, however, only holds if Tsk1 elements have been inactive since the ancestral hybrid, which seems improbable considering the nucleotide conservation of LTR, or if the GC content and/or the chromatin modification of C-left (see next paragraph) are a barrier for Tsk1 transposition. The replication delay of C-left would, under this hybrid hypothesis, be the consequence of an ancestral compositional heterogeneity. Hence, the primary cause of the replication delay could be that replication origins with a high GC content are not properly recognized by the replication machinery of an organism whose genome is predominantly GC-poor.
The alternative hypothesis is that the GC content difference of C-left is being progressively acquired. The loss of the silent cassettes HML and HMR could be the primary cause of this intrinsic mechanism. L. kluyveri is unique in Saccharomycetaceae in having lost HML and HMR. In L. thermotolerans and L. waltii, these cassettes are present in the subtelomeres of the chromosomal arms orthologous to C-left (Fig. 4). In S. cerevisiae, these cassettes are silenced through the deacetylation of histones H3 and H4 (Kayne et al. 1988). Loss of the barriers associated with silent cassettes could have resulted in an unscheduled chromatin modification spreading over C-left and being stopped by the presence of the centromere. The complete absence of any traces of Tsk1 from C-left could be explained by such chromatin modification, as previously reported for Ty1 in S. cerevisiae (Nyswaner et al. 2008).
In S. cerevisiae, the deacetylation of histones associated with silent cassettes induced their delayed replication (Stevenson and Gottschling 1999). It is also well known that replication origins located in subtelomeric regions are activated late during S phase (Ferguson et al. 1991; Ferguson and Fangman 1992). In L. kluyveri, the hypothetical chromatin modification could be the cause of C-left delayed replication and be also indirectly responsible for a higher and biased mutational rate. Indeed, three recent studies indicate an elevated number of SNPs in subtelomeric regions in the Saccharomyces sensu stricto species and in the genome of Drosophila melanogaster (Diaz-Castillo and Golic 2007; Anderson et al. 2008; Teytelman et al. 2008). These elevated SNP densities are consistent with an increased mutation rate in these regions. In addition, it has been recently shown that mutation rate is markedly increased in later replicating regions of the human genome (Stamatoyannopoulos et al. 2009). Furthermore, the error-prone activity of a translesion DNA polymerase, such as Rev1p, is known to be cell cycle-regulated: It is induced in late S phase and reaches its maximum at G2/M (Waters and Walker 2006). The REV1 gene encodes a deoxycytidyl transferase that incorporates dCMPs opposite abasic sites (Haracska et al. 2002). Such catalytic activity could promote a progressive enrichment in G+C bases.
Yet another possible mechanism that tends to fix AT-to-GC mutations is biased gene conversion (Brown and Jiricny 1988), presumably through GC-biased repair of mismatches in heteroduplexed recombination intermediates. Meiotic recombination is the major determinant of the evolution of GC content in primates (Duret and Arndt 2008). The need for at least one crossover per chromosomal arm in meiosis increases the recombination rate for short chromosome arms compared with longer ones. This explanation has been advanced to explain the positive correlation observed between GC content and crossover rate and their negative correlation with chromosome size in the chicken genome (Galtier and Duret 2008), but this is not relevant in L. kluyveri, as C-left is among the longest chromosomal arms in L. kluyveri. Several lines of evidence also suggest that the isochore structures observed in mammals and birds may be a consequence of the process of recombination (for review, see Duret et al. 2006). One particularly clear example of correlation between recombination and GC content is the pseudoautosomal region of the sex chromosomes of mammals, in which the third codon position of genes reaches 85% GC (Montoya-Burgos et al. 2003). At a smaller scale, the MAT locus of Cryptococcus neoformans shows a correlation between high GC content and meiotic recombination hotspots (Hsueh et al. 2006). The same correlation is observed between meiotic recombination hotspots and coldspots in the genome of S. cerevisiae (Gerton et al. 2000). In L. kluyveri, the MAT locus on C-left corresponds to an AT-rich region, suggesting that this locus is a recombination coldspot. The loss of the silent cassettes, and, thus, the necessity for both genes (a1 and a2 or alpha1 and alpha2) in the MAT locus not to be separated in order to preserve the sexual cycle, could have favored the establishment of recombination hotspots in the flanking regions and therefore have participated in a global increase of the GC content in C-left. Given that L. kluyveri diploids sporulate efficiently in laboratory conditions, it would be desirable to measure the recombination rate on C-left and compare it with the rate over the rest of the genome.
In conclusion, L. kluyveri appears as a tractable unicellular organism that provides the unique opportunity to further investigate and to understand the functional and structural properties of a region known as an “isochore” in more complex genomes. Additional experiments are obviously needed to resolve the origin of the intriguing base compositional heterogeneity observed in the genome of L. kluyveri. First, it would be of major interest to determine the chromatin structure in C-left. Second, exploratory genomics of the species of the Lachancea clade will refine the phylogeny of this clade and narrow the range of possible ancestors of L. kluyveri. Finally, sequencing different isolates of L. kluyveri using high-throughput resequencing methods or measuring the forward mutational spectrum using markers inserted in different locations would indicate whether the putative mechanisms of GC-content increase are maintained or if this GC-content heterogeneity is being resolved at the population level.
Methods
Yeast strains and growth conditions
The L. kluyveri strains used in this study are listed in Supplemental Table S1. They were provided by the CBS, Netherlands. Cells were routinely grown in YPD medium (1% yeast extract, 1% peptone, 1% glucose) at 30°C with shaking.
Cells from yeast colonies grown on YPD medium were tested for their sporulation capacity by plating cells on SPO medium (for 1 L: 2.5 g of Bacto yeast extract [Difco], 1 g of glucose, 10 g of potassium acetate). This plate was then incubated for 5 d at 30°C and the presence of tetrads was checked under an Olympus microscope.
For pheromone sensitivity assays, 50 μL of a 200-μg/mL solution of S. cerevisiae α-factor (Genepep) was spotted on YPD plates and, when dry, covered with a cell lawn containing 5 × 104 cells. Pictures were taken after 2 d of growth at 30°C.
Synchronization of the cell cycle
LAKL001 is a haploid strain obtained by random sporulation from strain FM479 (=CBS 3082, type strain of L. kluyveri). LAKL001 cultures were grown in YPD at 30°C. To synchronize cells in G1, α-factor of S. cerevisiae was added to exponential growth cultures at a final concentration of 16 μg/mL for 3 h at 23°C. The treated cells were harvested by centrifugation and washed twice with cold water, and then resuspended in YPD. The cell culture was incubated at 23°C and samples were collected for flow cytometry and isolation of genomic DNA.
Flow cytometry
Yeast cells were fixed overnight with a 70% (v/v) ethanol solution, then harvested and resuspended in 50 mM sodium citrate pH 7 and treated with 1 μg/mL RNase A (Sigma-Aldrich) for 1 h at 37°C and overnight at 4°C. Flow cytometry was performed after staining the cells with 30 μg/mL propidium iodide (Sigma-Aldrich) and analyzed with a FACScan and by CellQuest software (Becton-Dickinson).
Yeast genomic DNA preparation, labeling, and microarray experiments
Genomic DNA was isolated from cells using the genomic-tip 100/G isolation kit (Qiagen) and labeled with either Cy3 or Cy5 fluorescent dyes (GE Healthcare) using the BioPrime DNA labeling system kit (Invitrogen) according to the manufacturer's recommendations. Four micrograms of genomic DNA in 20 μL of sterile water was first heated for 10 min at 95°C. After addition of 20 μL of 2.5× random primers solution, the samples were heated again for 5 min at 95°C and chilled on ice. We then added the indicated compounds at the following final concentrations: 0.12 mM dATP, dGTP, and dTTP; 0.06 mM dCTP; 0.02 mM Cy3- or Cy5-dCTP (Amersham Biosciences); 1 mM Tris-HCl (pH 8.0); 0.1 mM EDTA; and 40 units of Klenow fragment (Invitrogen). The reaction mixtures were incubated for 2 h at 37°C. The reactions were then stopped by adding 0.5 M EDTA (pH 8). For each sample, the fluorescently labeled DNA was purified using the purification module (Invitrogen) and dissolved in 50 μL of sterile water. Hybridization and microarray washing were performed in accordance with the manufacturer's instructions (Agilent). Each experiment was run as a competitive hybridization by using Cy3-labeled DNA from G1 arrested cells and Cy5-labeled DNA from T2 or T4. Arrays scans were performed with a GenePix 4000A dual-channel (635 nm and 532 nm) laser scanner (genePix) with a resolution of 5 nm per pixel. The laser power was set at 100%, and the photomultiplier tension was adjusted between 680 and 800 V according to the average intensity of the hybridization of each slide in order to optimize the dynamic range of measurements. All the slides were analyzed using R (R Development Core Team; http://www.R-project.org) and Bioconductor (http://www.bioconductor.org) with the snapCGH package (M Smith, J Marioni, NP Thorne, Simon Tavare Hutchison/MRC Research Center, Department of Oncology, University of Cambridge, England). The ratio of the two fluorescent dyes was log2 transformed and normalized using intensity-dependent normalization (Picard et al. 2007).
Design of the microarrays
Oligos for the FM479 sequence were designed with the program OligoArray 2.1 (Rouillard et al. 2003). The oligonucleotide length range was set to 35–60 nt, the melting temperature (Tm) range to 86°C–99°C, and the GC content range to 40%–85%. These arrays were produced on Agilent 60 mer oligonucleotide high-density arrays 2 × 105 K. A total of 52,468 features designed on both strands were spotted on the microarrays; this represents one probe every 216 bp on average.
DNA extraction, PCR, and sequencing
To check the GC content of various L. kluyveri strains, total genomic DNA was prepared with the genomic TIP20 isolation kit (Qiagen). PCR amplification was performed with an Eppendorf thermocycler using Taq DNA polymerase in the recommended buffer and ∼100 ng of genomic DNA as a template. The following conditions were used: 30 cycles of denaturation for 10 sec at 95°C, annealing for 30 sec at 58°C, and elongation for 2 min at 72°C, followed by a final elongation for 10 min at 72°C. The sequencing reactions were performed in Abgene AB-1100 plates using ABI BigDye reagents and analyzed in an ABI 3700 sequencer. Additional sequences were determined by Genome Express and GATC Biotech. The GC content of each sequence was calculated using geecee software (http://emboss.sourceforge.net/). PCR amplifications with primers internal to Tsk1 were also performed to detect the presence of this element in the genome of the different L. kluyveri strains. The primers (Operon) used for all PCR amplifications are listed in Supplemental Table S4.
Genomic sequences
The genome of the L. kluyveri type strain (CBS 3082; NRRL Y-12651) was fully sequenced and assembled at the Washington University School of Medicine. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the project accession AACE03000000. The sequenced strain CBS 3082 is diploid, but only the MATa information was assembled and annotated; the MATα locus is positioned in a small additional contig (SKLU-Cont10304—accession AACE03000032). The full annotation has been realized by The Génolevures Consortium (http://www.genolevures.org/sakl.html).
In silico analyses
All DNA sequences between genetic features (protein-coding genes, transposable elements, tRNA genes, ncRNA, etc.) were considered as intergenic, which includes the 5′ UTR and 3′ UTR. Gaps (N nucleotides) were skipped during the GC-content computations. GC-content plots (Fig. 1) were computed with a window of 5 kb and a step of 2 kb. RSCU (Relative Synonymous Codon Usage) values were computed according to Sharp and Li (1987).
Genes were regarded as putative orthologs in pairwise comparisons if their products were reciprocal best hits (Rivera et al. 1998) with at least 40% sequence similarity and if their sequences were <30% different in length (Supplemental Table S5). Pairwise comparisons were computed for L. kluyveri/L. thermotolerans and L. kluyveri/L. waltii, and the map of conserved synteny was generated with the AutoGRAPH algorithm (Derrien et al. 2007)
Alignments of amino acid sequences from conserved gene families with only one member per species were performed using the MUSCLE algorithm (Edgar 2004) and further cleaned with Gblocks (Castresana 2000) before concatenation. Phylogenetic trees were constructed by maximum likelihood using PhyML (Guindon and Gascuel 2003) with a JTT substitution model corrected for heterogeneity among sites by a Γ-law distribution using four different categories of evolution rates. The proportion of invariable sites and the α-parameter of the Γ-law distribution were optimized according to the data. Trees were visualized with either TreeView (Page 1996) or NJ-Plot (Perriere and Gouy 1996).
Microsatellites were detected using the algorithm of Benson and Waterman (1994), Tandem Repeat Finder. The following parameters were used: match weight, +1; mismatch weight, −2, −3, and −4 (for di-, tri-, and tetranucleotide repeats, respectively); insertion/deletion weight −9; threshold to report, 10, 15, 20 (for di-, tri-, and tetranucleotide repeats, respectively); pattern size and lookcount, 2, 3, 4 (for di-, tri-, and tetranucleotide repeats, respectively); no short period, 1. This allowed us to detect microsatellites containing at least five (for tri- and tetranucleotide repeats) or six (for dinucleotide repeats) perfect repeat units (without mismatch) with one mismatch tolerated in each additional repeat unit. χ2 and Fisher's exact tests were used for numeration data.
Acknowledgments
This work was made possible thanks to privileged access to unpublished annotation data provided by The Génolevures Consortium coordinated by Jean-Luc Souciet. We thank Lionel Frangeul for his help in sequence analysis of the different L. kluyveri strains and the “Plateforme de Cytométrie” of the Institut Pasteur for the flow cytometry experiments. We also thank Guy-Franck Richard for his valuable help in the analysis of the microsatellites. We thank Claude Gaillardin and our colleagues from the Génolevures network and the Unité de Génétique Moléculaire des Levures for fruitful discussions. This work was supported by the GDR CNRS 2354 “Génolevures-3” and the ANR “Genarise” (ANR-05-BLAN-0331). C. Payen is a recipient of a predoctoral fellowship of the Fondation pour la Recherche Médicale (FRM) through the University Pierre and Marie Curie - Paris 6. B.D. is a member of the Institut Universitaire de France.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under the project accession no. AACE03000000.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.090605.108.
References
- Anderson JA, Song YS, Langley CH. Molecular population genetics of Drosophila subtelomeric DNA. Genetics. 2008;178:477–487. doi: 10.1534/genetics.107.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G, Waterman MS. A method for fast database search for all k-nucleotide repeats. Nucleic Acids Res. 1994;22:4828–4836. doi: 10.1093/nar/22.22.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G. The neoselectionist theory of genome evolution. Proc Natl Acad Sci. 2007;104:8385–8390. doi: 10.1073/pnas.0701652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TC, Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988;54:705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc Natl Acad Sci. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cliften PF, Hillier LW, Fulton L, Graves T, Miner T, Gish WR, Waterston RH, Johnston M. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. [DOI] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Andre C, Galibert F, Hitte C. AutoGRAPH: An interactive web server for automating and visualizing comparative genome maps. Bioinformatics. 2007;23:498–499. doi: 10.1093/bioinformatics/btl618. [DOI] [PubMed] [Google Scholar]
- Diaz-Castillo C, Golic KG. Evolution of gene sequence in response to chromosomal location. Genetics. 2007;177:359–374. doi: 10.1534/genetics.107.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Eyre-Walker A, Galtier N. A new perspective on isochore evolution. Gene. 2006;385:71–74. doi: 10.1016/j.gene.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Muller H, Therizols P, Lafontaine I, Dujon B, Fairhead C. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol Biol Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- Fan H, Chu JY. A brief review of short tandem repeat mutation. Genomics Proteomics Bioinformatics. 2007;5:7–14. doi: 10.1016/S1672-0229(07)60009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BM, Fangman WL. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Brewer BJ, Reynolds AE, Fangman WL. A yeast origin of replication is activated late in S phase. Cell. 1991;65:507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- Fudal I, Ross S, Gout L, Blaise F, Kuhn ML, Eckert MR, Cattolico L, Bernard-Samain S, Balesdent MH, Rouxel T. Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: Map-based cloning of AvrLm6. Mol Plant Microbe Interact. 2007;20:459–470. doi: 10.1094/MPMI-20-4-0459. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Encyclopedia of life sciences (ELS) Wiley; Hoboken, NJ: 2008. Biased gene conversion and its impact on human genome evolution. [Google Scholar]
- The Génolevures Consortium. Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 2009 doi: 10.1101/gr.091546.109. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Inaugural article: Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojkovic Z, Rislund L, Andersen B, Sandrini MP, Cook PF, Schnackerz KD, Piskur J. Dihydropyrimidine amidohydrolases and dihydroorotases share the same origin and several enzymatic properties. Nucleic Acids Res. 2003;31:1683–1692. doi: 10.1093/nar/gkg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout L, Fudal I, Kuhn ML, Blaise F, Eckert M, Cattolico L, Balesdent MH, Rouxel T. Lost in the middle of nowhere: The AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol. 2006;60:67–80. doi: 10.1111/j.1365-2958.2006.05076.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- Hisatomi T, Yanagishima N, Sakurai A, Kobayashi H. Interspecific actions of alpha mating pheromones on the a mating-type cells of three Saccharomyces yeasts. Curr Genet. 1988;13:25–27. doi: 10.1007/BF00365752. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Idnurm A, Heitman J. Recombination hotspots flank the Cryptococcus mating-type locus: Implications for the evolution of a fungal sex chromosome. PLoS Genet. 2006;2:e184. doi: 10.1371/journal.pgen.0020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEM Yeast Res. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Macaya G, Thiery JP, Bernardi G. An approach to the organization of eukaryotic genomes at a macromolecular level. J Mol Biol. 1976;108:237–254. doi: 10.1016/s0022-2836(76)80105-2. [DOI] [PubMed] [Google Scholar]
- Møller K, Sharif MZ, Olsson L. Production of fungal alpha-amylase by Saccharomyces kluyveri in glucose-limited cultivations. J Biotechnol. 2004;111:311–318. doi: 10.1016/j.jbiotec.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Montoya-Burgos JI, Boursot P, Galtier N. Recombination explains isochores in mammalian genomes. Trends Genet. 2003;19:128–130. doi: 10.1016/S0168-9525(03)00021-0. [DOI] [PubMed] [Google Scholar]
- Muller H, Hennequin C, Dujon B, Fairhead C. In sex and fungi: Molecular determination and evolutionary implication. In: Heitman J, et al., editors. Comparing MAT in the genomes of hemiascomycetous yeasts. ASM Press; Washington, D.C: 2007. pp. 247–263. [Google Scholar]
- Naumova ES, Serpova EV, Naumov GI. Molecular systematics of Lachancea yeasts. Biochemistry. 2007;72:1356–1362. doi: 10.1134/s0006297907120097. [DOI] [PubMed] [Google Scholar]
- Neuvéglise C, Bon E, Lepingle A, Wincker P, Artiguenave F, Gaillardin C, Casaregola S. Genomic exploration of the hemiascomycetous yeasts: 9. Saccharomyces kluyveri. FEBS Lett. 2000;487:56–60. doi: 10.1016/s0014-5793(00)02280-8. [DOI] [PubMed] [Google Scholar]
- Neuvéglise C, Feldmann H, Bon E, Gaillardin C, Casaregola S. Genomic evolution of the long terminal repeat retrotransposons in Hemiascomycetous yeasts. Genome Res. 2002;12:930–943. doi: 10.1101/gr.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 2008;178:197–214. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-query: An on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Phaff HJ, Miller MW, Shifrine M. The taxonomy of yeasts isolated from Drosophila in the Yosemite region of California. Antonie Van Leeuwenhoek. 1956;22:145–161. doi: 10.1007/BF02538322. [DOI] [PubMed] [Google Scholar]
- Picard F, Robin S, Lebarbier E, Daudin JJ. A segmentation/clustering model for the analysis of array CGH data. Biometrics. 2007;63:758–766. doi: 10.1111/j.1541-0420.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: Design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Eisen MB, Rine J. Silent but not static: Accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 2008;4:e1000247. doi: 10.1371/journal.pgen.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G2/M phase rather than S phase. Proc Natl Acad Sci. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock KG, Strathern JN. Molecular genetics in Saccharomyces kluyveri: The HIS3 homolog and its use as a selectable marker gene in S. kluyveri and Saccharomyces cerevisiae. Yeast. 1993;9:351–361. doi: 10.1002/yea.320090405. [DOI] [PubMed] [Google Scholar]
- Wu Q, James SA, Roberts IN, Moulton V, Huber KT. Exploring contradictory phylogenetic relationships in yeasts. FEM Yeast Res. 2008;8:641–650. doi: 10.1111/j.1567-1364.2008.00362.x. [DOI] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]