Abstract
The origin of new genes is extremely important to evolutionary innovation. Most new genes arise from existing genes through duplication or recombination. The origin of new genes from noncoding DNA is extremely rare, and very few eukaryotic examples are known. We present evidence for the de novo origin of at least three human protein-coding genes since the divergence with chimp. Each of these genes has no protein-coding homologs in any other genome, but is supported by evidence from expression and, importantly, proteomics data. The absence of these genes in chimp and macaque cannot be explained by sequencing gaps or annotation error. High-quality sequence data indicate that these loci are noncoding DNA in other primates. Furthermore, chimp, gorilla, gibbon, and macaque share the same disabling sequence difference, supporting the inference that the ancestral sequence was noncoding over the alternative possibility of parallel gene inactivation in multiple primate lineages. The genes are not well characterized, but interestingly, one of them was first identified as an up-regulated gene in chronic lymphocytic leukemia. This is the first evidence for entirely novel human-specific protein-coding genes originating from ancestrally noncoding sequences. We estimate that 0.075% of human genes may have originated through this mechanism leading to a total expectation of 18 such cases in a genome of 24,000 protein-coding genes.
New genes are a rich substrate for evolution to act upon. New genes frequently arise through duplication of existing genes, or through fusion, fission, or exon shuffling between genes (Long et al. 2003). Origination of genes from noncoding DNA is extremely rare: A few eukaryotic examples are known in yeast and Drosophila (Levine et al. 2006; Begun et al. 2007; Cai et al. 2008; Zhou et al. 2008) and a very recent paper reported initial evidence for this process in a primate ancestor (Toll-Riera et al. 2009). No cases have been previously reported in human.
Analysis of the differential presence and absence of genes in different genomes is hampered by incomplete genome sequence and annotation artifacts (Clamp et al. 2007). We undertook a rigorous and systematic analysis of the human genome to identify protein-coding genes with no counterpart in the chimp and macaque genomes. Essential to this analysis is an extremely strict and conservative set of criteria to exclude artifacts due to annotation errors or sequencing gaps. The central pillar of this analysis is a synteny framework to examine candidate novel genes. The synteny approach allowed us to pinpoint the expected location of the gene in other primate genomes and meticulously examine that region for evidence of protein-coding capacity. After careful exclusion of all cases where there might be an ortholog in another genome or where the annotated human gene is unreliable, we identified three novel human protein-coding genes that have originated from noncoding DNA since the divergence with chimp.
Results and Discussion
Identification of human genes with no protein-coding match in protein database or syntenic chimp genomic region
We built blocks of conserved synteny between human and chimp using unambiguous 1:1 orthologs identified as reciprocal best BLASTP hits with no other similarly strong hits. The synteny blocks we produced span 91% and 85% of the human and chimp genomes, respectively, and 21,195 (94%) of the 22,568 human protein-coding genes annotated by Ensembl are located within these blocks. Because we only used 1:1 orthologous regions, lineage-specific segmental duplications are excluded from this analysis.
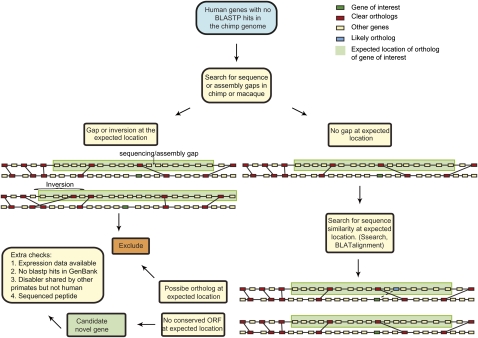
We exploited the extremely high gene order conservation between human and chimp to infer the expected location in chimp of all candidate novel genes and to scrutinize that region of genome for any evidence of the capacity to produce an orthologous protein. We defined the expected location of a chimp ortholog of a human gene to be within 10 genes on either side of the location of the human gene where the location was projected from the human genome to chimp along the most closely located 1:1 orthologs (Fig. 1).
Figure 1.
Schematic of analysis pipeline. The expected location of genes with no BLASTP hit was scrutinized for any evidence of a homologous protein-coding gene. The expected location of a gene is indicated by green shading and was defined as a 10 gene window on either side of the gene of interest projected onto the syntenic location in the other genome. Candidate genes were excluded if there was a sequencing gap in the expected location (or local inversions that rendered the expected location ambiguous) or similar sequence at the expected location with no clear exclusion from producing a protein.
We initially identified 644 human proteins with no BLASTP hit in chimp. For 425 of these there was a sequence or assembly gap, of at least the size of the human gene, within the expected location of the ortholog in the chimp genome. These cases were excluded from further analysis because we cannot exclude the trivial explanation that they are absent from the chimp genome simply because they have yet to be sequenced. For the remaining cases we used BLAT and Ssearch to examine the expected location of the gene for nucleotide similarity indicative of an undetected but valid ortholog. For 150 cases we found a similar annotated protein that had been missed in the initial BLASTP due to low sequence complexity or that the open reading frame (ORF) was present intact in chimp or macaque with no clear exclusion from producing a protein, though it is not annotated as a gene, so we infer that the ortholog is likely to be present. We also excluded human genes with an annotated and plausible ortholog in any other species (see Methods).
To minimize the chance that the gene of interest is itself an annotation artifact, we only considered human genes that are classified as “known” by Ensembl (i.e., they are also annotated in databases other than Ensembl) and that have expressed sequence tag (EST) support for transcription.
Finally, we searched the syntenic region in chimp and macaque to identify the orthologous DNA. All of these stringent filtering steps left three human protein-coding genes (CLLU1, C22orf45, and DNAH10OS) which have no apparent ortholog in any other species' genome, but where there is sequence similarity at the nucleotide level at the expected location of the gene in chimp and macaque (Table 1). Although the chimp and macaque sequence from the syntenic location is highly similar, there is no potential ORF from the same start codon or in the same reading frame aligning to at least half of the human protein. Furthermore, a BLASTP similarity search against all of GenBank confirms the absence of annotated paralogs or orthologs of these genes in any sequenced genome. We hypothesize that these genes have originated de novo in the human lineage, since the divergence with chimp from ancestrally noncoding sequence.
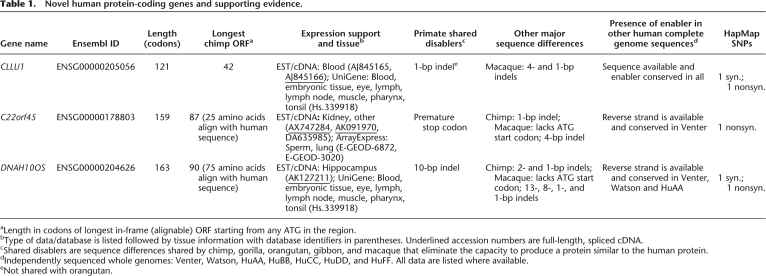
Table 1.
Novel human protein-coding genes and supporting evidence.
aLength in codons of longest in-frame (alignable) ORF starting from any ATG in the region.
bType of data/database is listed followed by tissue information with database identifiers in parentheses. Underlined accession numbers are full-length, spliced cDNA.
cShared disablers are sequence differences shared by chimp, gorilla, orangutan, gibbon, and macaque that eliminate the capacity to produce a protein similar to the human protein.
dIndependently sequenced whole genomes: Venter, Watson, HuAA, HuBB, HuCC, HuDD, and HuFF. All data are listed where available.
eNot shared with orangutan.
Sequence characteristics and expression evidence
Each of these three genes is coded for by an ORF uninterrupted by introns, though they do contain introns in the untranslated regions (UTRs). All of the predicted proteins are short with lengths ranging from 121 to 163 amino acids. Both the short length and the lack of introns within the coding sequence are expected properties of newly arisen genes because of the improbability of the evolution of a long ORF and the complexity of intron splicing signals. UTR introns are likely to be more easily acquired than coding region introns due to lower constraints (Hong et al. 2006). Little is known about these proteins and none of them has any complex protein domains annotated.
The expression of each of these genes is supported by several lines of evidence, including at least one complete, spliced cDNA sequence (Table 1). There are many examples in the literature of new genes with functionality in brain and testis (Burki and Kaessmann 2004; Emerson et al. 2004; Begun et al. 2007; Potrzebowski et al. 2008; Rosso et al. 2008; Zhou et al. 2008). One of the novel genes is expressed in male reproductive tissue and one was identified in brain tissues, but they were also identified in many other tissues (Table 1) and there is no statistical trend.
Human-specific mutations alter protein-coding capacity
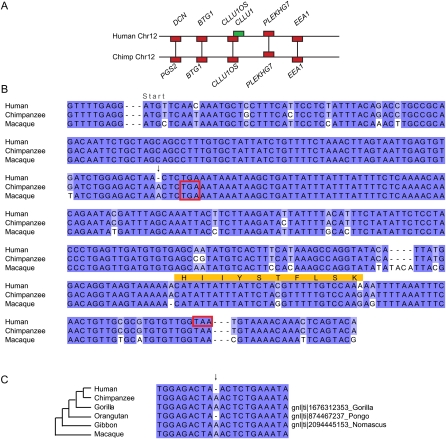
To further investigate the hypothesis of de novo origins in the human lineage we examined the nature of the nucleotide sequence differences between human, chimp, and macaque in the homologous regions of genome corresponding to the location of the gene. In particular, we focused on “disablers”—sequence differences that cause the inferred protein to be truncated or not translated at all. We examined the corresponding chimp and macaque sequences for the presence or absence of an ATG start codon, frameshift-inducing indels that result in an early stop codon, or nucleotide differences which result in an early stop codon (Figs. 2–4). In most cases there are multiple disabling sequence differences in both chimp and macaque. Several of these disablers are indels in chimp or macaque that result in a drastically different hypothetical protein sequence, as well as early termination, which alone do not prove that the ancestral sequence is noncoding because we cannot orient the changes (the available data are uninformative of the ancestral sequence), but which lend credence to the inference that the sequences are noncoding. Critically, for all three of the human genes we found that the chimp and macaque sequences shared one disabler and that the critical sequence difference is supported by high-quality sequence traces in all three genomes (Figs. 2–4; Table 1). To further confirm this we resequenced the DNA in the three orthologous regions in one chimp individual and verified the critical, shared sequence differences (GenBank accession numbers FJ713693, FJ713696, FJ713697). We also searched the NCBI trace databases of all other primates for sequence matches to the gene spanning the shared disabler. Each of the disablers shared by chimp and macaque is also shared with gorilla (Gorilla gorilla) and gibbon (Nomascus leucogenys), and two are also shared with orangutan (Pongo pygmaeus albelii; one was not shared) (Table 1). In all cases there is high sequence quality (Supplemental Fig. S1). Shared sequence differences between chimp, macaque, and other primates are likely to be ancestral rather than independent parallel mutations, and so, we infer that the ancestral sequence was noncoding.
Figure 2.
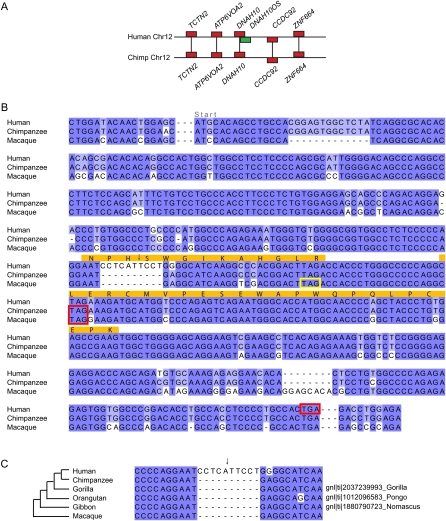
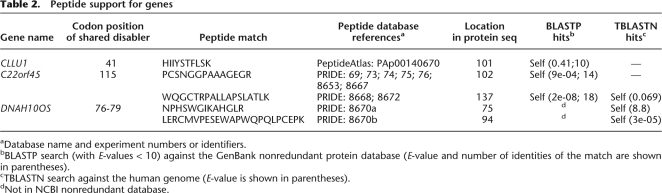
Sequence changes in the origin of CLLU1 from noncoding DNA. (A) Region of conserved synteny between human and chimp chromosomes 12. Genes are indicated by rectangular boxes and the region of chromosome is indicated by a horizontal line. Unambiguous 1:1 orthologs that were used to infer the synteny block are shown in red. One gene in this region, chronic lymphocytic leukemia up-regulated gene 1 (CLLU1), had no BLASTP hits in any other genome and is shown in green. (B) Multiple sequence alignment of the gene sequence of the human gene CLLU1 and similar nucleotide sequences from the syntenic location in chimp and macaque. The start codon is located immediately following the first alignment gap, which was inserted for clarity. Stop codons are indicated by red boxes. The sequenced peptide identified from this locus is indicated in orange. The critical mutation that allows the production of a protein is the deletion of an A nucleotide, which is present in both chimp and macaque (indicated by an arrow). This causes a frameshift in human that results in a much longer ORF capable of producing a 121-amino acids-long protein. Both the chimp and macaque sequences have a stop codon after only 42 potential codons. (C) Alignment of the region around the critical human enabler-mutation with similar nucleotide sequences from the syntenic regions in chimp, and macaque and sequence traces from gorilla, gibbon, and orangutan. For gorilla, gibbon, and orangutan the trace database accession number is shown on the right. The disabler is also shared by gorilla and gibbon indicating it is ancestral.
Figure 4.
Sequence changes in the origin of DNAH10OS from noncoding DNA. As in Figures 2 and 3: (A) region of conserved synteny between human and chimp chromosomes 12. One gene in this region, DNAH10OS, had no BLASTP hits in any other genome and is shown in green. (B) Multiple sequence alignment of the gene sequence of DNAH10OS and similar nucleotide sequences from the syntenic location in chimp and macaque. If the ORF began at the same position as the human start codon (note the start codon is present in chimp but absent in macaque), the macaque hypothetical protein sequence would be very different from the human protein due to frameshifts and would terminate at the stop codon indicated in yellow. The arrow indicates the location of a 10-bp indel shared by chimp and macaque that would result in premature termination irrespective of the other disablements. (C) The disabler is also shared by gorilla, orangutan, and gibbon indicating that this is a human-specific 10-bp insertion.
Human–chimp sequence divergence
We measured the sequence divergence of these human ORFs compared to chimp to search for clues as to the presence and nature of constraints acting on their evolution. We examined the alignment of the human and chimp nucleotide sequences (Figs. 2–4) and identified a total of 12 nucleotide substitutions between human and chimp (pooled over all three genes). Using macaque to orient the changes, we observed that seven of the substitutions occurred in the chimp lineage and five in the human lineage. Of the human-specific substitutions, three are synonymous changes and two are nonsynonymous. The chimp DNA is noncoding so it is not strictly possible to consider synonymous or nonsynonymous changes; however, we can say what the effect of that mutation would be in human (i.e., in an intact ORF). In this way we can infer that of the seven chimp substitutions, four are synonymous-like and three are non-synonymous-like. The amount of sequence divergence between human and chimp in these regions is low (just under 1%), which is not surprising given the close relationship of the two species. The number of substitutions (and of non-synonymous-like substitutions) is higher in the chimp lineage, which is consistent with the hypothesis that these regions are noncoding DNA in chimp. However, there is no statistical power to measure the significance of this observation.
Support from peptide databases
Even though each of these genes has good expression evidence, we sought further support for the veracity of these protein-coding genes because of the possibility that they are noncoding RNA or the possibility of contamination of transcription databases with genomic sequence and expressed pseudogenes (The ENCODE Project Consortium 2007). Many proteomics studies extract proteins from healthy cells, tissues, or fluids and survey the complement of proteins by sequencing short peptides through various methods (Roe and Griffin 2006). These data are thus a direct verification of the presence of a translated gene product. We searched the PRIDE (Martens et al. 2005) and PeptideAtlas (Deutsch et al. 2005) databases of short sequenced peptides with the gene names and found that all of the three genes have peptide matches indicating true protein-coding activity (Table 2). In all cases the peptides were sequenced from blood plasma samples. Each of these peptide matches is unique to these genes in that they do not display significant similarity to any other proteins in all of GenBank or to any hypothetical translation of the human genome, other than themselves, even with a very loose E-value threshold (Table 2). C22orf45 and DNAH10OS have two sequenced peptides each, and in the case of C22orf45, peptides uniquely matching this protein were identified in nine different experiments (Table 2). The peptide matching CLLU1 was detected a total of 903 times in three different samples (PeptideAtlas accession number PAp00140670).
Table 2.
Peptide support for genes
aDatabase name and experiment numbers or identifiers.
bBLASTP search (with E-values < 10) against the GenBank nonredundant protein database (E-value and number of identities of the match are shown in parentheses).
cTBLASTN search against the human genome (E-value is shown in parentheses).
dNot in NCBI nonredundant database.
Importantly, not only do these peptide sequences confirm the presence of a protein product, they also confirm that the human protein coding sequence extends beyond the critical shared disablers of other primates, i.e., that the unique coding sequence granted by the enabling human-specific sequence differences is actually translated (Figs. 2–4; Table 2). In particular, the human-specific enablers are spanned by the sequenced peptides in C22orf45 and DNAH10OS (Figs. 3, 4).
Figure 3.
Sequence changes in the origin of C22orf45 from noncoding DNA. As in Figure 2: (A) Region of conserved synteny between human and chimp chromosomes 22. One gene in this region, C22orf45, had no BLASTP hits in any other genome and is shown in green. (B) Multiple sequence alignment of the gene sequence of C22orf45 and similar nucleotide sequences from the syntenic location in chimp and macaque. The arrow indicates the location of an in-frame stop codon shared by chimp and macaque that would result in premature termination (red box) irrespective of the other disablements. The codons highlighted with a yellow box indicate the stop codon including all disablements (indels) in chimp and macaque for the reading frame starting from the same location as the human start (note the ATG start codon is absent in macaque and that the frameshifts mean the hypothetical protein sequence is drastically altered). (C) The disabler is also shared by gorilla, orangutan, and gibbon indicating it is ancestral.
Nonsense mediated decay (NMD) requires one round of translation in order to recognize any premature termination codons (Stalder and Muhlemann 2008). Therefore, even “nonsense” genes will produce at least one protein. However, the chances of sequencing this aberrant protein are slight. The identification of each of these peptides in multiple experiments demonstrates that the proteins have been translated and produced in sufficient abundance to be detectable in the protein sequencing survey, and so we infer they are present at ample levels to have an impact on the cell.
Human population polymorphism
In addition to the standard human genome sequence, several individual genomes have been completely sequenced (Levy et al. 2007; Wang et al. 2008; Wheeler et al. 2008). We examined each of the human genome sequences available through Ensembl for the presence of the critical enabling sequence difference that we had identified in the standard genome sequence (Table 1). There was no polymorphism at this site in any of the available data for any of the genes.
We also checked HapMap for single nucleotide polymorphisms (SNPs) within each of these ORFs (Table 1). A total of five SNPs were identified in the coding regions of these genes, three of them nonsynonymous. With such small numbers of SNPs there is no statistical power to test alternative evolutionary models, such as the action of selection or constraint.
Evidence of a selective sweep around these loci would further support the functionality of these genes in human if it were found. However, we did not detect any such evidence. We queried the analysis of the HapMap phase II data available through Happlotter (Voight et al. 2006) and found that none of these loci was detected by genome-wide screening of HapMap data for evidence of recent positive selection. Williamson et al. (2007) conducted a genome-wide search for evidence of complete selective sweeps and listed the top 101 regions with the strongest evidence for a recent selective sweep. None of the genes discussed in this letter falls within 100 kb of any of the proposed centers of these sweeps (the reporting threshold adopted by the authors), but CLLU1 is about 250 kb away from a sweep detected in the Chinese samples. Neither of the other genes was within 10 Mb of any of the detected sweeps.
De novo origins of at least three human protein-coding genes from ancestrally noncoding DNA
The genes coding for the three proteins, CLLU1, C22orf45, and DNAH10OS, are novel human-specific genes supported by several lines of evidence: Their expression has been verified by high quality data; their translation has been confirmed by the sequencing of short peptides unique to these proteins; the longest alignable chimp ORF is less than 50% of the length of the human ORF; the absence of coding capacity in the ancestral sequence is confirmed by the sharing of a disabler between chimp, gorilla, gibbon, and macaque; and multiple additional disabling sequence differences are present in macaque. There are also no known disabling human polymorphisms at these loci.
These novel genes are not well characterized and only chronic lymphocytic leukemia up-regulated gene 1 (CLLU1) has been discussed in the literature. It was originally recognized as a highly expressed gene in chronic lymphocytic leukemia (CLL) (Buhl et al. 2006). The authors also noted that the coding capacity is not conserved in mouse and chimp (Buhl et al. 2006). The role of this gene in chronic lymphocytic leukemia is not clear, but it has been suggested as a therapeutic target (Buhl et al. 2006). The sequenced peptide from this locus was identified in plasma pooled from dozens of healthy individuals (according to the database notes). Our analysis has shown that the CDS is disabled in chimp, gorilla, gibbon, and macaque and is enabled in human by a 1-base-pair (bp) deletion, which shifts the reading frame and extends the potential protein with respect to the ancestral state (Fig. 2). Surprisingly, orangutan also shares this 1-bp deletion due to a probable parallel mutation. If the ancestral primate sequence was coding, then we would need to infer that an identical 1-bp insertion occurred in four lineages independently, whereas if we infer the presence of the disabler in the ancestral sequence, then we must infer two independent 1-bp deletions. The inference that the ancestral sequence was noncoding is a more parsimonious explanation of the data, even without considering that the parallel insertion of a specific base into an identical location is probably less likely than the parallel deletion of one base. Furthermore, the macaque orthologous DNA harbors several other indels, which support the inference that the ancestral sequence was noncoding.
Mechanism of de novo gene origin
We hypothesize that there are at least two steps in the evolution of a novel protein-coding gene from ancestrally noncoding DNA. The DNA must become transcribed and it must also gain a translatable ORF. These steps may occur in either order so that a transcribed locus that does not originally encode a protein, perhaps even an RNA gene, may acquire an ORF. Alternatively, a new ORF, once created by mutation, may become transcribed, for example, through the serendipitous use of a nearby existing gene promoter.
Here we have documented particular DNA sequence changes in the evolution of three human-specific ORFs and have demonstrated in each case that at least one critical mutation that enables the ORF is human-specific because an identical disabled state is found in chimp, gorilla, gibbon, and macaque. We cannot, at present, determine whether the ORF originated before or after expression was acquired because EST coverage is so low for chimp. However, such an analysis would not be informative in any case because we are sure that chimp cannot produce any of these proteins; so, irrespective of RNA expression, the protein-coding gene can only be present in human.
It has previously been noted that lineage-specific, presumably novel, genes have a greater tendency to overlap existing genes (Makalowska et al. 2007). Furthermore, functional retrogenes, which are duplicate genes generated by reverse transcription of mRNA, but include none of the original untranscribed regulatory signals, may acquire transcription through recruitment of promoters of fortuitous neighbors or through de novo promoter evolution (Kaessmann et al. 2009). All three novel genes discussed here, CLLU1, C22orf45, and DNAH10OS, are overlapping other genes on the opposite strand. This close proximity to other genes probably allows the novel genes to exploit existing expression machinery, though potential promoter regions are not well annotated at present. The region around the CLLU1 gene on chromosome 12 has a high number of ESTs from B cells, indicating that this region is particularly accessible to transcription (Buhl et al. 2006) and this property is likely to have facilitated the expression of the new ORF. Furthermore, the ENCODE project results showed that a high fraction of the genome is likely to be transcribed (The ENCODE Project Consortium 2007), so acquisition of transcription may not be a significant hurdle in the evolution of new genes.
Concluding remarks
This is the first rigorous and genome-wide search for evidence of new protein-coding human genes, which have evolved de novo from ancestrally noncoding sequence. Prior to this study, there were few reports of novel gene origination by this mechanism and none identified human-specific genes (Levine et al. 2006; Begun et al. 2007; Cai et al. 2008; Zhou et al. 2008; Toll-Riera et al. 2009). The novel proteins identified in this study are all short, encoded by an uninterrupted ORF, are supported by expression data, and the corresponding regions of chromosome where the ortholog is expected to be found in chimp and macaque harbor disabling mutations, which mean that the protein cannot be produced. For all three of these genes, one disabler is shared between chimp, gorilla, gibbon, and macaque indicating that the primate ancestor did not have the protein-coding gene. Translation of the genes is also confirmed by the detection of short sequenced peptides.
Although we also performed the complementary analysis looking for novel chimp genes, no reliable cases were identified, possibly due to lower genome sequence quality and human-genome–centric-genome annotation practices.
Because of the extremely strict criteria used in this study to avoid false positive results, the number of newly arisen human protein-coding genes is probably higher than found here. We identified three reliable cases of de novo gene origination in the human genome where the syntenic region in chimp and macaque was not disrupted by inversions or sequencing gaps and did not have the capacity to produce a similar protein. From our results we estimate that only about 4000 human genes were amenable to this analysis (i.e., the syntenic region was identifiable, intact, and without unidentified ORFs). We identified three reliable cases of de novo gene origination in these 4000 genes. Without considering the requirement for expression support for the human genes, we can therefore estimate that the frequency of novel protein-coding genes in the human genome is about 0.075%. If the human genome contains ∼24,000 genes, then we expect that close to 18 genes have originated de novo since the divergence with chimp. As the data become more complete it will be possible to search for more cases.
The three genes reported here are the first well-supported cases of protein-coding genes that arose in the human lineage and are not found in any other organism. It is tempting to infer that human-specific genes are at least partly responsible for human-specific traits and it will be very interesting to investigate the functions of these novel genes.
Methods
We performed an all-against-all BLASTP search of all human, chimp, and macaque proteins from Ensembl (Hubbard et al. 2007) v 46 with an E-value threshold of 1 × 10−4. We defined unambiguous orthologs as reciprocal best hits between any pair of genomes where there was no other hit with an E-value within a range of 1 × 103. Synteny blocks were constructed, anchored on these unambiguous orthologs, where the gap between anchors was no more than 10 genes in either genome. Local differences in gene order were permitted within this range.
Likely orthologous ORFs at the expected location were defined as BLAT (Kent 2002) or SSearch (Pearson and Lipman 1988) sequence matches, where the translated sequence had ≥90% identity with the human protein in each of the exons and no in-frame stop codons in the first half of the alignment, and where any inferred introns were at least 18 nucleotides (nt) long (very short introns of 1–5 bp are frequently inferred by automated pipelines to avoid frameshifts and to force a match, but there is no evidence for splicing of introns of less than 18 nt [Gilson and McFadden 1996]).
In some cases an ortholog was annotated by Ensembl in more distantly related vertebrates. We examined these cases to determine if these may be old genes that were inactivated in some primates. Some of these proposed orthologs had multiple implausibly small introns, and we discarded these as potential orthologs. For example, the current Ensembl release proposes a Mouse lemur ortholog of CLLU1, but the gene sequence includes many disablers (indels and stop codons), which were dealt with by the automated gene prediction pipeline by inferring five introns of less than 3 bp long in this “gene.” These are not plausible introns and we conclude that this locus cannot produce a protein in this organism. Otherwise, where Ensembl proposes a plausible ortholog we inferred that the human gene is an old gene with several parallel inactivations in vertebrate genomes.
The breakdown of the candidate genes was as follows: 644 human genes had no BLASTP hit in chimp, these are the initial candidates; 425 had a sequence or assembly gap (as large as the gene) in the chimp expected location; 150 had a plausible ortholog in the chimp expected location; 36 had a gap in the macaque expected location; six had smaller gaps in chimp or macaque that appeared to overlap the gene (i.e., we observed partial nucleotide similarity ending in a gap and the gene may be present though only partially sequenced); seven human genes were deemed to be possible annotation artifacts (e.g., absence of methionine or implausibly small introns); and one candidate had a possible ortholog in Xenopus. This leaves 19 candidates of which 16 had an uninterrupted (though unannotated) ORF in chimp or macaque of at least 50% of the length of the human ORF.
The DNA sequence of the human genes was aligned with DNA from the syntenic location in chimp and macaque using MultiPipMaker (Schwartz et al. 2000) and manually curated and visualized using JalView (Clamp et al. 2004).
Peptide matches in PRIDE and PeptideAtlas databases were identified by searching with the gene name. The search returns experiment details (experiment numbers are listed in Table 2) where each experiment involves the fractionation and sequencing (by mass spectroscopy or other methods) of short peptides. One experiment might identify peptides from thousands of different proteins. We extracted the peptides from the database and confirmed that they match the protein sequence of the gene of interest and we also used BLASTP and TBLASTN to confirm their specificity.
Acknowledgments
We thank Henrik Kaessmann for supplying chimpanzee DNA; Ken Wolfe, Laurent Duret, and Mario Fares for helpful suggestions; and all of the members of the McLysaght laboratory for discussions. This work is supported by Science Foundation Ireland.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ713693, FJ713696, and FJ713697.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.095026.109.
References
- Begun DJ, Lindfors HA, Kern AD, Jones CD. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics. 2007;176:1131–1137. doi: 10.1534/genetics.106.069245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl AM, Jurlander J, Jorgensen FS, Ottesen AM, Cowland JB, Gjerdrum LM, Hansen BV, Leffers H. Identification of a gene on chromosome 12q22 uniquely overexpressed in chronic lymphocytic leukemia. Blood. 2006;107:2904–2911. doi: 10.1182/blood-2005-07-2615. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics. 2008;179:487–496. doi: 10.1534/genetics.107.084491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Eng JK, Zhang H, King NL, Nesvizhskii AI, Lin B, Lee H, Yi EC, Ossola R, Aebersold R. Human Plasma PeptideAtlas. Proteomics. 2005;5:3497–3500. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson PR, McFadden GI. The miniaturized nuclear genome of eukaryotic endosymbiont contains genes that overlap, genes that are cotranscribed, and the smallest known spliceosomal introns. Proc Natl Acad Sci. 1996;93:7737–7742. doi: 10.1073/pnas.93.15.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: Mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: Glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Makalowska I, Lin CF, Hernandez K. Birth and death of gene overlaps in vertebrates. BMC Evol Biol. 2007;7:193. doi: 10.1186/1471-2148-7-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R. PRIDE: The proteomics identifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe MR, Griffin TJ. Gel-free mass spectrometry-based high throughput proteomics: Tools for studying biological response of proteins and proteomes. Proteomics. 2006;6:4678–4687. doi: 10.1002/pmic.200500876. [DOI] [PubMed] [Google Scholar]
- Rosso L, Marques AC, Weier M, Lambert N, Lambot MA, Vanderhaeghen P, Kaessmann H. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol. 2008;6:e140. doi: 10.1371/journal.pbio.0060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Toll-Riera M, Bosch N, Bellora N, Castelo R, Armengol L, Estivill X, Mar Alba M. Origin of primate orphan genes: A comparative genomics approach. Mol Biol Evol. 2009;26:603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang G, Zhang Y, Xu S, Zhao R, Zhan Z, Li X, Ding Y, Yang S, Wang W. On the origin of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]