Abstract
In eukaryotic cells the stability and function of many proteins are regulated by the addition of ubiquitin or ubiquitin-like peptides. This process is dependent upon the sequential action of an E1-activating enzyme, an E2-conjugating enzyme, and an E3 ligase. Different combinations of these proteins confer substrate specificity and the form of protein modification. However, combinatorial preferences within ubiquitination networks remain unclear. In this study, yeast two-hybrid (Y2H) screens were combined with true homology modeling methods to generate a high-density map of human E2/E3-RING interactions. These data include 535 experimentally defined novel E2/E3-RING interactions and >1300 E2/E3-RING pairs with more favorable predicted free-energy values than the canonical UBE2L3–CBL complex. The significance of Y2H predictions was assessed by both mutagenesis and functional assays. Significantly, 74/80 (>92%) of Y2H predicted complexes were disrupted by point mutations that inhibit verified E2/E3-RING interactions, and a ∼93% correlation was observed between Y2H data and the functional activity of E2/E3-RING complexes in vitro. Analysis of the high-density human E2/E3-RING network reveals complex combinatorial interactions and a strong potential for functional redundancy, especially within E2 families that have undergone evolutionary expansion. Finally, a one-step extended human E2/E3-RING network, containing 2644 proteins and 5087 edges, was assembled to provide a resource for future functional investigations.
Protein ubiquitination is mediated by the sequential action of an E1 activating enzyme, an E2 conjugating enzyme, and a range of E3 proteins, which are thought to confer substrate specificity (Hershko and Ciechanover 1998). Two main forms of E3 proteins have been characterized: HECT domain ligases, which act as enzymatic intermediates in protein ubiquitination and E3-RING proteins, which appear to be nonenzymatic recognition factors, although their exact role in catalysis remains to be established (Ozkan et al. 2005; Petroski et al. 2006). Although the sequence of events that facilitate the addition of ubiquitin (Ub) or ubiquitin-like (Ubl) proteins is conserved in all eukaryotic cells, the extent and form of Ub and Ubl modification can be diverse, ranging from the addition of single Ub or Ubl residues at one or more sites within a target protein (mono- and multi-ubiquitination), to the assembly of a range of structurally distinct polyubiquitin chains (Peng et al. 2003), which may confer different functional properties (Welchman et al. 2005; Ikeda and Dikic 2008).
Although E2 and E3 proteins are thought to work in a combinatorial manner to generate different forms of substrate modification (Weissman 2001; Christensen et al. 2007), very little is known about the specificity or combinatorial nature of human E2/E3-RING interactions. As such, the potential for redundancy or antagonism within the human ubiquitome remains unclear, as does the degree of connectivity between different network components. As Ub and Ubl proteins are known to play a key role in the regulation of both physiological and pathological processes, there is a growing need to develop a better understanding of the complex ways in which E2 and E3 proteins work together in human cells.
While isolated biochemical studies and unbiased global interactome projects continue to provide invaluable and exciting data, coverage of the human ubiquitome, and, in particular, E2/E3 interactions, remains limited. To provide new insights into partner preferences and the degree of redundancy within this combinatorial process, the density of network coverage must be significantly increased using techniques that define or predict binary interactions. Analysis of data contained within the MINT, IntAct, BioGRID, and HPRD databases revealed 60 human E2/E3-RING interactions (Fig. 1A; Supplemental Files 3 and 4), including data from both binary and co-complex isolation studies. This lack of experimental data is compounded by the expansion in E2- and E3-RING protein numbers, which has occurred during eukaryotic evolution. Searches performed using data from the Inparanoid and Homologene databases show that 34 of the 39 human E2-related proteins have clearly identifiable orthologs in yeast, fly, or worm. However, 48% (146 out of 304) of E3-RING proteins do not (Supplemental File 1). Consequently, methods developed to predict interactions between orthologous proteins in different species (Interolog interactions) (Matthews et al. 2001; Lehner and Fraser 2004) cannot be used to assign combinatorial preferences for all human E2/E3-RING complexes.
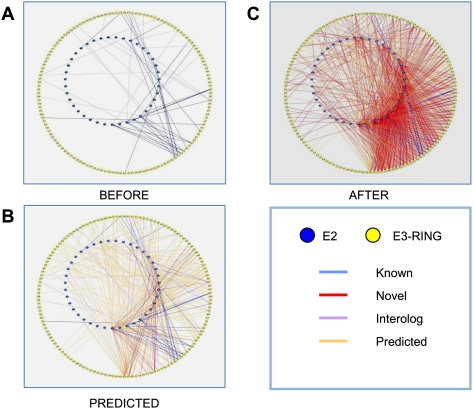
Figure 1.
Binary human E2/E3-RING protein interaction networks. (A) Previously known interactions derived from the MINT, IntAct, BioGRID, and HPRD databases at the time of this study. (B) Predicted human E2/E3-RING interactions including Interologs (purple edges) or non-Interolog predicted interactions from Hi-map and IntNet databases (orange edges). (C) Increased coverage within the human E2/E3-RING interaction space as a result of this study. Novel interactions are shown as red edges. Bold edges represent interactions confirmed by our data. Blue nodes represent E2 ubiquitin conjugating enzymes, while yellow nodes represent E3-RING proteins. To aid network analysis and node identification, all networks are provided as ready-to-view Cytoscape files (Supplemental File 3).
To address these problems we have assembled a high-density binary protein interaction map containing >1810 human E2- or E3-RING interactions. Initially, two stringent Y2H methods were used to investigate the spectrum of human E2 protein interactions and the combinatorial nature of human E2/E3-RING complexes. In addition, a structure-based true homology modeling method was also used to provide an independent prediction of interactions between 3180 human E2/E3-RING pairs. Finally, experimental data from this study were combined with known human E2- or E3-RING interactions and available Interolog data to generate a one-step extended map, which provides an initial insight into the gross topology and modular organization within the highly combinatorial human E2/E3-RING network.
Results and Discussion
Constructing a high-density binary human E2/E3-RING interaction network
To initiate a systematic analysis of human E2 protein interactions, 39 annotated human E2 proteins, including five E2-like proteins plus five transcriptional variants were cloned and used as “baits” to screen two high-complexity human Y2H “prey” libraries, one derived from fetal brain and a second from K562 cells. Following retesting of all positive interactions and reconfirmation of bait-prey specificity (Lehner et al. 2004), 229 reproducible positive interactions were identified (Supplemental File 2), of which >90% are novel. Although this form of library screen may not provide a comprehensive insight into all E2 partners, it is interesting to note that almost 30% of the detected interactions involve E3 proteins, and a further 21 prey proteins may also function in the addition or removal of Ub or Ubl proteins. To expand the human E2/E3-RING interaction network, a series of targeted Y2H studies were performed in which ∼5700 potential E2/E3-RING combinations were individually tested. After repeating each matrix screen, 557 reproducible positive E2/E3-RING interactions were identified. These interactions were then combined with data from Y2H library screens to generate an integrated set of 568 experimentally defined positive human E2/E3-RING interactions (Fig. 1C; Supplemental File 4). Images shown in Figure 1 can be enlarged and analyzed by accessing the Cytoscape-ready files provided in Supplemental File 3.
Analysis of Y2H data
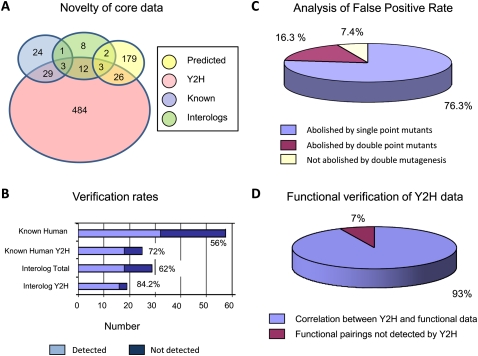
Comparison of our data with that contained in public databases shows that >94% of the E2/E3-RING interactions reported in this study are novel. Furthermore, ∼97% of the observed interactions could not have been predicted from existing Interolog data (Fig. 2A). These results reflect the paucity of information relating to the combinatorial nature of protein ubiquitination networks in all eukaryotic systems. Interestingly, our ability to validate known or predicted interactions was highly dependent upon the source of the original data (Fig. 2B). Of the 29 testable Interolog interactions, 18 (>62%) were validated in this study. However, when Interolog predictions were derived from other Y2H studies, our validation rate increased significantly to >84% (16/19). As for other predicted interactions, the Hi-map and IntNet databases predicted 35 and 181 interactions, of which 10 (28.6%) and 21 (11.6%) were verified, respectively. However, higher confidence predictions from each database were more frequently verified in our screens.
Figure 2.
Analysis of data from targeted Y2H experiments. (A) The novelty of Y2H data was determined relative to known interactions contained within the MINT, IntAct, BioGRID, and HPRD databases, known Interolog interactions, or non-Interolog predicted interactions (derived from the Hi-map or IntNet databases). (B) Reconfirmation of known or predicted E2/E3-RING interactions is highly dependent upon the source of the predicted data. (C) Strategic mutagenesis studies were performed to establish the proportion of positive Y2H interactions that conform to the known molecular/structural requirements for E2/E3-RING complex formation (see experimental data presented in Supplemental File 5). (D) To assess the efficiency with which Y2H data can predict functionally active E2/E3-RING complexes, 51 different E2/E3-RING combinations were systematically tested for ubiquitination activity in vitro (see experimental data presented in Supplemental File 6). Activity profiles for specific E2/E3-RING complexes were then compared with Y2H data to establish the percentage correlation between the two data sets.
Of the 60 human E2/E3-RING interactions contained within the MINT, IntAct, BioGRID, and HPRD databases at the time of this study, 32/57 tested interactions (>56%) were reconfirmed (Fig. 2B), which is comparable to previous Y2H studies (Lehner and Sanderson 2004; Rual et al. 2005; Stelzl et al. 2005). However, these databases contain interactions identified in co-complex isolation studies. As such, this data will include indirect interactions, which we would not expect to detect in Y2H studies. Significantly, 72% (18/25) of binary human protein interactions derived from Y2H studies were reconfirmed in our study (Fig. 2B).
To assess the potential false-positive rate of E2/E3-RING interactions detected in our Y2H experiments, a structure-based mutagenesis strategy was used to establish the proportion of predicted E2/E3-RING complexes that conform to the known molecular requirements for E2/E3-RING protein interactions. Previous analysis of X-ray and NMR structures of E2/E3-RING and E2/E3-RING-like complexes (Zheng et al. 2000; Dodd et al. 2004) revealed conserved amino acids within E3-RING or E3-RING-like proteins, which are required for E2 protein binding. Significantly, these residues are not required for the overall structural integrity of the RING protein (Joazeiro et al. 1999; Hewitt et al. 2002).
Focusing on a subset of 12 highly connected E3-RING proteins, we generated a series of single- and double-point mutants, in which two conserved amino acids that are known to be involved in E2 binding were mutated (Supplemental File 5). The ability of the resulting E3-RING mutants to interact with their original E2 partners was then retested. Using this strategy, 80 different E2/E3-RING pairings (>14% of the core E2/E3-RING data set) were systematically retested. Of these interactions, >76% were abolished by the introduction of a single-point mutation and a further 16% were eliminated by double-point mutations (Fig. 2C). Therefore, >92% of E2/E3-RING complexes predicted by targeted Y2H screens conform to the known structural requirements for previously verified E2/E3-RING interactions. Significantly, a selection of known interactions between E3-RING proteins and non-E2 protein partners, such as homo- and heterotypic E3-RING interactions, were not disrupted by the introduction of mutations that abolished E2 protein binding (Supplemental File 5). Also, it is important to note that both “strong” and “weak” two-hybrid interactions were equally susceptible to disruption by this mutagenesis strategy (see experimental data presented in Supplemental File 5), thereby indicating that in our assays, reproducible “weak” positive Y2H interactions are equally as relevant as interactions that exhibit strong growth under “higher stringency” conditions. Although accurate extrapolation between growth under different selection conditions and “interaction strength” is difficult, our data suggests that there may be a significant range of interaction affinities between different E2/E3-RING complexes. If true, this could be an important factor in establishing a functional hierarchy within combinatorial E2/E3-RING networks.
Predicting functional E2/E3-RING complexes
To establish the accuracy with which our Y2H data can predict functional E2/E3-RING complexes, 51 different E2/E3-RING combinations were systematically tested for ubiquitination activity. Significantly, all 18 of the 51 E2/E3-RING complexes found to be positive in Y2H assays were also functionally active in vitro. In addition, >94% of E2/E3-RING combinations that were negative in Y2H studies were also inactive in in vitro ubiquitination assays (see Supplemental File 6 for experimental data). Overall, this represents a 93% correlation between Y2H predictions and functional activity (Fig. 2D). Significantly, both “strong” and “weak” Y2H interactions were found to be functionally active in vitro. Therefore, based on this evidence it appears that the majority of E2/E3-RING complexes identified in Y2H studies do represent functionally active ubiquitination complexes.
Primary analysis of the high-density binary E2/E3-RING interaction network
Comparison of the human E2/E3-RING interaction network before and after this study shows a dramatic increase in network complexity (Fig. 1A–C). Analysis of the new high-density network reveals an immense potential for the formation of combinatorial complexes, in which most E3-RING proteins appear to interact with a distinct selection of different E2 partners. Of particular interest are examples where subgroups of E3-RING proteins exhibit common patterns of interaction, with the same complement of E2 proteins. For example, several E3-RING proteins interact with members of the UBE2D family and UBE2N (see Supplemental File 5) as previously observed for the Kaposi's sarcoma-associated herpesvirus K3 protein (Dodd et al. 2004). The fact that we observe a number of similar combinatorial modules involving host E3-RING proteins may indicate that similar ubiquitination events could also be common in host Lys-63 ubiquitin chain-related regulatory processes. Another striking property of the human E2/E3-RING network is the high proportion of E3-RING proteins that interact with members of the UBE2D and UBE2E families of E2 proteins. From the emerging topology it appears that these families may carry the main burden of ubiquitination events in human cells. Alternatively, the UBE2D and UBE2E families may serve to prime a broad range of proteins for more specialized modifications, which are then mediated by E2 proteins with more restricted partner profiles. Although such predictions remain speculative, it is fascinating to see that similar topological trends are also implied when E2/E3-RING complexes are predicted by purely computational methods (Supplemental File 9).
True homology modeling of human E2/E3-RING interactions
To complement experimental data generated in Y2H studies, a structure-based true homology modeling approach was also used to independently predict the likelihood of E2/E3-RING interactions. Assuming that similar amino acid sequences have a similar fold and that domains within a similar fold interact in a similar way (Aloy and Russell 2006), it is possible to predict novel protein–protein interactions based on existing structural information (Kiel et al. 2005, 2007). By generating homology models of proteins with similar sequences, protein design algorithms (Kiel et al. 2007) can then be used to calculate the relative interaction energies of related binary interactions. Using this approach, predicted free-energy values for >3000 possible E2/E3-RING parings were calculated (data provided in Supplemental File 4).
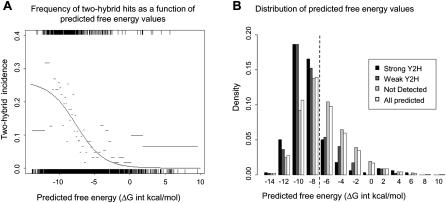
Comparison of data from true homology modeling and Y2H studies show common and informative trends. For example, the probability of detecting a positive Y2H interaction increases in concordance with more favorable (lower) free-energy predictions (Fig. 3A). In addition, the overall distribution of predicted free-energy values for complexes detected in Y2H studies is more favorable than free-energy values predicted for tested complexes that were not detected in Y2H assays (Fig. 3B). Also, the majority of E2/E3-RING complexes identified in Y2H studies have predicted free-energy values that are more favorable than either the functionally defined UBE2L6/RNF40 complex (−6.65 ΔG int kcal/mol) (Chin et al. 2002) or the canonical UBE2L3–CBL crystal structure (−7.87 ΔG int kcal/mol; Fig. 3B). As such, we predict that this subset of interactions may contain many functionally relevant E2/E3-RING pairs. Comparing the range of free-energy values assigned to complexes that were, or were not, detected for individual E2 protein, the mean ΔG int value of detected complexes is generally lower than that of complexes that were not detected (Supplemental File 9A). In these cases, the higher (least favorable) ΔG int value of detected complexes may provide a useful working confidence limit for complexes involving that particular E2 protein. Although some E2 families (UBE2V1, UBE2V2, TSG101) are predicted to have consistently unfavorable predicted ΔG int values, interactions with these proteins were frequently detected in Y2H studies. This dichotomy may reflect the inherent challenges of modeling complexes that involve more distant members of the E2 family. Equally, not all complexes predicted to have favorable ΔG int values were detected in Y2H assays. However, no single interaction assay can detect all possible interactions, and the Y2H assay may miss interactions that are hindered by large N-terminal fusions. Also, some E2/E3-RING complexes may only assemble in response to conditional signals or modifications, which cannot be reproduced in yeast. Finally, our Y2H studies identified very few interactions involving UBE2G1, UBE2DNL, UBE2F, UBE2S, or UBE2M proteins (Supplemental File 9B), all of which were also considered to be unfavorable for true homology predictions. Also, E2 proteins that were predicted to form complexes with lower ΔG int values (Supplemental File 9A) have the highest number of detected interaction partners (Supplemental File 9B). Phylogenetic expansion of E2 proteins has predominantly occurred within the two most highly connected subfamilies (UBE2D and UBE2E). Interestingly, individual members of these subfamilies appear to have maintained a very similar spectrum of E3-RING partners. If these E2 proteins regulate a range of important biological processes, then there could be a physiological advantage in maintaining a high degree of redundancy among E2 proteins. This would protect ubiquitination networks from genetic perturbation and also confer favorable gene dosage effects. Also, subtle variations in protein sequence may confer different substrate preferences, or facilitate interactions with a different range of non-E3-RING partners, all of which would increase combinatorial diversity within ubiquitination networks.
Figure 3.
Comparison of data from targeted Y2H studies and true homology modeling methods. (A) To assess the ability of true homology modeling methods to predict the probability of detecting E2/E3-RING complexes in Y2H assays, the predicted free-energy values for 3150 E2/E3-RING complexes were compared with experimental profiles detected in Y2H studies. A binary score (1/0 = interaction was/was not observed) was assigned for each complex tested in Y2H assays. SigmaPlot 10.0 (Systat) was then used to fit the set of free-energy/binary score data pairs to the logistic model: p(Y2H interaction | free-energy score = x) = a / (1 + exp (−(x + b) / c)), with values for the parameters a, b, and c obtained by nonlinear regression. The resulting regression curve is shown in black. Black vertical lines indicate the distribution of the predicted free-energy values for interacting and noninteracting pairs on the upper and lower horizontal axes, respectively. Predicted free-energy values were ordered and split into bins of 200, and the frequency of Y2H interactions within each bin is shown as a gray horizontal bar (bar width indicates the free-energy values covered by each bin). (B) Distribution of free-energy values for all predicted human E2/E3-RING complexes (open bars); “strong” Y2H interactions selected on −Ade and −His + 2.5 mM 3AT plates (black bars); “weaker” Y2H interactions selected on −His + 2.5 mM 3AT plates (dark gray bars); interactions tested but not detected in Y2H studies (light gray bars). Dashed line indicates the approximate predicted free-energy values for the structurally defined UBE2L3–CBL complex (−7.87 ΔG int kcal/mol).
Analysis of a one-step extended human E2/E3-RING interaction network
To investigate the organization of E2- and E3-RING proteins within the broader context of the human interactome, a one-step extended network was assembled (Supplemental Files 10 and 11). This network includes all known (or Interolog) partners for each protein included in the core E2/E3-RING network (Fig. 1C). This extended network (containing 2644 proteins and 5087 interactions) was then used to investigate patterns of degree distribution, to perform a GO term enrichment analysis, and to identify recurrent network modules.
To assess the spectrum of biological processes that may be regulated by E3-RING proteins, a GO term enrichment analysis was performed on proteins within the outer shell of the extended human E2/E3-RING network. This data reveals a broad spectrum of statistically enriched biological processes (Supplemental File 12), which emphasizes the fundamental importance of E2/E3-RING interactions in almost all biological processes. Further analysis of the extended E2/E3-RING network reveals recurrent modules that provide new insights into the functional organization of components within ubiquitination networks. These include modules in which E3-RING proteins are indirectly linked to E2 proteins, either via heterotypic E3-RING interactions (Supplemental File 10Ba) or via a non-E2/E3 protein bridge (Supplemental File 10Bb). Further analysis is required to establish whether these modules contain novel adaptor proteins or simply lack edges, which would otherwise link orphan E3-RING proteins to E2 partners. Also, it is significant to note that most (37/42) E2 proteins represented in the extended network also interact directly with non-E3-RING proteins. This may imply that direct E2-substrate ubiquitination (Hoeller et al. 2007) may be more frequent than expected. Alternatively, this subclass of E2 proteins may represent cofactors that effect catalytic activity or contribute to the recognition/retention of substrate proteins.
With respect to E3-RING proteins, many peripheral proteins were found to interact with multiple E3-RING proteins (Supplemental File 10Bc). This is consistent with a model in which different E2/E3-RING combinations may confer different modifications on the same substrate. Another recurrent module is the “RING-junction” (Supplemental File 10Bd), in which multiple E2 proteins link to a broad spectrum of peripheral proteins via a single E3-RING. It is not yet clear how these modules may operate. As both E2–E3 interactions and E3–substrate interactions may function via a system of competitive exclusion, it is possible that hierarchical binding properties or regulated expression patterns may impose conditional or temporal restrictions on E2/E3-RING or E3-RING/substrate interactions. In this way, junction modules may work in a manner similar to “railway points,” coordinating the flow of regulatory information to different biological processes.
Although the extended human E2/E3-RING network already appears complex, this is only the first stage of an ongoing process of analysis, reiteration, and verification, which will be required to generate a comprehensive annotated map of the human ubiquitome. However, this primary network reveals many novel interactions and provides a new framework, which can be used to drive forward future hypothesis-driven studies in many areas of biomedical research and network biology.
Methods
Construction of Y2H bait and prey clones
Human E2 proteins were generated by proofreading PCR using templates obtained from the mammalian gene collection (Strausberg et al. 2002) or from Matchmaker cDNA libraries (Clontech). In each case, primers were designed in accordance with RefSeq annotations for the full-length protein-coding region, including the endogenous stop codon. Each primer contained forward or reverse flanking sequences to facilitate transfer of the PCR products into Y2H bait (pGBAD-B) or prey (pACTBD-B) vectors, respectively (Semple et al. 2005). Bait vectors were constructed by gap repair recombination cloning, as described previously (Estevez et al. 2003). E3-RING proteins were obtained from two independent sources (as indicated in Supplemental File 4). Initially, 91 human E3-RING proteins were amplified from Matchmaker human fetal brain or K562 cDNA libraries (Clontech). As for E2 proteins, amplification of human E3-RING proteins was performed by proofreading PCR using primers designed in accordance with gene-specific RefSeq annotations in order to generate full-length protein-coding regions (including stop codon). Gene-specific PCR products with GATEWAY compatible flanking sequences were then transferred into the pACTBD-B Y2H prey vector (Semple et al. 2005) by in vivo gap repair recombination cloning. In addition, a collection of 90 E3-RING proteins was obtained from the Human Orfeome collection as GATEWAY entry clones in the pDONR223 vector. Each of these clones was transferred into a GATEWAY-converted pGAD vector by conventional GATEWAY cloning methods and into the GATEWAY-compatible pACTBE-B vector (Semple et al. 2005) by in vivo recombinant gap repair cloning. All clones were sequenced following auto-activation tests to verify insert identity. In all Y2H studies, bait constructs were transfected into the PJ69-4a yeast strain, while prey constructs were transferred into the complementary PJ69-4α strain.
Yeast two-hybrid library screens
Library screens were performed as described previously (Lehner et al. 2004). In brief, ∼109 bait cells were mated with ∼109 cells from either a Matchmaker human K562 cell line cDNA prey library (Clontech) or with a comparable number of cells derived from a Matchmaker human fetal-brain prey cDNA library (Clontech). Following mating, positive colonies were selected on media lacking Ade for 7–10 d. −Ade positive colonies were also tested for activation of the lac-Z reporter and all prey inserts were identified by PCR and sequencing. All positive interactions were retested in fresh yeast, and specificity was confirmed by testing for interaction with an irrelevant bait clone (LSM2). The identity of prey inserts was established by using the BLAST algorithm to search the UniGene database.
Yeast two-hybrid matrix screens
E2 bait clones were systematically mated against arrays of E3-RING prey clones on YPAD media for a period of 12 h. Diploids were then selected following replication and growth on −Leu / −Trp plates for 24 h. Activation of the ADE2 (stringent) or HIS3 (less stringent) reporters were independently assayed by replication to media lacking either adenine or histidine (+2.5mM 3−AT). Growth on selective plates was then scored over a period of 11 d. Only reproducible interactions detected in at least two independent assays were counted as positive interactions. Singleton interactions that were not reproducibly detected were not recorded as positive interactions
Mutagenesis and reconfirmation of E2/E3-RING interactions
Strategic mutagenesis of a selection of E2-interacting E3-RING proteins was performed as follows. MATα prey clones containing the E3-RING proteins to be mutated were grown in SD-L overnight and pACTBD-B plasmids were isolated using the Wizard plus SV plasmid purification system following the manufacturer's instructions (Promega). In each case, primers were designed in accordance with the RefSeq annotation for specific E3-RING sequences at the sites around the residues corresponding to CBL W(408) and I(383), but with an altered primer sequence, which introduced a change to an alanine at the respective codons. Primers were designed to be between 25 and 35 bases (optimally 10–15 bases either side of the mutation) with a melting temperature ≤78°C. Mutagenesis was then performed using the QuikChange Site-directed mutagenesis kit and PCR cycling parameters in accordance with the QuikChange Site-directed mutagenesis protocol. Mutated E3-RING prey pACTBD-B plasmids were retransformed into PJ69-4α MATα yeast and transformants were selected on media lacking leucine (preys). Colonies were picked and subjected to YCPCR and full-length bidirectional sequence verification to check for accuracy of the mutagenesis strategy. Mutated E3-RING prey clones were then retested for interaction against E2 baits as described above.
Functional ubiquitination assays
Recombinant GST-RING Ub-ligases were expressed in Rosetta 2(DE3)pLysS cells (Novagen). One-liter cultures of cells were induced at OD600 of 0.8 with 150 μM IPTG, and proteins were expressed at 20°C overnight. For increased solubility of the Zn-binding RING proteins, 200 μM zinc sulphate was added at the point of induction. Cells were harvested and flash-frozen. A total of 50 mL lysis buffer containing 270 mM sucrose, 50 mM Tris (pH 8.0), 50 mM NaF, 1 protease inhibitor cocktail tablet (Roche) (0.1% v/v β-mercaptoethanol, 1 mg/mL lysozyme, and 0.1 mg/mL DNase) was added per liter of culture. After sonication, cell lysates were cleared using a Sorvall SS-34 rotor (18,000 rpm, 30 min, 4°C), and supernatants were incubated with Glutathione Sepharose 4B (GE Healthcare) for 1 h to immobilize soluble GST fusion proteins. Subsequently, the Sepharose beads were washed with 500 mL high-salt buffer (500 mM NaCl, 25 mM Tris [pH 8.5], 5 mM DTT) and 300 mL low-salt buffer (150 mM NaCl, 25 mM Tris [pH 8.5], 5 mM DTT). To elute GST-RING Ub-ligases, beads were incubated with 30 mM glutathione in lysis buffer, and after the concentration was measured by NanoDrop, samples were flash-frozen in liquid nitrogen. All RING domains were >95% pure after purification.
Auto-ubiquitination assays were carried out in 30-μL reactions at 37°C containing 25 ng of Ub-activating enzyme (E1), 1 μg of Ub-conjugating enzyme (E2), 2 μg of RING Ub-ligase (E3), 5 μg of ubiquitin, 10 mM ATP, 40 mM Tris (pH 7.5), 10 mM MgCl2, and 0.6 mM DTT. After 1 h, the reaction was stopped by addition of 10 μL of 4× SDS sample buffer (Invitrogen), resolved by SDS-PAGE on 4%–12% precast gels, and subjected to immunoblotting analysis using polyclonal anti-ubiquitin antibody (Upstate).
True homology modeling
Prediction of free-energy values for different human E2/E3-RING complexes was performed in accordance with previously published methods (Kiel et al. 2005, 2007). As the pdb:1fbv structure was the only available template for modeling the E2/E3-RING with a range of isolated UBC (1i7k, 1x23, 2csv, 2cyx, and 1a3s) or E3-RING (1rmd, 1z6u, and 2ckl) structures. All isolated structures exhibited good matches to E2/UBC and E3-RING domains within the 1fbv complex, especially at interaction surfaces. Therefore, homology modeling using the single UBE2L–CBL structure as a template should provide reliable insights into potential E2/E3-RING protein-interaction profiles.
The template structure for true homology modeling studies was generated by deleting secondary structural elements and loops that were not involved in interactions within the 1fbv complex. Homology modeling was then achieved by replacing side-chain residues in accordance with structure-based alignments (Supplemental Files 7 and 8) using the BuildModel command of FoldX (Guerois et al. 2002; Schymkowitz et al. 2005a,b) vs 2.7. Using this approach, global and interaction energies for >3000 E2/E3-RING pairings were calculated (Supplemental File 4). Visual inspection of modeled complexes identified several E2- and E3-RING proteins with strong van der Waal clashes (vdWCs), which would be expected to prevent interactions. As the UBC domain of 1zdn had strong vdWCs with E3-RING finger domains and UBC domains with a Leu at position 1006 of the UBE2L3–CBL complex (pdb:1fbv) had strong vdWCs with Ile383 in the backbone of E3-RING proteins, these E2 proteins were eliminated from modeling studies. Also, it was noted that UBC domains with either Leu or Ile at position 1005 of the 1fbv structure (UBE2G1, UBE2S, and UB2M) could have small vdWCs with Glu386 of E3-RING proteins. Complexes with predicted van der Waals clashes >10 kcal/mol were excluded from homology modeling studies, whereas complexes with mild (<3 kcal/mol) clashes were retained. All complexes with predicted van der Waal clashes are annotated in Supplemental File 4.
With respect to E3-RING proteins 1rmd, 1z6u, 1weo, 1chc, 1g25, 1x3j, 1ldj, 1rmd, 2hod, 2ckl, and 2csy, all superimpose well onto the E3-RING structure within the 1fbv complex. However, other structures (1v87, 1jm7, 2d8s, and 2d8t), although similar to the 1fbv complex, exhibit local differences within E2-binding loops, which may reduce the efficacy of structural predictions. Also, structures including 1iym, 1wim, le4u, 1bor, 1vyx, 1f62, 1wfk, and 2csz exhibited considerable variations within UBC interaction domains, while E3-RING proteins PHF7, UHRF2, RFWD3, MDM2, and MDM4 all showed variations within conserved RING domain motifs. Consequently, E2 proteins UBE2G1, UBE2M, UBE2S, UEVLD, and UBE2U, and E3-RING proteins PHF7, UHRF2, RFWD3, MDM2 and MDM4, RNF144A, MARCH8, RNF181, DZIP3, RNF126, RNF13, RNF133, RNF167, BIRC2, BIRC3, PML, BIRC7, MUL1, LRSAM1, and RNF166 were all eliminated from structural free-energy predictions. Modeled interactions that have moderate predicted vdWCs >3 kcal/mol are indicated in Supplemental File 4.
GO-term enrichment analysis
Gene lists were prepared for the E2s and RINGs used in the current screen, and interaction partners were obtained from the human interactome or from the predicted/Interolog databases. Fisher's exact test was used to calculate the probability of observing at least as many (few) genes annotated with each GO term as were observed in the list of genes. This was performed for each GO term in the Biological Process, Molecular Function, or Cellular Component GO hierarchy. The calculations could only be performed for GO-annotated genes. Hence, those genes that are present in the interactome, but which do not have an annotation for the relevant GO hierarchy (BP/MF/CC), were not included in the calculation, nor were genes that are not present in the interactome. For GO terms that had a P-value < 0.05, we determined whether the GO term was over- or underrepresented within the gene list by calculating the P-value expected, had one less gene been annotated with the GO term (where this was possible). GO annotations were obtained from Bioconductor V2.0 (www.bioconductor.org), and all calculations were performed in R V2.5.1 (http://www.r-project.org/).
Acknowledgments
We acknowledge Phil James for the donation of yeast strains PJ69-4a and PJ69-4α and the pGAD vector. We are also grateful to David Markie for providing the pGBAD-B, pACTBD-B, and pACTBE-B vectors, which were invaluable for our studies, and to Philip Cohen for E2 enzymes and expression constructs used in functional assays. The work described in this manuscript was in part funded by the U.K. Medical Research Council and the Wellcome Trust. K.S-A. and M.V. were supported by a gift from the Ellison Foundation and a grant from the U.S. National Human Genome Research Institute (2R01HG001715) to M.V. M.V. is a “Chercheur Qualifié Honoraire” from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium). Work performed by C.K. and L.S. was supported by EU grant number LSHG-CT-2003-505520 (INTERACTION PROTEOME).
Footnotes
[Supplemental material is available online at http://www.genome.org. The protein interactions from this study have been submitted to the IMEx Consortium (http://imex.sf.net) through IntAct (http://www.ebi.ac.uk/intact) under identifier no. IM-11696.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093963.109.
References
- Aloy P, Russell RB. Structural systems biology: Modelling protein interactions. Nat Rev Mol Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- Chin LS, Vavalle JP, Li L. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem. 2002;277:35071–35079. doi: 10.1074/jbc.M203300200. [DOI] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific-polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Dodd RB, Allen MD, Brown SE, Sanderson CM, Duncan LM, Lehner PJ, Bycroft M, Read RJ. Solution structure of the Kaposi's sarcoma-associated herpesvirus K3 N-terminal domain reveals a novel E2-binding C4HC3-type RING domain. J Biol Chem. 2004;279:53840–53847. doi: 10.1074/jbc.M409662200. [DOI] [PubMed] [Google Scholar]
- Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. J Biol Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: New molecular signals. “Protein Modifications: Beyond the Usual Suspects” review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, Serrano L. Recognizing and defining true Ras binding domains II: In silico prediction based on homology modelling and energy calculations. J Mol Biol. 2005;348:759–775. doi: 10.1016/j.jmb.2005.02.046. [DOI] [PubMed] [Google Scholar]
- Kiel C, Foglierini M, Kuemmerer N, Beltrao P, Serrano L. A genome-wide Ras-effector interaction network. J Mol Biol. 2007;370:1020–1032. doi: 10.1016/j.jmb.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Lehner B, Fraser AG. A first-draft human protein-interaction map. Genome Biol. 2004;5:R63. doi: 10.1186/gb-2004-5-9-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Semple JI, Brown SE, Counsell D, Campbell RD, Sanderson CM. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/s0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein–protein interactions or “interologs.”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Kleiger G, Deshaies RJ. Evaluation of a diffusion-driven mechanism for substrate ubiquitination by the SCF-Cdc34 ubiquitin ligase complex. Mol Cell. 2006;24:523–534. doi: 10.1016/j.molcel.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: An online force field. Nucleic Acids Res. 2005a;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci. 2005b;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple JI, Prime G, Wallis LJ, Sanderson CM, Markie D. Two-hybrid reporter vectors for gap repair cloning. Biotechniques. 2005;38:927–934. doi: 10.2144/05386RR03. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein–protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]