Abstract
We describe a new method, Tag-seq, which employs ultra high-throughput sequencing of 21 base pair cDNA tags for sensitive and cost-effective gene expression profiling. We compared Tag-seq data to LongSAGE data and observed improved representation of several classes of rare transcripts, including transcription factors, antisense transcripts, and intronic sequences, the latter possibly representing novel exons or genes. We observed increases in the diversity, abundance, and dynamic range of such rare transcripts and took advantage of the greater dynamic range of expression to identify, in cancers and normal libraries, altered expression ratios of alternative transcript isoforms. The strand-specific information of Tag-seq reads further allowed us to detect altered expression ratios of sense and antisense (S-AS) transcripts between cancer and normal libraries. S-AS transcripts were enriched in known cancer genes, while transcript isoforms were enriched in miRNA targeting sites. We found that transcript abundance had a stronger GC-bias in LongSAGE than Tag-seq, such that AT-rich tags were less abundant than GC-rich tags in LongSAGE. Tag-seq also performed better in gene discovery, identifying >98% of genes detected by LongSAGE and profiling a distinct subset of the transcriptome characterized by AT-rich genes, which was expressed at levels below those detectable by LongSAGE. Overall, Tag-seq is sensitive to rare transcripts, has less sequence composition bias relative to LongSAGE, and allows differential expression analysis for a greater range of transcripts, including transcripts encoding important regulatory molecules.
A key first step in understanding cellular processes is a quantitative representation of gene expression profiles, including those relevant to cancer. As part of the Cancer Genome Anatomy Project (CGAP), gene expression profiles of a wide variety of cancer tissues and cells were measured using LongSAGE libraries, created and sequenced using conventional Sanger sequencing methods (Lal et al. 1999). Prior to completion of the project, the advent of new massively parallel sequencing technologies made feasible an improvement in the efficiency and sensitivity with which tag-based gene expression can be measured. We thus sought to develop and apply a next-generation sequencing approach for tag-based gene expression profiling to complete the CGAP database.
Several recently developed sequencing technologies, such as the 454 Life Sciences (Roche) pyrosequencing platform (Margulies et al. 2005), the Illumina Genome Analyzer (Bentley 2006), and Applied Biosystems SOLiD platform (http://solid.appliedbiosystems.com), offer massively parallel production of short reads. Using these technologies, thousands to millions of isolated and amplified DNA molecules can be attached to a solid surface (such as a flowcell or microbeads), and sequenced by synthesis in parallel. Such technologies offer up to two orders of magnitude increase in per base cost efficiency compared to capillary sequencing (von Bubnoff 2008). These platforms have made feasible previously cost-prohibitive projects such as genome resequencing (Green et al. 2006; Bentley et al. 2008; Ley et al. 2008; Wang et al. 2008b) and deep transcriptome and noncoding RNA sequencing (Nielsen et al. 2006; Weber et al. 2007; Marioni et al. 2008; Morin et al. 2008; Rosenkranz et al. 2008), as well as genome-wide protein binding-site surveys (ChIP-seq) (Jothi et al. 2008; Wederell et al. 2008).
The high-throughput methods preceding the massively parallel sequencing approaches mentioned above are diverse but can generally be classified either as sequence-based or hybridization-based. The former are often termed “digital” because they reflect the number of individual observations of a transcript, while the latter, typically in the form of microarrays, are termed “analog” as they provide a surrogate hybridization-based measure of individual transcript abundance. Digital gene expression profiling using expressed sequence tags (ESTs) (Adams et al. 1991; Hillier et al. 1996) was cost-restrictive, and more cost-efficient tag-based techniques such as serial analysis of gene expression (SAGE) were developed (Velculescu et al. 1995). Despite increases in cost efficiency compared to EST profiling, the expense and specialized facilities required for high-throughput capillary sequencing prevented SAGE from becoming as widespread as its microarray counterparts.
Our goal was to implement a tag sequencing protocol on the Illumina platform, analogous to LongSAGE (Saha et al. 2002), and to use this protocol to measure transcript abundance in human cancers. The Illumina (Bentley et al. 2008) sequence-by-synthesis technology currently offers ∼80 million reads (10 million reads per lane; eight-lane flow cell) from a single run of the instrument. This makes possible gene expression profiling experiments with much improved dynamic range and considerable cost savings compared to capillary sequencing of LongSAGE. Our approach, called Tag-seq, generates 21–base pair (bp) tags, generally from the 3′ ends of transcripts. The method is similar to the LongSAGE approach (Saha et al. 2002) but forgoes the need for ditag production, concatenation, and cloning. Deep sequencing of tags is achieved using only a single lane of a flow cell, and typical yields are in the range of 5–10 million sequences.
Compared to conventional microarrays, Tag-seq has no cross-hybridization of related sequences and in principle offers dynamic range limited only by sequencing depth. Compared to RNA-seq, Tag-seq performs comparably in terms of gene discovery and dynamic range. While Tag-seq does not provide information regarding the internal structure of transcripts, it can distinguish between transcripts originating from both DNA strands. There are advantages in using a strand-specific gene expression platform, for example to measure the prevalent antisense transcription in the human genomes (Katayama et al. 2005). Here, we conduct an analysis of Tag-seq data from the CGAP collection to illustrate the utility of the method in addressing questions of relevance to cancer biology.
Results
Data generation and filtering
The Tag-seq protocol is similar to the LongSAGE approach (Saha et al. 2002), in which a restriction endonuclease (NlaIII) cleaves each individual transcript in a sample, and a type II restriction endonuclease (MmeI) is used to generate a 21-bp tag from the 3′-most NlaIII site. In LongSAGE, tags from individual transcripts are ligated together to form ditags that are concatenated, cloned, and sequenced using capillary sequencing. The Tag-seq method, in contrast, forgoes ditag production and concatenation, and allows the direct sequencing of tags using massively parallel sequencing on the Illumina Genome Analyzer (see Methods; Fig. 1). Typically, a Tag-seq library is sequenced to a depth of 10 million tags, which represents an increase of two orders of magnitude over the sequencing depth of a typical LongSAGE library. Our expectation was that the added depth of the Tag-seq method would improve representation of important low-abundance transcripts at the limits of or beyond LongSAGE sensitivity.
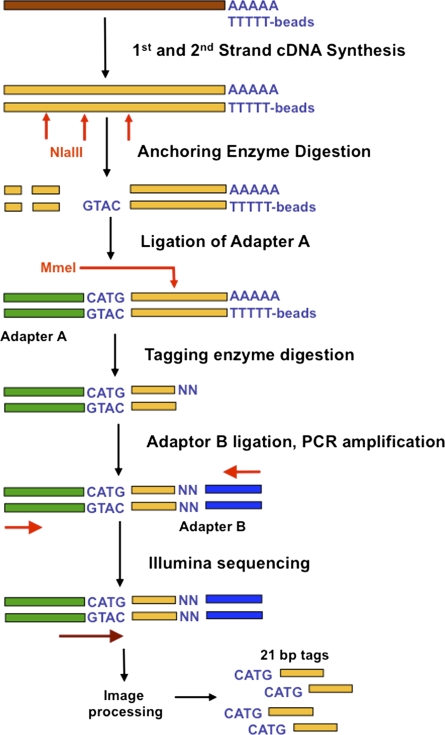
Figure 1.
Outline of Tag-seq library generation. Each mRNA (brown) underwent double-stranded cDNA synthesis using oligo(dT) beads, to capture polyadenylated RNA. cDNA (gold) is digested with the NlaIII anchoring restriction enzyme (vertical red arrows), leaving a 4-bp overhang (GTAC). Only cDNA fragments anchored to oligo(dT) beads are retained. Adapter A (green) is ligated to the overhang and adds a recognition site for the TypeIIS tagging enzyme MmeI. Following MmeI digestion (red vertical arrow), a second adapter is ligated (Adapter B, blue) to the resulting 2-bp overhang. PCR primers (horizontal red arrows) annealing to adapters A and B are used to enrich tags. Cluster generation and sequencing (horizontal brown arrow) is performed on the Illumina cluster station and analyzer. The resulting image files are processed to extract the read sequences, and 21-bp SAGE tags are further extracted from the reads. Tags consist of the 4-bp NlaIII recognition sites and 17 bp of unique sequence, and constitute a total of 21 bases that can be mapped back to the original mRNA (brown).
We used the Tag-seq platform to complete the CGAP digital gene expression profiling project, by generating 35 libraries from cancer and normal tissue samples. To assess the similarities between the new Tag-seq data and the existing LongSAGE data, we compared the data from these 35 libraries to that from 77 LongSAGE libraries. In total, we produced two metalibraries, one containing 6.9 million LongSAGE tags from the 77 libraries (1.1 million distinct tag sequences), and one containing 170 million Tag-seq tags from the 35 quality filtered libraries (four million distinct tag sequences). These libraries are publicly available as part of the CGAP collection (Supplemental Table S1; Lal et al. 1999). The CGAP libraries also included two libraries, one Tag-seq library and one LongSAGE library, which were created from the same human embryonic stem cell (hESC) RNA source.
To ensure that we analyzed high quality data in the Tag-seq libraries, we removed potentially erroneous tags using a novel filtering algorithm (Supplemental Methods). Briefly, tags were removed if they occurred once (singletons), or if they differed by one base pair from more highly expressed tags (one-offs) unless they mapped to the genome or transcriptome. On average, 22.1% of filtered tags could be mapped to Ensembl transcripts, while only 1.2% of tags removed by the filter could be mapped to transcripts. While filtered tag sequences comprised an average of 7.5% of all tag sequences, their abundance corresponded to an average of 56.0% of the total library size, and they identified >97.5% of the total number of genes detected by all tags.
Effect of depth on tag sequence diversity and abundance
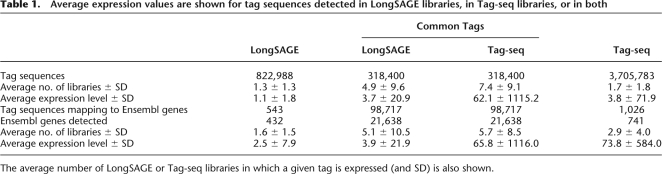
By comparing the Tag-seq and LongSAGE metalibraries, we sought to first determine whether differences in Tag-seq and LongSAGE protocols resulted in any significant bias in tag or gene representation. As expected, we found a significant overlap between these metalibraries, with >300,000 unique tag sequences detected using both methods. On average, these commonly detected tag sequences were expressed in a larger proportion of Tag-seq libraries than LongSAGE libraries, and had 17-fold higher expression in Tag-seq libraries (Table 1). A large number of tag sequences were detected by only one method; in general, these were expressed at lower levels than those tag sequences found by both methods, and in fewer libraries. The three million tag sequences detected only by Tag-seq were on average 1/16 the abundance of the tags detected in common by both methods (absolute counts, Table 1) and, therefore, were likely undetectable in the LongSAGE libraries due to their comparatively shallow sequencing depth. Thousands of Tag-seq tag sequences did not map to any unique or repetitive sites in the genome or the transcriptome. These may indicate the presence of either novel transcripts or novel isoforms of annotated genes that lead to the creation of novel tag sequences spanning splice sites (80,875 Tag-seq tag sequences and 63,166 LongSAGE tag sequences expressed over counts of 10; Supplemental Fig. S1).
Table 1.
Average expression values are shown for tag sequences detected in LongSAGE libraries, in Tag-seq libraries, or in both
The average number of LongSAGE or Tag-seq libraries in which a given tag is expressed (and SD) is also shown.
Nearly a third of the tags detected in both metalibraries mapped to 21,638 genes. A small proportion of tag sequences found solely in LongSAGE (8.1%) or Tag-seq (3.5%) mapped to Ensembl genes (Table 1). Although in general the tag sequences found only by Tag-seq had expression levels below those detectable by LongSAGE, the 741 genes found only in Tag-seq had an average expression level higher than that for the genes found in common. They are therefore likely to be genes specific to tissues not profiled by LongSAGE. With the exception of the hESC replicate, all LongSAGE and Tag-seq libraries represented diverse tissues, although the greater number of LongSAGE libraries doubled the diversity of tissues profiled by LongSAGE. The 430 genes found only by LongSAGE were on average less frequently expressed than genes detected by both methods, and may represent genes specific to tissues profiled using LongSAGE.
We next investigated the effect of depth on gene representation by comparing the Tag-seq and LongSAGE replicate libraries created from the same hESC RNA sample. The Tag-seq replicate (library id “hs0238”) had a total of 293,179 tag sequences (error tags removed; Supplemental Methods), of which 40,149 (13.7%) mapped to Ensembl genes, either in introns, exons, or on the opposite strand. The LongSAGE replicate (library id “1313” in Supplemental Table S1) had a total of 19,998 tag sequences, of which 13,983 (69.9%) mapped to Ensembl genes. The LongSAGE tag sequences mapped to 7,055 genes and the Tag-seq tag sequences mapped to 11,165 genes, which included 93.5% of the genes found by LongSAGE. Thus, added depth improved gene detection in this tissue 1.6-fold. Since each tag sequence mapping to a gene can represent an individual transcript isoform (Siddiqui et al. 2005), we analyzed the average expression of all transcript isoforms. The transcripts of the 6.5% of genes only found by LongSAGE were expressed at low levels (average of 4.0 counts) and may be underrepresented in the Tag-seq library due to variability in the replicate library creation. The detection of transcription factors (TFs) was 1.8-fold greater, with 429 TFs detected by LongSAGE, and 799 TFs detected by Tag-seq. The average expression of the 393 TFs detected in common was higher (69.8 in the Tag-seq replicate, 6.8 in the LongSAGE replicate) than that of the 36 TFs detected only in LongSAGE (5.9) and the 406 TFs detected only by Tag-seq (26.7).
To determine whether these additional genes found by Tag-seq were functionally different than those found by both methods, we conducted an assessment of Gene Ontology (GO) categories overrepresented in the Tag-seq versus the LongSAGE replicate (Ashburner et al. 2000). The most significantly overrepresented terms in this tissue were found by both methods. Thus, increased sequencing depth resulted in identification of thousands of additional genes that belonged to the same functional categories as moderately and highly abundant genes detected by LongSAGE tags.
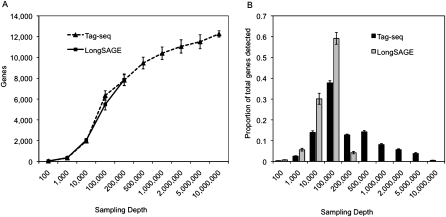
We next asked whether a Tag-seq library unambiguously identified a larger number of genes on average than a standard LongSAGE library. We performed a sampling simulation to estimate the number of genes represented by different “depths” of sequencing in each Tag-seq and LongSAGE library. Sampling up to 300,000 tags from individual LongSAGE libraries resulted in detection of up to 10,000 genes (Fig. 2A). Quality filtered Tag-seq libraries sampled at depths of up to 10 million tags detected up to 13,000 genes. This suggested that the added depth provided by the Tag-seq approach results in a more comprehensive interrogation of gene expression profiles, with 48.3% and 36.3% of expressed genes detectable at depths greater than those of a typical (100,000 tags) or large (200,000 tags) LongSAGE library, respectively. At every sampling depth level greater than one million tags in Tag-seq, the rate of gene detection was reduced (Fig. 2B).
Figure 2.
Average number (A) and proportion (B) of Ensembl genes unambiguously identified in Tag-seq and LongSAGE libraries as a function of sampling depth. Error bars represent the SD of the average number of identified genes in 77 LongSAGE libraries and 35 Tag-seq libraries. The largest LongSAGE libraries were ∼300,000 tags, while the largest Tag-seq libraries were ∼10 million tags.
Differences in gene abundance between Tag-seq and other gene expression platforms
Having established that the measured sampling depth of Tag-seq improved gene discovery, we evaluated the concordance of tag abundance between the two methods, by reanalyzing the Tag-seq and LongSAGE replicate hESC libraries. The LongSAGE replicate had a total of 272,465 tags, while the Tag-seq replicate had a total of 3,636,083 quality filtered tags. Tags expressed in common between these libraries had a Pearson coefficient of 0.60 (Supplemental Fig. S2). We analyzed another set of replicate Tag-seq and LongSAGE libraries created from the same mouse RNA (Supplemental Methods), and found they had a Pearson correlation of 0.64. This was comparable to the correlation between the LongSAGE library and a technical replicate generated with the SAGELite protocol (0.64). SAGELite is a variant of LongSAGE used to create libraries from samples that are too small to yield sufficient amounts of mRNA for standard LongSAGE library construction (Peters et al. 1999). We observed a lower Pearson coefficient between the Tag-seq technical replicate and the SAGELite replicate (0.43), indicating these methods have different biases relative to LongSAGE.
We generated Pearson correlations between three non-CGAP Tag-seq libraries and their respective technical replicates analyzed on Affymetrix exon arrays. Correlations were calculated for expressed tags which represented known transcripts and mapped uniquely or not at all to the genome, and their corresponding Affymetrix probes. Pearson coefficients for the three technical replicates were very similar to each other (0.59, 0.60, and 0.61), and to that of Tag-seq and LongSAGE replicates (Supplemental Fig. S2). An analysis of dynamic range between the Tag-seq and Affymetrix data showed that genes detected in common had a 13-fold greater dynamic range in Tag-seq versus Affymetrix (see Supplemental Results; Supplemental Fig. S3; and a twofold greater dynamic range when considering log-transformed expression values, Supplemental Table S2).
We also analyzed a pair of replicate RNA-seq/Tag-seq libraries created from the same RNA source, and found that, relative to RNA-seq, Tag-seq performed comparably in gene identification (see Supplemental Results; Supplemental Table S3) and gene expression measures (Pearson correlation of gene abundance: 0.54). Illumina does not currently distinguish between reads derived from opposing DNA strands, and RNA-seq reads were therefore not able to discriminate between sense and antisense transcription. For nearly a third (29.5%) of the genes detected by both methods in this replicate library set the Tag-seq replicate detected expression on the antisense strand (Supplemental Table S3). In the case of 613 loci detected by both methods, the Tag-seq reads clearly show that expression arises solely from the antisense strand. At these loci, correlations between gene expression levels measured by Tag-seq versus RNA-seq (0.50) were the same as those at loci with sense expression in both technologies (0.54).
GC-content bias
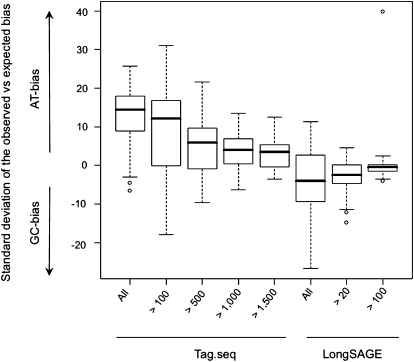
We next investigated whether there was any detectable bias in the sequence composition of tags profiled by the Tag-seq and LongSAGE platforms. The GC-bias of a platform can be calculated by comparing the number of standard deviations by which the observed bias in an individual library deviates from that of the expected bias (Siddiqui et al. 2006; Supplemental Methods). We found that Tag-seq libraries were significantly more AT-rich than LongSAGE libraries (Fig. 3). As previously observed, LongSAGE libraries had a weak GC-bias (−3.51 ± 8.08), while Tag-seq libraries had a stronger AT-bias (12.99 ± 5.39), comparable to that of the Affymetrix platform (HGU 133 GeneChip; Siddiqui et al. 2006). As observed for Affymetrix, this bias decreased in parallel with increasing expression level, such that highly expressed Tag-seq sequences were significantly less biased (all filtered tag sequences vs. those expressed over counts of 500, P = 2.1 × 10−10, t-test). This suggests that, as sequencing depth increases in sequencing-based technologies, a distinct class of genes with increasing AT-content is detected. We tested whether this was the case in Tag-seq by comparing the GC-content of the genes with high versus low frequency tags, and found that genes that expressed ≤100 tag counts were significantly more AT-rich than genes expressed ≥1,500 tag counts (P = 2.8 × 10−4, t-test; Supplemental Fig. S4). This was true of gene sequences that included introns, but not of cDNA sequences, indicating that the AT-content of the genomic regions in which these genes were encoded was correlated to their expression level. In LongSAGE bias also decreased with increasing expression level, such that tag sequences expressed over 20 and over 100 counts become significantly less biased (all tag sequences vs. those expressed over counts of 100, P = 1.9 × 10−3, t-test). This trend was also correlated to the GC-content level of the genes to which LongSAGE tags mapped to, indicating that the source for these observations was also biological in nature rather than a technical artifact (Supplemental Fig. S4).
Figure 3.
GC-bias of Tag-seq and LongSAGE libraries was calculated in units of the number of SDs by which the observed bias differed from the expected bias (see text). Positive units represent libraries with more AT-rich tag sequences than expected (AT-bias), while negative units represent libraries with more GC-rich tag sequences than expected (GC-bias). Calculated bias is shown for all quality filtered Tag-seq and all LongSAGE tag sequences, at increasing thresholds of tag expression (x-axis).
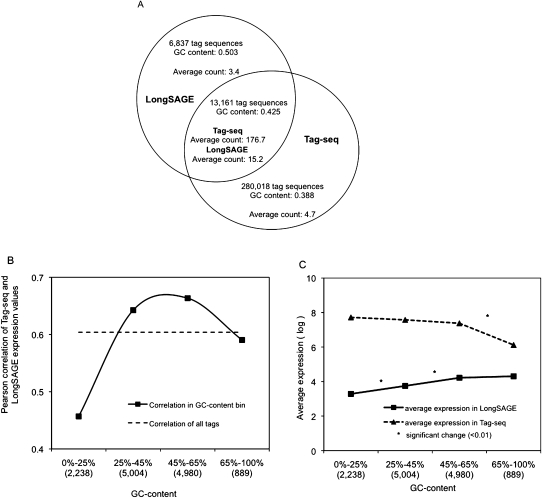
Next, we determined the extent to which tag representation was biased in Tag-seq versus LongSAGE, by reanalyzing the hESC replicate libraries made from the same RNA source. Tag sequences detected solely by LongSAGE had a greater GC-content than those detected solely by Tag-seq (0.50 vs. 0.39); however, both sets of tag sequences were on average very infrequently expressed (Fig. 4A). In contrast, the 13,161 tag sequences detected by both methods were highly expressed and had an intermediate GC-content (0.43) that was nearly identical to the average GC-content of all Ensembl transcript tag sequences (0.42). We looked at whether the correlation of expression of these common tag sequences differed as a function of tag GC-content. We divided the tags into four bins representing increasing proportions of tag GC-content (bin1: 0%–25%; bin2: 25%–45%; bin3: 45%–65%; bin4: 65%–100%), and found that the Pearson correlation changed as a function of GC-content, with AT-rich tags having the lowest correlation (Fig. 4B).
Figure 4.
GC-content biases in Tag-seq and LongSAGE technical replicate libraries. (A) Comparison of the GC-content and average count of tag sequences found either in common or by each of the Tag-seq and LongSAGE replicate libraries. (B) Pearson correlations were calculated for tags binned by GC-content. Bins are labeled with the range of the observed GC-content, and the number of binned tags (x-axis). (C) Average expression of tag sequences in each GC-content bin was calculated for both Tag-seq and LongSAGE, and the log of each average was plotted. An asterisk (*) denotes bins between which the expression of tag sequences was significantly different (measured using a t-test, P < 0.01).
We investigated the cause of the decreased correlation between AT-rich tag sequences in the two methods, and found a relationship between tag abundance and tag GC-content. In LongSAGE we observed a positive correlation between tag abundance and GC-content for the first three bins (bin1 vs. bin2, P = 1.6 × 10−3; bin2 vs. bin3, P = 1.4 × 10−3; t-test). In contrast, the abundance of the same tag sequences in the Tag-seq replicate did not correlate with GC-content, with the exception of the most GC-rich bin (bin3 vs. bin4, P = 9.4 × 10−8; Fig. 5C). This relationship between GC-content and tag abundance held for all Tag-seq and all LongSAGE libraries (Supplemental Fig. S5).
Figure 5.
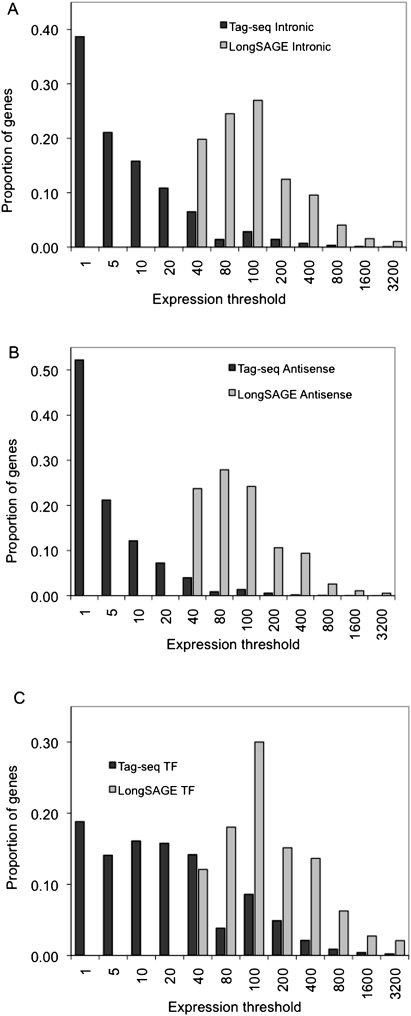
The proportion of the average number of genes detected by tags in LongSAGE and Tag-seq libraries is shown at a series of expression thresholds (tags per million). Bars represent the proportion of the average number of genes with intronic tags (A), antisense tags (B), and DNA-binding domains (transcription factors) (C) in Tag-seq and LongSAGE libraries.
Improved representation of low abundance LongSAGE transcripts in Tag-seq libraries
Given the increased depth of Tag-seq libraries, we expected to observe increased numbers of tags for transcripts at the limit of detection in LongSAGE (Siddiqui et al. 2005). Two such tag categories include antisense and intronic tags. Antisense tags originate from transcripts that are transcribed from the opposite strand (Supplemental Fig. S6), while intronic tags may represent unannotated exons and UTRs within known genes (Saha et al. 2002), or previously unannotated sequences transcribed from introns, such as embedded genes (e.g., HA_003240, Hirst et al. 2007) or miRNA genes (Kim 2005). Another class of generally low abundance transcripts of biological interest consists of transcription factors (TFs). To investigate the expression levels of TFs in Tag-seq and LongSAGE libraries, we downloaded the set of 2890 human genes that encoded DNA-binding domains (DBD) (http://dbd.mrc-lmb.cam.ac.uk/DBD/index.cgi?About), which should include all TFs, and searched for their presence in the CGAP libraries.
We enumerated tag sequences that mapped in the sense orientation to TF exons, antisense to known genes, and sense to gene introns, in each library, at increasing thresholds of expression. Overall, an average Tag-seq library detected 1.7 times as many TF genes as a LongSAGE library (849 vs. 504), 6.3 times as many genes with antisense (AS) tags (4999 vs. 795), and 2.8 times more genes with intronic tags (7651 vs. 2752). The majority of genes found by Tag-seq were at expression levels below those detectable in existing LongSAGE libraries (Fig. 5).
We confirmed the relationship between sequencing depth and the diversity and abundance of intronic and antisense tags by analyzing the Tag-seq and LongSAGE hESC replicate libraries. To ensure that the relationship between tag sequence diversity and tag abundance was due to no other factors except depth, we generated an in silico library of 272,465 randomly subsampled tags from the Tag-seq replicate. The in silico library, hereafter referred to as sub_Tag-seq, theoretically represents a random sample of the most highly expressed tags in the Tag-seq replicate and should, therefore, be very similar to the LongSAGE replicate. We found that sub_Tag-seq was moderately correlated with the LongSAGE replicate (Pearson correlation of 0.6), with most of the variation coming from low frequency tags (data not shown). Any differences in the abundance of intronic and antisense tags in the sub_Tag-seq library relative to the Tag-seq library would most likely be due to decreased depth.
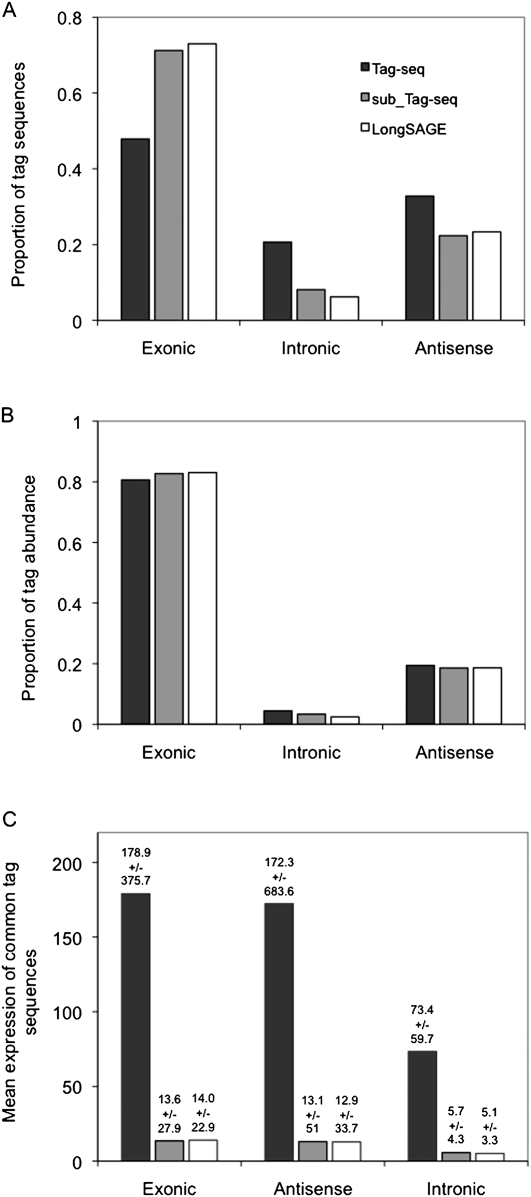
A comparison of the Tag-seq replicate, sub_Tag-seq, and the LongSAGE replicate supports the described increase in the diversity of intronic and antisense tags in deeper libraries. We compared the proportion of tag sequences in each library that mapped either to exons, introns, or to the antisense strand of Ensembl genes (Fig. 6A). In the Tag-seq replicate, the most abundant categories of mapped tag sequences were exonic tags (47.8%), closely followed by antisense tags (32.1%) and intronic tags (20.6%). In contrast, the LongSAGE replicate was far more likely to detect tags mapping to exons (73.0%) than antisense (23.4%) or intronic tags (6.2%). Thus, the Tag-seq replicate is enriched in antisense and intronic tag sequences; this enrichment is not observable at sampling depths <300,000 tags, since the tags in sub_Tag-seq library mapped in proportions similar to those of the LongSAGE replicate (differences were not significant). These observations held when comparing all Tag-seq to all LongSAGE libraries (Supplemental Fig. S7), indicating that low-frequency antisense and intronic tags were present in all the profiled human tissues and were not specific to hESCs. The altered proportion of antisense, intronic, and exonic tag sequences was highly significant (t-test between Tag-seq and LongSAGE tag sequence proportions: antisense P = 6.2 × 10−5, intronic P = 1.0 × 10−10, exonic P = 1.6 × 10−24).
Figure 6.
Detection of exonic, intronic, and antisense tags in the Tag-seq and LongSAGE hESC replicates. Tag sequences from the Tag-seq technical replicate, the in silico derived sub_Tag-seq, and the LongSAGE replicate were mapped to the introns, exons, and antisense strands of Ensembl genes. The proportions of distinct tag sequences (A) and tag abundance (B) are reported relative to all mapped quality-filtered tags. Average tag counts (±SD) are reported for all tag sequences found in common between the three libraries (C).
Interestingly, the abundance of exonic, intronic, and antisense tag sequences was almost identical between methods (Fig. 6B; Supplemental Fig. S7B). This suggests that the large numbers of low frequency tag sequences detected only in Tag-seq were expressed in the same relative ratios as higher frequency tag sequences detected by both methods. Thus, exonic tags were the most abundantly expressed (∼80%), followed by antisense tags (∼20%), and intronic tags (0.1%).
The additional depth in Tag-seq had a dramatic effect on the dynamic range of expression of moderate to abundantly expressed tags, which could be detected by both methods. On average, exonic tag sequences were detected at frequencies 12.7-fold higher in the Tag-seq versus the LongSAGE replicate, and antisense and intronic tag sequences were detected at levels 13.4- and 14.4-fold higher (Fig. 6C; Supplemental Fig. S7C). The range of expression was an order of magnitude higher in Tag-seq versus LongSAGE, indicating a significantly greater dynamic range of expression.
Sense–antisense transcripts in cancer libraries
Having assessed the technical differences between the LongSAGE and Tag-seq protocols, we undertook a biological analysis of the CGAP library collection. We first analyzed the AS tags with a focus on their differential expression in libraries representing cancerous and normal tissue samples. Previous studies have shown that the ratio of sense to antisense transcripts changes between normal and malignant tissue samples (Chen et al. 2005), and that antisense transcripts can be implicated in disease processes (Tufarelli et al. 2003; Reis et al. 2004). Our goal was to highlight the potential of the Tag-seq approach to identify known and novel antisense transcripts whose expression ratios changed significantly with respect to the sense gene, between normal and diseased states, between different stages of disease progression, or between cancer subtypes.
To achieve this, libraries were first grouped by tissue into 15 groups (Supplemental Table S1; Supplemental Methods). Libraries belonging to each tissue were segregated into groups representing normal and cancerous samples and, when possible, were further segregated into cancer stages (precancerous samples vs. malignant for instance; Supplemental Table S4). The ratio of sense to antisense transcription between each of the tissue groups was assessed at every relevant locus; either using pairs of sense tags mapping to known sense–antisense (S-AS) gene pairs, or using sense tags mapping to single genes with a corresponding novel tag mapping antisense to the same gene (abbreviated Single-AS; Supplemental Fig. S6).
Altered expression ratios between 389 S-AS gene pairs and between 2195 Single-AS pairs were found in the 15 tissue groups. Random assignment of tags to genes showed that real S-AS genes were, on average, 55 times more likely to have ratio changes than would be expected by chance, while Single-AS were 17.5 times more likely than expected by chance, suggesting a higher rate of false positives in these pairs. We developed a normalization protocol to identify pairs with large expression ratio changes (Supplemental Methods), and to ensure higher ranking of highly expressed gene pairs and of those pairs with lower variance in their ratios. Overall, tissues comprised solely of Tag-seq or LongSAGE libraries had equivalent numbers of gene pairs with ratio changes. Since the tissues profiled by the different methods were distinct, we could make no a priori predictions regarding the number of gene pairs with different ratios found by Tag-seq or LongSAGE. By definition, the genes targeted by this analysis are moderately to highly expressed, and could be found by both methods. Thus, in the absence of Tag-seq and LongSAGE replicates for a whole tissue, we conclude that both methods are capable of finding gene pairs whose abundance ratios change between cancerous and normal samples, and which therefore may be differentially regulated in cancer versus normal tissues.
To determine whether there was an enrichment of biological categories in these genes, we conducted a functional annotation clustering analysis (Dennis et al. 2003; Huang et al. 2007). In this analysis, annotations (such as GO terms; Ashburner et al. 2000) that share common genes are more likely to be grouped together. We found that genes with extreme ratio changes (in the top 10%) were highly enriched in GO terms relating to the regulation of developmental processes, to the regulation of cell death, and to cell proliferation (Supplemental Table S5), terms which are relevant to cancer biology.
To further evaluate the biological relevance of these pairs, we enumerated the number of Cancer Gene Census genes in the data set (Futreal et al. 2004). This is a catalog of genes with mutations that have been causally implicated in multiple cancers. Of the total 312 cancer census genes, expression was detected in the CGAP data set for 300. Interestingly, over one quarter of these genes (72 Single-AS and six S-AS) were also found to have significant ratio changes between normal and cancerous libraries in the studied tissues (Table 2; Supplemental Table S6). The pairs with ratio differences in the top 10% of the range of differences were identified, revealing a total of 30 of the cancer census genes remaining in this shortlist (27 Single-AS, three S-AS). Thus, 38% of the cancer genes were in the top 10% of differentially expressed genes with extreme ratio changes between cancer and normal tissues, which is a significant enrichment (P < 7.0 × 10−4, χ2 test).
Table 2.
The proportion of S-AS and Single-AS genes that were differentially expressed and belonged to the cancer census gene set
Transcript isoforms in cancer libraries
Differential expression of transcript isoforms was analyzed in 4237 genes with multiple expressed tags, since these tags potentially represent alternative 3′ polyadenylation sites (Siddiqui et al. 2005). A total of 1957 of these genes had tag pairs whose ratio of expression changed between libraries grouped by disease state (e.g., cancerous vs. normal). For 1304 (66.6%) of these genes, the sequence bounded by the two tags harbored predicted miRNA targeting sites (Grimson et al. 2007), suggesting that miRNAs may regulate isoform expression in one of the two states (Hirst et al. 2007). The proportion of miRNA-targeted genes in this list was nearly three times greater than the proportion of miRNA-targeted genes in the human genome (22.0%, P < 2.2 × 10−16, χ2 test; Table 3). Of the 772 genes with transcript pairs that had the 10% most extreme expression ratio changes, we found an additional enrichment of transcripts harboring miRNA targeting sites (72.5%; Table 3). For 33.1% of these genes, the longer isoform was consistently more abundant in cancers; for 41.0% of these genes, the shorter isoform was consistently more abundant in cancer; for the remaining 26.9% of genes, either isoform was more abundant in cancer in some sample.
Table 3.
Genes with isoforms that were differentially expressed between two disease states were enriched in miRNA targeting sites relative to all genes
The enrichment of miRNA targeting sites increased further for those genes with differential expression values in the top 20% and even further for those in the top 10%.
We found 93 miRNA targeting sites with enriched frequencies in the set of genes with the top 10% most extreme expression isoform ratio changes (versus the frequencies in the set of all genes with isoform ratio changes, P < 0.05, hypergeometric distribution test; Supplemental Table S7). A closer look at the most enriched sites showed that these miRNAs have been previously observed to have altered expression in cancers (e.g., miR-124 in glioblastoma multiforme [Silber et al. 2008]; miR-181 and miR-15/16 in B-cell chronic lymphocytic leukemia [Calin et al. 2002; Pekarsky et al. 2006]; miR-224 in thyroid tumors and in hepatocellular carcinoma [Nikiforova et al. 2008; Wang et al. 2008a]).
Discussion
To complete the CGAP digital gene expression profiling project, we developed Tag-seq as an efficient and cost-effective alternative to LongSAGE. Tag-seq library construction is similar to the LongSAGE protocol, but sequencing employs Illumina's massively parallel sequencing by synthesis protocol in place of conventional Sanger sequencing. Every read in a sequenced Tag-seq library represents a 17-bp sequence tag adjacent to the 3′ most NlaIII site of an individual transcript and, therefore, represents a digital count of that transcript.
Relative to another Illumina-based transcript profiling technology, RNA-seq (Marioni et al. 2008; Rosenkranz et al. 2008), Tag-seq performs comparably in terms of gene discovery and measured dynamic range. For gene expression profiling experiments where accurate profiling of transcripts from both strands of the genome is required, Tag-seq data are superior since, unlike RNA-seq, it allows discrimination of sense and antisense transcripts. Sense and antisense genes are encoded on the opposite strands of the same genomic locus and yield transcripts that have sequence complementarity. Their genomic arrangement and sequence complementarity increase the likelihood that their regulation is affected by common factors (such as chromatin state) and their relative expression (such as transcriptional interference), at both the transcriptional and post-transcriptional level (Vanhee-Brossollet and Vaquero 1998; Dahary et al. 2005). To date, antisense transcripts have been observed for up to 75% of the mammalian transcriptome in data sets generated by both sequence-based and hybridization-based methods (Katayama et al. 2005). Given the high prevalence of antisense transcription in the mammalian genome, and the link between antisense transcripts and disease (Tufarelli et al. 2003; Reis et al. 2004), Tag-seq was well suited to the study of cancer-relevant gene expression in the context of the CGAP project. We found known and novel S-AS gene pairs for which the ratio of expression changed significantly between cancer subtypes or between cancer and normal states. These were enriched in known cancer-related genes, supporting a role for antisense transcription in cancer biology. For instance, we found evidence for antisense transcription at the BCL6 locus, which encodes a repressor of transcription known to be involved in lymphomas. Antisense ESTs have previously been observed at this locus, lending support to our observations of antisense transcription (Supplemental Fig. S8). The ratio between the sense and antisense tags at this locus was significantly up-regulated in the subset of libraries from grade II carcinoma epithelium and associated myofibroblast samples, leading to reduced sense-to-antisense ratio in those samples. These libraries represented cell types sampled from one breast cancer patient, implicating the relationship between BCL6 and its antisense transcript in the biology of this individual breast cancer. While carcinoma-associated myofibroblasts are not necessarily cancer cells per se, they have epigenetic alterations similar to those seen in malignant carcinoma epithelium, and are globally hypomethylated (Jiang et al. 2008). One plausible explanation for the increase in antisense expression at this locus is increased hypomethylation at CpG islands downstream from the BCL6 gene (Supplemental Fig. S8).
While Tag-seq is able to distinguish transcript strand of origin, it only provides limited information regarding transcript structure. Thus, to gather data on expressed transcript isoforms, exon arrays or RNA-seq would be the more suitable technologies. However, Tag-seq is still informative on the expression of the subset of gene isoforms that lead to a different 3′ NlaIII tag sequence as a consequence of alternative 3′ end formation. We were able to analyze >4200 genes with such transcript isoforms and expression in CGAP, and to find differential expression of isoforms between cancer and normal states. Intriguingly, we found an enrichment of transcripts harboring miRNA targeting sites in the sequence unique to one of two differentially expressed isoforms (Hirst et al. 2007; Ghosh et al. 2008), implicating their regulation in cancer biology.
Compared to Affymetrix microarrays, Tag-seq is capable of de novo gene discovery without the requirement of genome-wide probe design, does not suffer from cross-hybridization of related sequences, and achieves essentially unlimited dynamic range simply by increasing sequencing depth. At the current level of sampling (∼10 million tags), genes detected by Tag-seq had a 13-fold greater measurable fold change than the same genes detected by Affymetrix.
Relative to LongSAGE, the additional depth of sampling provided by Tag-seq led to a greater number of genes identified in a given tissue, and improved the measurable dynamic range of those genes. One other report has thus far shown that Tag-seq surpasses LongSAGE in sequencing depth (Hanriot et al. 2008). We extend these findings by reporting for the first time that, with increasing depth, Tag-seq also allowed detection of a distinct subset of transcriptome space, enriched in AT-rich genes, intronic tags, antisense tags, and novel intergenic tags. The enhanced detection of low-frequency AT-rich tag sequences in Tag-seq was similar to previous observations made in Affymetrix arrays (Siddiqui et al. 2006), although the detection of AT-rich sequences was in that case interpreted as a technological bias. These new results suggest that this AT-rich class of tag sequences do not represent technical bias in either method, but rather a biological difference in the types of transcripts present at lower frequencies, which is detectable using both sequencing-based and hybridization-based technologies. The depth of sampling achieved by LongSAGE is not large enough to detect this subset of the transcriptome. Furthermore, we found that Tag-seq has less GC-bias, leading to a more accurate interpretation of the abundance of tags spanning the range of GC-content.
Overall, Tag-seq identifies more genes than LongSAGE, detects a greater dynamic range of expression, and thus allows differential expression analysis for a greater range of transcripts. Tag-seq libraries provide an excellent resource for the discovery of known and novel transcripts with expression changes relevant to disease processes, and highlight the applicability of next-generation tag sequencing to gene expression profiling.
Methods
Tag-seq library construction
All libraries were constructed using one of two protocols: Tag-seq or Tag-seqLite. Tag-seq is a variant of LongSAGE as described (Siddiqui et al. 2005; Khattra et al. 2007), with modifications forgoing the requisite production of ditags and concatemers and allowing direct sequencing on the Illumina Genome Analyzer (Fig. 1). Typically 500–2000 ng of DNase I-treated total RNA was used in library construction. Briefly, after double-stranded cDNA synthesis using oligo(dT) beads (Invitrogen) the cDNA was digested with an anchoring restriction enzyme (NlaIII) and ligated to Illumina specific adapter, Adapter A, containing a recognition site for the TypeIIS tagging enzyme MmeI (New England Biolabs). Following MmeI digestion, dephosphorylation with shrimp alkaline phosphatase (USB Corp), and purification, a second Illumina adapter, Adapter B, containing a 2-bp degenerate 3′ overhang was ligated (Fig. 1). Tags flanked by both adapters were enriched by PCR using Phusion DNA polymerase (Finnzymes) and Gex PCR primers 1 and 2 (Illumina) following the manufacturer's instructions. Separate 15 and 17 cycle reactions were run using the following program: 98°C for 30 sec, followed by 15–17 cycles of 98°C for 10 sec, 60°C for 30 sec and 72°C for 15 sec, and then 72°C for 5 min. The PCR products were run on a 12% PAGE gel and the ∼85-bp DNA band was excised and purified using Spin-X filter column (Costar) followed by ethanol precipitation. The DNA quality was assessed and quantified using an Agilent DNA 1000 series II assay (Agilent) and Nanodrop 7500 spectrophotometer (Nanodrop), and the DNA sample was diluted to 10 nM. Cluster generation and sequencing was performed on the Illumina cluster station and analyzer (Illumina) following the manufacturer's instructions. Raw sequences were extracted from the resulting image files using the open source Firecrest and Bustard applications (Illumina) on a 32 CPU cluster running Red Hat Enterprise Linux 4 (Red Hat) and Sun Grid Engine 6 (Sun Microsystems). For samples with RNA amount ranging 4–50 ng, Tag-seqLite was applied. Briefly, first strand cDNA was synthesized with Superscript II Reverse Transcriptase (Invitrogen) and was amplified by a 20-cycle PCR according to the SAGE-Lite method. SAGE-Lite biochemistry is based upon the SMART (switching mechanism at the 5′ end of RNA transcripts) cDNA synthesis strategy (Clontech) for the generation of full-length cDNA. Following the amplification, 500 ng of cDNA was processed according to the standard Tag-seq protocol as described above, except that the final PCR amplification was 13–15 cycles.
Tag extraction
Sequencing of a Tag-seq amplicon starts at the first base following the Adapter A sequence. Thus, the first 17 to 18 bases of a read are the transcript-derived tag sequence, and the remaining bases are the Adapter B sequence. As expected, 99% of adapters found in a Tag-seq library occur in positions 18 and 19 of the read. The “Raw” Tag-seq library is then constructed by truncating all reads at length 17.
All statistical analyses and methods used for tag analysis are as previously described for LongSAGE (Siddiqui et al. 2005).
Ensembl data
Full gene sequences (including introns), cDNA sequences, and gene boundary coordinates were downloaded from the Ensembl version 47 release (Birney et al. 2004), based on the NCBI human genome build 36, using the Ensembl API (www.ensembl.org). Virtual sense and antisense tag sequence databases were generated for both full gene and cDNA sequences using in-house Perl scripts. Briefly, all NlaIII sites were identified for each sequence, and the adjoining 17 bp in the 3′ direction were designated the sense tags, while the 17 bp in the 5′ direction were designated the antisense tags. The human genome sequence was downloaded from NCBI (ftp://ftp.ncbi.nih.gov), and the complete sequence, including repeat regions, was used to create virtual sense and antisense tag databases. Sense and antisense tag sequences mapping to unique locations in the genome were distinguished from those mapping in multiple locations.
Acknowledgments
We thank Don Riddle and Peter Ruzanov for helpful discussions, Malachi Griffith for providing Affymetrix array data, Asim Siddiqui for insights into GC-bias, and Scott DiGuistini for providing Blue Stain fungus data. We thank Daniela Gerhard for helpful discussions and guidance, and the anonymous reviewers for comments that strengthened the manuscript. This study was funded in part by the National Institutes of Health (contract no. N01-SO-12400), the National Cancer Institute Office of Cancer Genomics (contract no. 27xS053), and the Genome Canada/Genome BC MORGEN Project. A.S.M. is a trainee funded by the Canadian Institutes of Health Research, and M.A.M. is a Michael Smith Foundation for Health Research Senior Scholar and a Terry Fox Young Investigator.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094482.109.
References
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Dahary D, Elroy-Stein O, Sorek R. Naturally occurring antisense: Transcriptional leakage or real overlap? Genome Res. 2005;15:364–368. doi: 10.1101/gr.3308405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B. MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse cytoplasmic β-actin gene. Nucleic Acids Res. 2008;36:6318–6332. doi: 10.1093/nar/gkn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanriot L, Keime C, Gay N, Faure C, Dossat C, Wincker P, Scote-Blachon C, Peyron C, Gandrillon O. A combination of LongSAGE with Solexa sequencing is well suited to explore the depth and the complexity of transcriptome. BMC Genomics. 2008;9:418. doi: 10.1186/1471-2164-9-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Hirst M, Delaney A, Rogers S, Schnerch A, Persaud D, O'Connor M, Zeng T, Moksa M, Fichter K, Mah D, et al. LongSAGE profiling of nine human embryonic stem cell lines. Genome Biol. 2007;8:R113. doi: 10.1186/gb-2007-8-6-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Gonda TA, Gamble MV, Salas M, Seshan V, Tu S, Twaddell WS, Hegyi P, Lazar G, Steele I, et al. Global hypomethylation of genomic DNA in cancer-associated myofibroblasts. Cancer Res. 2008;68:9900–9908. doi: 10.1158/0008-5472.CAN-08-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Khattra J, Delaney AD, Zhao Y, Siddiqui A, Asano J, McDonald H, Pandoh P, Dhalla N, Prabhu AL, Ma K, et al. Large-scale production of SAGE libraries from microdissected tissues, flow-sorted cells, and cell lines. Genome Res. 2007;17:108–116. doi: 10.1101/gr.5488207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Lal A, Lash AE, Altschul SF, Velculescu V, Zhang L, McLendon RE, Marra MA, Prange C, Morin PJ, Polyak K, et al. A public database for gene expression in human cancers. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu A-L, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KL, Hogh AL, Emmersen J. DeepSAGE—digital transcriptomics with high sensitivity, simple experimental protocol and multiplexing of samples. Nucleic Acids Res. 2006;34:e133. doi: 10.1093/nar/gkl714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu C-G, Rassenti L, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- Peters DG, Kassam AB, Yonas H, O'Hare EH, Ferrell RE, Brufsky AM. Comprehensive transcript analysis in small quantities of mRNA by SAGE-Lite. Nucleic Acids Res. 1999;27:e39. doi: 10.1093/nar/27.24.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- Rosenkranz R, Borodina T, Lehrach H, Himmelbauer H. Characterizing the mouse ES cell transcriptome with Illumina sequencing. Genomics. 2008;92:187–194. doi: 10.1016/j.ygeno.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20:508. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, et al. A mouse atlas of gene expression: Large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc Natl Acad Sci. 2005;102:18485–18490. doi: 10.1073/pnas.0509455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui AS, Delaney AD, Schnerch A, Griffith OL, Jones SJM, Marra MA. Sequence biases in large scale gene expression profiling data. Nucleic Acids Res. 2006;34:e83. doi: 10.1093/nar/gkl404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Lim D, Petritsch C, Persson A, Maunakea A, Yu M, Vandenberg S, Ginzinger D, James CD, Costello J, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- Vanhee-Brossollet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- von Bubnoff A. Next-generation sequencing: The race is on. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lee ATC, Ma JZI, Wang J, Ren J, Yang Y, Tantoso E, Li K-B, Ooi LLPJ, Tan P, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008a;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008b;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]