Abstract
Mobile genetic elements play important roles in the evolution and diversification of bacterial genomes. In enterohemorrhagic Escherichia coli O157, a major factor that affects genomic diversity is prophages, which generate most of the large-size structural polymorphisms (LSSPs) observed in O157 genomes. Here, we describe the results of a systematic analysis of numerous small-size structural polymorphisms (SSSPs) that were detected by comparing the genomes of eight clinical isolates with a sequenced strain, O157 Sakai. Most of the SSSPs were generated by genetic events associated with only two insertion sequence (IS) elements, IS629 and ISEc8, and a number of genes that were inactivated or deleted by these events were identified. Simple excisions of IS629 and small deletions (footprints) formed by the excision of IS629, both of which are rarely described in bacteria, were also detected. In addition, the distribution of IS elements was highly biased toward prophages, prophage-like integrative elements, and plasmids. Based on these and our previous results, we conclude that, in addition to prophages, these two IS elements are major contributors to the genomic diversification of O157 strains and that LSSPs have been generated mainly by bacteriophages and SSSPs by IS elements. We also suggest that IS elements possibly play a role in the inactivation and immobilization of incoming phages and plasmids. Taken together, our results reveal the true impact of IS elements on the diversification of bacterial genomes and highlight their novel role in genome evolution.
Insertion sequence (IS) elements are the smallest autonomously mobile genetic elements (generally 700–2500 base pairs [bp] in size) and are widely distributed in both eukaryotic and bacterial genomes. Most bacterial IS elements are of the DNA type (Chandler and Mahillon 2002). They only encode transposases (TPases) and thus are phenotypically cryptic by themselves, but the insertion/transposition of IS elements can result in gene inactivation and/or affect the expression of adjacent genes. IS elements can also induce a variety of genomic rearrangements, including deletions, inversions, and duplications (Wei et al. 2003; Kothapalli et al. 2005; Iguchi et al. 2006). Thus, it is generally assumed that IS elements play important roles in bacterial genome evolution. During long-term experimental evolution of Escherichia coli strain B in vitro (Papadopoulos et al. 1999; Schneider et al. 2000), several IS-mediated mutations were detected; some of these mutations contributed to adaptation to or improved fitness in the medium used for the experiment (glucose minimal medium). Several comparative genome analyses have also revealed that IS elements induce large genomic inversions and translocations in some bacterial species (Jin et al. 2002; Parkhill et al. 2003; Yang et al. 2005; Chain et al. 2006; Rohmer et al. 2007; Naito et al. 2008; Salzberg et al. 2008). However, it remains largely unknown how and to what extent IS elements affect genome diversification among closely related bacterial strains or species.
IS elements are classified into about 20 families based on various features, such as the sequences of the TPases and the terminal inverted repeats. The mechanism of transposition differs between IS families, but most IS elements transpose through either the nonreplicative (cut-and-paste) or the replicative (copy-and-paste) mechanisms (Chandler and Mahillon 2002). The IS3 family is one of the largest IS families, and its molecular mechanism of transposition has been well characterized. In several IS3 family members, such as IS2, IS3, IS150, and IS911, transposition has been shown to be achieved through the formation of a “figure eight” intermediate (Lewis and Grindley 1997; Sekine et al. 1999; Haas and Rak 2002; Rousseau et al. 2002; Ohtsubo et al. 2004). One study revealed that in IS911, a circular form that integrates into a new insertion site is replicatively generated through the formation of a “figure eight” intermediate (Duval-Valentin et al. 2004). On the other hand, very little is known about the fate of the donor DNA in IS transpositions, although some kind of genomic alteration could occur in the chromosomal regions that serve as the donor DNA. A detailed structural comparison of closely related genomes that contain a number of active copies of IS3 family members may shed some light on this unresolved scientific issue.
E. coli comprise a wide variety of strains with different phenotypes. Most are commensal inhabitants of vertebrates and rarely cause diseases, except in immunocompromised hosts. However, several E. coli clones or lineages have acquired specific sets of virulence-related genes that cause a range of diseases in humans. Among the various types of pathogenic strains, enterohemorrhagic E. coli (EHEC) O157:H7, which causes diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome, is one of the most important food-borne pathogens in many industrialized countries (Mead and Griffin 1998).
The chromosome of the sequenced O157:H7 strain RIMD 0509952 (referred to as O157 Sakai) contains 1.4 Mb of sequences that are not present in the benign laboratory E. coli strain K-12. These Sakai-specific sequences, or S-loops, are scattered throughout a 4.1-Mb chromosome backbone that is shared with K-12 (Hayashi et al. 2001). Most of the large Sakai-specific sequences are prophages and prophage-like integrative elements (Sp1-Sp18 and SpLE1-SpLE6, respectively), and many of the virulence-related genes in O157 have been introduced into the O157 genome by these genetic elements or a large plasmid, pO157 (Makino et al. 1998; Hayashi et al. 2001). K-12 also contains 11 prophages and prophage-like elements, implying that bacteriophages are major contributors to the genomic diversification of E. coli (Ohnishi et al. 2001; Hayashi et al. 2008). The O157 Sakai genome is also rich in IS elements; 25 types of IS elements (98 copies in total) have been identified, and 37 of the 98 copies are apparently intact (Hayashi et al. 2001). These intact IS elements, including IS629, an IS3 family member (19 copies), ISEc8 (eight copies), IS677 (three copies), IS1-Nuxi and IS609 (two copies each), and three other types (one copy each), may also contribute to genomic diversification in the O157 lineage.
We previously compared the genomic structures and compositions of eight O157 clinical strains isolated in Japan in 1998 with those of O157 Sakai by whole genome PCR scanning (WGPS), a long-range PCR-based method for fine comparison of genome structures (Ohnishi et al. 2002), and comparative genomic hybridization (CGH) using an O157 oligo DNA microarray (Ogura et al. 2006). The results of these analyses and the data from another laboratory (Wick et al. 2005) revealed that an unexpectedly high degree of genomic diversity exists in the O157 lineage. Among the numerous structural polymorphisms identified, the longer polymorphisms were exclusively observed in regions harboring prophages, indicating that prophages are a major contributor to genomic diversity within the O157 lineage (Ohnishi et al. 2002). This conclusion is supported by the results of genomic comparisons of O157 substrains obtained from long-term subculturing in vitro (Iguchi et al. 2006). In our WGPS analysis of the eight O157 strains, we also identified numerous genomic segments that exhibited small size differences, ranging from a few hundred bases to several kilobases. However, it is not known how these small-size structural polymorphisms (referred to as SSSPs) are generated.
In the present study, we systematically analyzed SSSP-containing genomic segments found in the eight O157 strains. Most were generated by genetic events associated with two IS elements, IS629 and ISEc8, and we identified several genes that were inactivated or deleted by such events. We also found simple excisions of IS629 and small deletions that were generated upon the excision of IS629, both of which have rarely been described in bacteria. These results indicate that these two IS elements should be regarded as major factors contributing to the genomic diversification of O157. The distribution of IS elements in O157 is highly biased toward prophages, prophage-like integrative elements, and plasmids, suggesting that these elements may play a role in the inactivation and immobilization of incoming phages and plasmids.
Results
Systematic analysis of SSSP-containing genomic regions
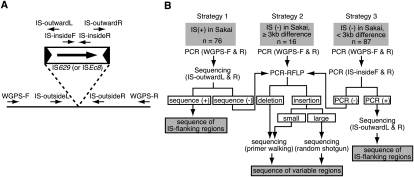
We examined 179 SSSP-containing genomic segments that were identified in the eight O157 strains (hereafter referred to as strains #2–#9) by WGPS (Ohnishi et al. 2002). We first randomly selected 20 SSSP-containing segments and amplified them by long-range PCR with the primers used in our previous WGPS analysis (Ohnishi et al. 2002). The PCR products were digested with one or two restriction enzymes, and the resulting digestion patterns were compared with those of the reference strain O157 Sakai (restriction fragment length polymorphism analysis of the PCR products [PCR-RFLP]) to identify subregions of each segment that exhibit altered digestion patterns compared with the reference. We then determined the sequences of these subregions by primer walking based on the Sakai genome sequence. This analysis revealed that most of the 20 SSSPs were introduced by small genomic rearrangements related to two IS elements, IS629 and ISEc8 (15 of IS629 and two of ISEc8), suggesting that the two IS elements are frequently associated with SSSPs. Therefore, we categorized the 179 segments into three groups (Fig. 1B): those in which IS629 and/or ISEc8 were present in O157 Sakai (Group 1) and those in which neither IS element was present in O157 Sakai with size differences of >3 kb (Group 2) or <3 kb (Group 3) compared with O157 Sakai in the WGPS analysis. We then systematically analyzed each group using three different strategies (Fig. 1B, Strategies 1–3).
Figure 1.
Experimental scheme for the analysis of SSSP-containing regions. (A) The locations of the PCR primers specific to IS629 or ISEc8 (IS-insideF and -insideR and IS-outwardL and -outwardR) and to each IS-flanking genomic region (IS-outsideL and -outsideR). (B) The three strategies used to analyze the three types of SSSP-containing regions.
All strategies involved amplification of each segment by long-range PCR with the primer sets prepared in our previous WGPS analysis (Ohnishi et al. 2002). In Strategy 1 (for Group 1), we directly sequenced the long-range PCR products with IS-outward primers (Fig. 1A) to determine whether the IS629 or ISEc8 elements had inserted into the same locus as in O157 Sakai. In Strategy 2 (for Group 2), we identified subregions displaying SSSPs in each segment by PCR-RFLP analysis and determined the DNA sequences of these subregions. In Strategy 3 (for Group 3), we searched for the presence of IS629 or ISEc8 in each segment by IS629-specific or ISEc8-specific PCRs and, if the segment contained either IS element, determined the sequence of the IS insertion site by direct sequencing of the long-range PCR products with IS-outward primers. When Strategy 1 or Strategy 3 did not work for a segment, the segment was further analyzed using Strategy 2.
These strategies allowed us to define the genetic events that generated SSSPs in 165 segments. The results are summarized in Table 1 and Figure 2. The 14 segments that we failed to characterize were parts of lambda-like prophage genomes. O157 contains multiple lambda-like prophages that are similar to one another. All 14 segments are located within such prophage regions. Some structural alterations, such as partial replacement of the prophage genomes, have probably taken place in these segments.
Table 1.
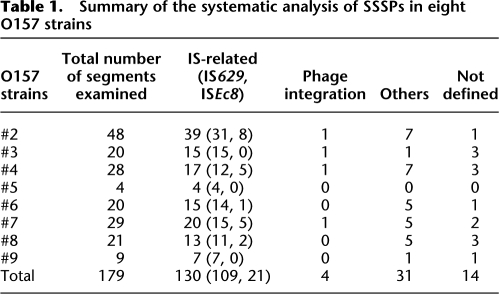
Summary of the systematic analysis of SSSPs in eight O157 strains
Figure 2.
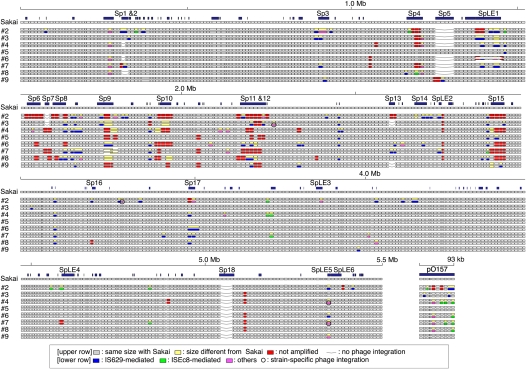
Systematic SSSP analysis results for eight O157 strains. (Upper row) For each strain, WGPS data (Ohnishi et al. 2002) are presented; (lower row) the results of the SSSP analysis. In the WGPS data, segments that yielded PCR products with lengths identical to or different from O157 Sakai are represented by gray or yellow rectangles, respectively; (red) those that were not amplified. In this study, we analyzed all of the yellow segments, which contain SSSPs. (Blue) Segments found to contain IS629-related genomic rearrangements; (light green) those containing ISEc8-related rearrangements; (magenta) segments containing other types of structural changes; of these, segments containing strain-specific phage integrations are circled; (blue rectangles) genomic regions >500 bp in length that are not present in K-12 (Sakai-specific regions or S-loops). Loci lacking prophages (Sp1–Sp18) or prophage-like integrative elements (SpLE1–SpLE6) are indicated by a line.
Of the 165 SSSPs, 130 (78.8%) were generated by IS-related genetic events, four (2.4%) by strain-specific prophage integration, 19 (11.5%) by small genomic deletion probably induced by homologous recombination, and 12 (7.3%) by unknown mechanisms. Because several segments exhibited different types of SSSPs between different strains, the total number of SSSP-containing genomic loci identified was 119 (105 in the chromosome and 14 in the pO157 plasmid). IS-related structural alterations were found in 98 of the 119 loci (82.4%), indicating that IS elements are the predominant way in which SSSPs are generated in O157 genomes.
IS-related genomic rearrangements
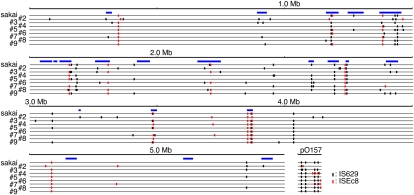
The 130 IS-related SSSPs detected in the 98 genomic loci described above were generated by two types of IS elements: IS629, which generated 109 SSSPs at 77 loci, and ISEc8, which generated 21 SSSPs at 20 loci. Although most of the ISEc8-related SSSPs (16 of 20 loci) were single strain-specific ISEc8 insertions, we identified eight additional genomic loci within which ISEc8 was inserted in all strains (and thus showed no structural polymorphisms). In contrast, the distribution of IS629 in the O157 genomes exhibited highly variable and complex patterns among the strains, and IS629 appeared to have induced various types of genomic rearrangements. The distributions of IS629 and ISEc8 in the nine O157 genomes, including that of O157 Sakai, are summarized in Figure 3.
Figure 3.
Distribution of IS629 and ISEc8 in the nine O157 genomes used in this study. The locations of IS629 and ISEc8 elements on each chromosome and plasmid are shown. (Black vertical lines) Insertion sites for IS629; (red lines) insertion sites for ISEc8; (blue rectangles) regions corresponding to prophages and phage-like integrative elements.
Simple insertion sites of IS629 and ISEc8
Most IS elements create 2–14 bp of duplicated sequences at the site of insertion, called a target site duplication (TSD), and the length of the TSD differs depending on what type of IS element is inserted. The presence of a TSD indicates that the locus where the IS inserted has not undergone any additional genome rearrangements after IS insertion. Thus, simple insertion sites containing TSDs can be used to determine the sequence specificity and other common insertion site features of each IS type.
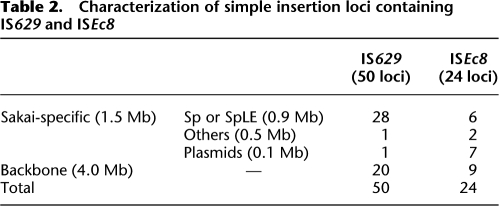
We identified a total of 50 and 24 simple insertion loci for IS629 and ISEc8, respectively, in the nine O157 strains used in this study (Table 2; see Supplemental Table S2 for more details). When IS629 or ISEc8 is inserted, TSDs of 3 or 4 (mainly 3) bp and 8 bp are created, respectively (Kusumoto et al. 1999; Schneiker et al. 1999). Comparison of the sequences of TSDs indicated that neither element favors a specific target sequence (Supplemental Table S2). By analyzing their genomic locations, we found that these IS elements inserted more frequently (about 4.3 times higher) in Sakai-specific regions (45 loci in 1.5 Mb) than in the chromosome backbone (29 loci in 4.1 Mb; Fig. 3). IS629 insertion is highly biased toward prophages and prophage-like integrative elements, whereas ISEc8 insertion is highly biased toward the pO157 plasmid.
Table 2.
Characterization of simple insertion loci containing IS629 and ISEc8
IS-associated genomic deletion
All IS-associated genomic deletions identified were associated with IS629, and none was associated with ISEc8 (Supplemental Table S3). Among the 20 deletions identified, most were larger than 2 kb, with the largest being 20 kb, which was detected in segment 276f/278r of strain #6. It is interesting that these deletions frequently occur (15 of 20, 75%) in the regions downstream from TPase genes, which are transcribed in the direction opposite that of chromosomal DNA replication (Supplemental Table S3).
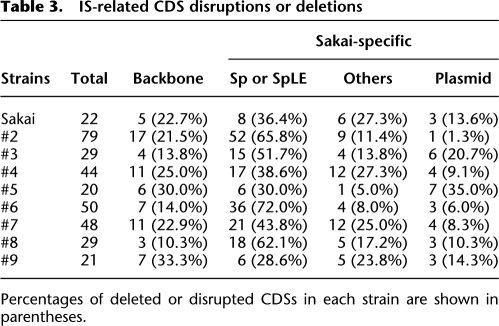
Genes disrupted or deleted by IS-related genetic events
We identified a total of 203 genes that were disrupted or deleted by IS insertions or IS-associated deletions in one or more O157 strains. As summarized in Table 3, 163 of these genes (80.3%) were within Sakai-specific sequences (see Supplemental Table S3 for more details), including many genes related to phage functions or unknown functions. There were also a significant number of genes of known or predicted functions, such as 10 genes for various transporters (substrates were unpredictable in most cases) and 11 transcriptional regulator genes. In addition, several virulence-related genes, such as those for curlin biosynthesis, a gene for catalase-peroxidase, and an AidA-I adhesin-like protein, were disrupted or deleted in some strains. This suggests that the IS-related genomic rearrangements may have significantly altered virulence and other phenotypes in O157 strains. We confirmed that curlin production was abolished specifically in strains that contained IS insertions in the csg operons that encode the curlin biosynthesis genes (strains #2, #4, and #5; Supplemental Fig. S2).
Table 3.
IS-related CDS disruptions or deletions
Percentages of deleted or disrupted CDSs in each strain are shown in parentheses.
Other types of structural changes
Other types of genomic rearrangements have been generated by strain-specific phage integrations and several other mechanisms. We identified four prophages that were not present in O157 Sakai: in segment 167f/168r of strain #3, in segment 256f/257r of strain #2, and in segment 448f/448.1r of strains #4 and #7 (Fig. 2). We determined the sequence of each of these prophages, except for a prophage in the 448f/448.1r segment of strain #7 (Supplemental Fig. S1). The first prophage (named PPO157_s3) was 12 kb in size and had integrated into the tqsA gene. This phage was very similar to Sp7 of O157 Sakai, a unique prophage with chimeric features of lambdoid and P4-like phages (Asadulghani et al. 2009). Because Sp7 had integrated into the ycfD/phoQ intergenic region in O157 Sakai but strain #3 did not contain a prophage in this region (Fig. 2), PPO157_s3 may have been derived from Sp7 through recombination with one or more other phages, followed by reintegration into the tqsA locus. The second prophage (PPO157_s2) was 11.4 kb in size and contained 17 protein-coding sequences (or CDSs). This prophage had integrated into the eutA/eutB intergenic region, the same region in which the CPZ-55 prophage had integrated in K-12 (Blattner et al. 1997). However, both prophages have largely degraded and only the integrase genes exhibited high sequence similarity (92.8% amino acid sequence identity) between the two prophages. The third prophage (PPO157_s4) was 11.7 kb in size and had integrated into the same tRNA gene (leuX), within which an integrative element, SpLE5, is present in O157 Sakai. PPO157_s4 is very similar to the satellite phage P4 (Halling et al. 1990), but encodes the EcoO109I restriction-modification system (Kita et al. 1999, 2002). Although O157 Sakai also harbors a P4-like prophage (Sp2), its chromosomal location and genome structure differ from those of PPO157_s4, indicating that the two phages independently invaded the O157 lineage. The prophage in segment 448f/448.1r of strain #7 was identical or very similar to PPO157_s4, because the 448f/448.1r segments of strains #4 and #7 exhibited identical PCR-RFLP patterns (data not shown).
Most of the other SSSPs (19 segments in 12 loci) were created by deletions that were probably introduced by homologous recombination between identical or highly homologous sequences of 3–141 bp. The observed deletion sizes ranged from 115 bp to 10,210 bp (Supplemental Table S4). The remaining SSSPs (12 segments in seven loci) were also generated by small genomic deletions, but the mechanisms are unknown because neither a homologous sequence (≥3 bp) nor an IS element was detected at the deletion boundaries.
Identification of footprints created by IS629 transposition
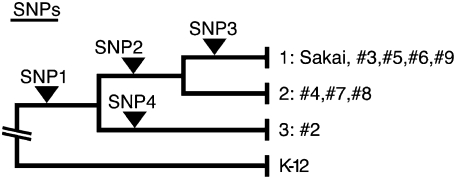
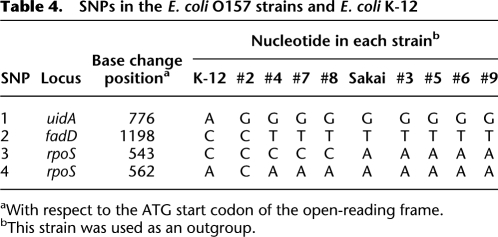
Through multi-locus sequencing analysis of 15 housekeeping genes (Wick et al. 2005), we identified four single-nucleotide polymorphisms (SNPs) that can be used to divide the nine O157 strains into three distinct clades (Fig. 4; Table 4). By reinspecting all IS629 and ISEc8 insertion loci (77 and 28, respectively) identified in the nine strains and considering phylogenetic relationships, we noted several genomic loci in which IS629 was absent in a strain, although it would be predicted to be present according to the phylogenetic position of the strain. For example, although all other strains contain an IS629 insertion in segment 207f/208r, IS629 is missing at this locus in strain #7. This suggests that IS629 was inserted into this locus, but was then deleted in strain #7 by an unknown mechanism. Although transposition mechanisms have been extensively studied for many IS elements, neither the fate of the donor DNA nor the structural changes induced upon IS transposition have been elucidated for any individual IS element. Therefore, we hypothesized that detailed structural analysis of such IS insertion loci could provide insight into the genomic structural changes induced by IS transposition.
Figure 4.
Phylogenetic tree of the nine O157 strains used in this study. The phylogenetic tree was constructed based on the sequencing and analysis of 15 housekeeping genes. The locations of SNPs identified in this analysis (Table 4) are indicated.
Table 4.
SNPs in the E. coli O157 strains and E. coli K-12
aWith respect to the ATG start codon of the open-reading frame.
bThis strain was used as an outgroup.
We amplified all of the genomic loci in which IS629 or ISEc8 was found in any of the eight O157 strains (other than the fully sequenced strain Sakai) by long-range PCR with the primer sets used in our previous WGPS analysis (Fig. 1) and sequenced each of the PCR products using IS-out-L and/or IS-out-R primers designed for each IS insertion site to directly determine the nucleotide sequence of each locus. For ISEc8, simple insertions with 8-bp TSDs or no insertions were found at 25 insertion loci. Three other loci contained disrupted ISEc8 insertions in all nine O157 strains. On the other hand, in 37 of the 77 IS629 insertion loci examined, a significant number of small deletions were found, which were not identified in the first series of SSSP analyses. Only simple insertions with 3-bp or 4-bp TSDs or no insertions were found in the remaining 40 loci. Thus, all IS629-related rearrangements identified in the nine O157 strains used in this study can now be classified as one of three types: (1) simple insertions with 3-bp or 4-bp TSDs, (2) IS629 transposition-mediated deletions of >100 bp, and (3) small deletions of <100 bp. Most of the deletions in the third group were identified by resequencing, as described in this section.
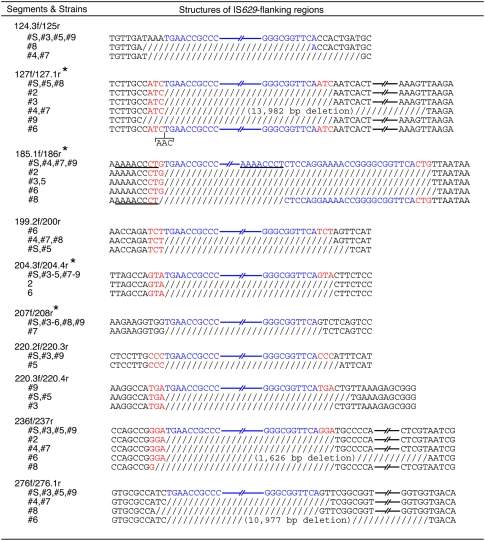
Considering the phylogenetic positions of each strain, we found four loci (in segments 127f/127.1r, 185.1f/186r, 204.3f/204.4r, and 207f/208r, indicated by asterisks in Fig. 5) in which IS629 was inserted once in the past and either was subsequently deleted by simple excision or left behind structural changes. For example, in segment 185.1f/186r, the insertion of an IS629 element occurred before the separation of Clades 1 and 2 (and probably after their separation from Clade 3). Subsequently, in strains #3 and #5, IS629 was deleted by simple excision, which reverted the genomic structure of the insertion sites to that of the originals. In strain #6, however, excision (or transposition) of IS629 appears to have left a 1-bp deletion. In strain #8, homologous recombination between two identical octamer sequences within and outside of the IS element induced an IS-associated deletion. Similarly, simple excisions or footprints of various sizes were detected in the other three segments. Deletions at six additional loci could also be regarded as footprints generated by IS629 excision (Fig. 5) because they occurred precisely at IS629 insertion sites and no sequence capable of inducing homologous recombination was found at the deletion points. These results indicate that IS629 has been deleted by simple excision and, more importantly, that the excision (or transposition) of IS629 causes deletions of various sizes at the insertion site, ranging from a single base pair to several thousand base pairs.
Figure 5.
Simple excision of IS629 and the footprints generated upon excision or transposition. (Blue) IS629 sequences; (red) target site duplications (TSDs). (Slash) Deleted sequences; the size of each deletion is shown in parentheses. Homologous sequences that probably induced deletions through homologous recombination are underlined. In the four genomic loci labeled with asterisks, the phylogenetic relationships among the strains indicate that the absence of IS629 and genomic deletions in some strains were generated by a simple excision and upon the excision (or transposition) of IS629. Deletions observed at six other loci were also likely generated by a similar mechanism because they occurred precisely at IS629 insertion sites.
Discussion
IS elements are generally regarded as genetic factors that significantly contribute to genome diversification and evolution. However, only a limited number of genome-wide studies have been conducted to compare IS distributions in closely related genomes, which are required to reveal how and to what extent IS elements actually contribute to genome diversification, even in bacteria (Schneider et al. 2002; Kawai et al. 2006; Rohmer et al. 2007). Fine-scale genomic comparison of naturally occurring closely related strains should provide the best opportunity to understand this process, but such an analysis has not yet been performed.
In the present study, we performed systematic genome-wide analysis of SSSP-containing genomic regions in the nine O157 strains previously identified by our laboratory through WGPS analysis (Ohnishi et al. 2002). We found that a large portion of the SSSPs (130 of 165) were generated by genetic events associated with only two types of IS elements, IS629 and ISEc8 (Figs. 2 and 3; Table 1). Further analysis revealed that, whereas most of the ISEc8 insertions identified were simple insertions (17 of 20), IS629 induced a wide variety of genomic rearrangements, including simple insertions, simple excisions, and various deletions of IS-flanking regions ranging from 1 bp to several kilobase pairs in size. This indicates that these IS elements served as important driving forces behind the genomic diversity of O157 strains. In our previous genomic comparisons of O157 strains by WGPS and CGH (Ohnishi et al. 2002; Ogura et al. 2006), most of the large structural polymorphisms were found in prophage regions. Combining our previous results with the results presented herein, we now conclude that IS629 and ISEc8, along with bacteriophages, are the primary contributors to genomic diversification in O157 strains and that LSSPs were generated mainly by bacteriophages and SSSPs by IS elements.
Our analysis also revealed that insertion sites for IS629 and ISEc8 exhibit a highly biased distribution in O157 genomes. These IS elements were located much more frequently in O157-specific regions than in the chromosome backbone (Table 2; Supplemental Table S2). The genomic locations of other types of IS elements in the O157 Sakai genome indicate that their distribution is also highly biased toward the O157-specific regions, and thus this trend is not specific to IS629 and ISEc8 (Supplemental Table S5). Although IS elements are randomly inserted into the O157 genome, it is most likely that clones with IS insertions into genes with essential functions have been selectively removed from the population and those with insertions into regions that are less or nonessential to growth, such as many of the Sakai-specific regions, have been able to survive.
Prophages, prophage-like integrative elements, and plasmids comprise a large portion of the O157-specific regions. Accordingly, IS insertion and IS-mediated deletion have induced the inactivation or deletion of various foreign genes that were carried into the O157 lineage by these mobile elements, along with genes required for their mobility (Table 3; Supplemental Table S3). This suggests that IS insertion and IS-mediated deletion could generate various phenotypic differences among the O157 strains, including differences in potential virulence, as we have shown for curlin production. However, we need to analyze a wider range of O157 strains collected over a longer span of time to examine how much of this variation would be fixed in the population. More interestingly, the results suggest the possibility that the IS elements play a role in immobilizing newly incoming phages and plasmids, leading to their fixation in the O157 genome. The IS elements (at least IS629) may also hasten their departure from the genome because many of these IS elements cause deletion upon excision. The presence of IS elements in genomic regions is often discussed in the context of the incorporation of foreign DNA into the genome during and/or following horizontal gene transfer. However, our findings may imply novel roles for IS elements in bacterial genome evolution.
Of the IS elements identified in O157 Sakai, IS629 has the highest copy number. Our present results confirmed that IS629 transposes very actively in O157 genomes (Fig. 3). In this regard, it may be worth noting that a report by Kusumoto et al. (2004) showed that IS1203 (a synonym of IS629) transposes at a much higher frequency in O157 cells than in K-12 cells. Although the molecular mechanism underlying this phenotype is not known, the O157 lineage may contain some specific factor that facilitates IS629 transposition.
Another notable finding related to IS629 transposition is the identification of a significant number of genomic loci for which a footprint was generated upon the simple excision (or transposition) of IS629 (Fig. 5). Both events are rarely described in bacteria (Chain et al. 2006). IS629 is a member of the IS3 family (Matsutani et al. 1987), and therefore it is very likely that IS629, like other IS3 family members, also forms a “figure eight” intermediate upon transposition (Lewis and Grindley 1997; Sekine et al. 1999; Haas and Rak 2002; Rousseau et al. 2002; Ohtsubo et al. 2004), but it is not yet known whether it transposes replicatively or nonreplicatively. In either case, however, some specific mechanism(s) appears to be required to induce simple excision and the formation of footprints. By elucidating the molecular mechanisms behind these phenomena, we will be able to better understand the fate of donor DNA in IS transposition, which would, in turn, lead to a deeper understanding of the processes responsible for genome diversification.
Methods
Bacterial strains, growth conditions, and DNA extraction
O157 Sakai was isolated during a large outbreak in 1996 in the city of Sakai, Osaka Prefecture, Japan (Watanabe et al. 1996). The strain has already been sequenced (Hayashi et al. 2001) and was used as a reference strain in this study. The eight other O157 strains examined in this study, 980938 (referred to as #2), 980706 (#3), 990281 (#4), 980551 (#5), 990570 (#6), 981456 (#7), 982243 (#8), and 981795 (#9), were all human clinical isolates. They were selected from 1798 clinical O157 strains isolated in Japan in 1998 based on their XbaI-digestion patterns in pulsed-field gel electrophoresis (PFGE) analysis and were previously analyzed by WGPS and CGH (Ohnishi et al. 2002; Ogura et al. 2006). Bacterial cells were grown to stationary phase at 37°C in Luria-Bertani (LB) medium. Genomic DNA was purified using the Genomic-tip 100/G and Genomic DNA buffer set (QIAGEN K.K.), according to the manufacturer's instructions.
Analysis of SSSP-containing regions
Long-range PCR amplification of the O157 chromosomal segments was performed as previously described (Ohnishi et al. 2002) using an LA PCR kit (TaKaRa Bio, Inc.) and the primers previously prepared for WGPS analysis (WGPS-F and WGPS-R primers). All primer sequences are available on our website (http://genome.bio.titech.ac.jp/bacteria/o157/genome.html). DNA sequencing of PCR products was performed using the ABI 3710 autosequencer (Applied Biosystems, Inc.), as previously described (Ogura et al. 2008).
All 179 segments exhibiting SSSPs in our previous WGPS analysis (Ohnishi et al. 2002) were divided into groups based on the type of SSSP and analyzed using the three strategies shown in Figure 1 (for details, see the main text). If an IS629 or ISEc8 element was inserted in at least one of the nine O157 strains, IS-outsideL and IS-outsideR primers designed for each IS insertion site were used to determine the DNA sequence of that locus in all eight O157 strains. All primer sequences used in this analysis are listed in Supplemental Table S1.
Multi-locus sequence analysis
To determine the phylogenetic relationships of the O157 strains, a multi-locus sequence analysis was carried out using 15 housekeeping genes (arcA, aroE, aspC, clpX, cyaA, dnaG, fadD, grpE, icd [also known as icdA], lysP, mdh, mtlD, mutS, rpoS, and uidA). These genes were sequenced and analyzed according to the protocol provided on the STEC website (http://www.shigatox.net/mlst).
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on the Priority Areas “Applied Genomics” (to T.H.); Grants-in-Aid 17019058 (to T.H.) and 20790338 (to T.O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and a Grant-in-Aid (H17-Sinkou-ippan-019) from the Ministry of Health, Labor, and Welfare of Japan (to T.H.). We thank Eiichi Ohtsubo, Hisako Ohtsubo, and Yasuhiko Sekine for helpful discussions and Akemi Yoshida, Yumiko Takesita, Noriko Kanemaru, Akiko Magishi, and Saori Kajiwara for technical assistance.
Footnotes
[Supplemental material is available online at http://www.genome.org. The nucleotide sequences from this study have been submitted to DDBJ/GenBank/EMBL database under accession nos. AB471314–AB471489 (IS629 insertion sites), AB471490–AB471530 (ISEc8 insertion sites), AB471531–AB471555 (other types of structural changes), AB471556 (prophage PPO157_s3), AB471557 (prophage PPO157_s2), and AB471558 (prophage PPO157_s4).]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.089615.108.
References
- Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, Nakayama K, Hayashi T. The defective prophage pool of Escherichia coli O157: Prophage–prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009;5:e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Chain PS, Hu P, Malfatti SA, Radnedge L, Larimer F, Vergez LM, Worsham P, Chu MC, Andersen GL. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: Evidence of gene reduction in an emerging pathogen. J Bacteriol. 2006;188:4453–4463. doi: 10.1128/JB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M, Mahillon J. Insertion sequences revisited. In: Craig N, et al., editors. Mobile DNA II. American Society for Microbiology Press; Washington, DC: 2002. pp. 305–336. [Google Scholar]
- Duval-Valentin G, Marty-Cointin B, Chandler M. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J. 2004;23:3897–3906. doi: 10.1038/sj.emboj.7600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, Rak B. Escherichia coli insertion sequence IS150: Transposition via circular and linear intermediates. J Bacteriol. 2002;184:5833–5841. doi: 10.1128/JB.184.21.5833-5841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling C, Calendar R, Christie GE, Dale EC, Deho G, Finkel S, Flensburg J, Ghisotti D, Kahn ML, Lane KB, et al. DNA sequence of satellite bacteriophage P4. Nucleic Acids Res. 1990;18:1649. doi: 10.1093/nar/18.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Ooka T, Ogura Y, Asadulghani MD. Genomic view on the evolution of enterohemorrhagic Escherichia coli. In: Baquero F, et al., editors. Evolutionary biology of bacterial and fungal pathogens. American Society for Microbiology Press; Washington, DC: 2008. pp. 407–419. [Google Scholar]
- Iguchi A, Iyoda S, Terajima J, Watanabe H, Osawa R. Spontaneous recombination between homologous prophage regions causes large-scale inversions within the Escherichia coli O157:H7 chromosome. Gene. 2006;372:199–207. doi: 10.1016/j.gene.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, et al. Genome sequence of Shigella flexneri 2a: Insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Nakao K, Uchiyama I, Kobayashi I. How genomes rearrange: Genome comparison within bacteria Neisseria suggests roles for mobile elements in formation of complex genome polymorphisms. Gene. 2006;383:52–63. doi: 10.1016/j.gene.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Kita K, Tsuda J, Kato T, Okamoto K, Yanase H, Tanaka M. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J Bacteriol. 1999;181:6822–6827. doi: 10.1128/jb.181.21.6822-6827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K, Tsuda J, Nakai SY. C.EcoO109I, a regulatory protein for production of EcoO109I restriction endonuclease, specifically binds to and bends DNA upstream of its translational start site. Nucleic Acids Res. 2002;30:3558–3565. doi: 10.1093/nar/gkf477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli S, Nair S, Alokam S, Pang T, Khakhria R, Woodward D, Johnson W, Stocker BA, Sanderson KE, Liu SL. Diversity of genome structure in Salmonella enterica serovar Typhi populations. J Bacteriol. 2005;187:2638–2650. doi: 10.1128/JB.187.8.2638-2650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto M, Nishiya Y, Kawamura Y, Shinagawa K. Identification of an insertion sequence, IS1203 variant, in a Shiga toxin 2 gene of Escherichia coli O157:H7. J Biosci Bioeng. 1999;87:93–96. doi: 10.1016/s1389-1723(99)80014-0. [DOI] [PubMed] [Google Scholar]
- Kusumoto M, Suzuki R, Nishiya Y, Okitsu T, Oka M. Host-dependent activation of IS1203v excision in Shiga toxin-producing Escherichia coli. J Biosci Bioeng. 2004;97:406–411. doi: 10.1016/S1389-1723(04)70227-3. [DOI] [PubMed] [Google Scholar]
- Lewis LA, Grindley ND. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol Microbiol. 1997;25:517–529. doi: 10.1046/j.1365-2958.1997.4871848.x. [DOI] [PubMed] [Google Scholar]
- Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, et al. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 1998;5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- Matsutani S, Ohtsubo H, Maeda Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- Mead PS, Griffin PM. Escherichia coli O157:H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, Nakayama K, Toh H, Yoshimura F, Kuhara S, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Kurokawa K, Ooka T, Tashiro K, Tobe T, Ohnishi M, Nakayama K, Morimoto T, Terajima J, Watanabe H, et al. Complexity of the genomic diversity in enterohemorrhagic Escherichia coli O157 revealed by the combinational use of the O157 Sakai OligoDNA microarray and the whole genome PCR scanning. DNA Res. 2006;13:3–14. doi: 10.1093/dnares/dsi026. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Abe H, Katsura K, Kurokawa K, Asadulghani M, Iguchi A, Ooka T, Nakayama K, Yamashita A, Hattori M, et al. Systematic identification and sequence analysis of the genomic islands of the enteropathogenic Escherichia coli strain B171-8 by the combined use of whole-genome PCR scanning and fosmid mapping. J Bacteriol. 2008;190:6948–6960. doi: 10.1128/JB.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi M, Kurokawa K, Hayashi T. Diversification of Escherichia coli genomes: Are bacteriophages the major contributors? Trends Microbiol. 2001;9:481–485. doi: 10.1016/s0966-842x(01)02173-4. [DOI] [PubMed] [Google Scholar]
- Ohnishi M, Terajima J, Kurokawa K, Nakayama K, Murata T, Tamura K, Ogura Y, Watanabe H, Hayashi T. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc Natl Acad Sci. 2002;99:17043–17048. doi: 10.1073/pnas.262441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo E, Minematsu H, Tsuchida K, Ohtsubo H, Sekine Y. Intermediate molecules generated by transposase in the pathways of transposition of bacterial insertion element IS3. Adv Biophys. 2004;38:125–139. [PubMed] [Google Scholar]
- Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci. 1999;96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, Guina T, Svensson K, Hayden HS, Jacobs M, et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau P, Normand C, Loot C, Turlan C, Alazard R, Duval-Valentin G, Chandler M. Transposition of IS911. In: Craig N, et al., editors. Mobile DNA II. American Society for Microbiology Press; Washington, DC: 2002. pp. 367–383. [Google Scholar]
- Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, et al. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204. doi: 10.1186/1471-2164-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156:477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Depeyrot J, Coursange E, Lenski R, Blot M. Genomic comparisons among Escherichia coli strains B, K-12, and O157:H7 using IS elements as molecular markers. BMC Microbiol. 2002;2:18. doi: 10.1186/1471-2180-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S, Kosier B, Puhler A, Selbitschka W. The Sinorhizobium meliloti insertion sequence (IS) element ISRm14 is related to a previously unrecognized IS element located adjacent to the Escherichia coli locus of enterocyte effacement (LEE) pathogenicity island. Curr Microbiol. 1999;39:274–281. doi: 10.1007/s002849900459. [DOI] [PubMed] [Google Scholar]
- Sekine Y, Aihara K, Ohtsubo E. Linearization and transposition of circular molecules of insertion sequence IS3. J Mol Biol. 1999;294:21–34. doi: 10.1006/jmbi.1999.3181. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Wada A, Inagaki Y, Itoh K, Tamura K. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet. 1996;348:831–832. doi: 10.1016/s0140-6736(05)65257-9. [DOI] [PubMed] [Google Scholar]
- Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, III, Rose DJ, Darling A, et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun. 2003;71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick LM, Qi W, Lacher DW, Whittam TS. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol. 2005;187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]