Abstract
Our knowledge of yeast genomes remains largely dominated by the extensive studies on Saccharomyces cerevisiae and the consequences of its ancestral duplication, leaving the evolution of the entire class of hemiascomycetes only partly explored. We concentrate here on five species of Saccharomycetaceae, a large subdivision of hemiascomycetes, that we call “protoploid” because they diverged from the S. cerevisiae lineage prior to its genome duplication. We determined the complete genome sequences of three of these species: Kluyveromyces (Lachancea) thermotolerans and Saccharomyces (Lachancea) kluyveri (two members of the newly described Lachancea clade), and Zygosaccharomyces rouxii. We included in our comparisons the previously available sequences of Kluyveromyces lactis and Ashbya (Eremothecium) gossypii. Despite their broad evolutionary range and significant individual variations in each lineage, the five protoploid Saccharomycetaceae share a core repertoire of approximately 3300 protein families and a high degree of conserved synteny. Synteny blocks were used to define gene orthology and to infer ancestors. Far from representing minimal genomes without redundancy, the five protoploid yeasts contain numerous copies of paralogous genes, either dispersed or in tandem arrays, that, altogether, constitute a third of each genome. Ancient, conserved paralogs as well as novel, lineage-specific paralogs were identified.
Yeasts have played a critical role in our understanding of molecular function and evolution in eukaryotes. Their small, compact genomes, their importance in a variety of fermentation processes, and the facility of manipulating them in the laboratory have led to the determination and analysis of the genome sequences of several yeast species (for review, see Dujon 2005, 2006; Kurtzman and Piskur 2006; Scannell et al. 2007a). Molecular phylogenies suggest that unicellular yeasts arose from ancestral fungal lineages several times independently, from Basidiomycota leading, for example, to the Cryptococcus and Malassezia species (Loftus et al. 2005; Xu et al. 2007), and from Ascomycota leading to the well-studied Schizosaccharomyces species, on one hand (Wood et al. 2002; Aslett and Wood 2006), and to the large, homogeneous class known as hemiascomycetes, or budding yeasts, on the other hand. The hemiascomycetes have enjoyed most of the attention of genomic studies, owing in part to their large number of species (more than a thousand are described [Kurtzman and Fell 2006] and many more likely exist [Boekhout 2005]), but mostly because they include Saccharomyces cerevisiae, the first eukaryote sequenced (Goffeau et al. 1996), and a favored experimental model for functional genomics (Hofmann et al. 2003; Ooi et al. 2006).
An exploration of 13 hemiascomycete genomes revealed their broad evolutionary range (Souciet et al. 2000), and several internal subdivisions exist, three of which have been characterized by complete sequencing of the genomes of a number of species (Dietrich et al. 2004; Dujon et al. 2004; Jones et al. 2004; Kellis et al. 2004). Yarrowia lipolytica, with a GC-rich genome roughly twice as large as that of other hemiascomycetous yeasts, is the representative species of the first subdivision. It contains more protein-coding genes and more introns than other yeasts and has a larger variety of transposable elements. It also has several peculiarities, such as multiple subtelomeric rDNA loci and dispersed 5S RNA coding genes, half of which are transcriptionally fused to tRNA genes (Acker et al. 2008). A second subdivision of hemiascomycetes consists of species that translate CTG codons as serine rather than leucine, a reassignment believed to have occurred more than 170 million years ago (Miranda et al. 2006). This subdivision has been intensely studied because it contains the pathogenic yeast Candida albicans (Jones et al. 2004; Noble and Johnson 2007) and related Candida species (Magee et al. 2008). It also contains Debaryomyces hansenii, whose genome shows numerous tandem gene arrays (TGAs) that contribute to gene family expansion (Dujon et al. 2004). A third subdivision consists of yeasts of the “Saccharomyces complex” or Saccharomycetaceae, the subdivision of hemiascomycetes with the most genome sequences available. This subdivision contains a large variety of species that, although sharing a number of common physiological and genomic properties, represent a broad phylogenetic range. They were recently classified into 14 distinct clades (Kurtzman 2003; Kurtzman and Robnett 2003) whose genomes remain unequally explored.
The importance of S. cerevisiae as a model system (for review, see Barnett 2007; Replansky et al. 2008) has resulted in most genomic studies focusing on closely related species leaving other clades of Saccharomycetaceae relatively unexplored, with only few exceptions (Dietrich et al. 2004; Dujon et al. 2004; Kellis et al. 2004). Today, genome sequences are available (at various coverages) for Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces kudriavzevi, Saccharomyces bayanus, Saccharomyces exiguus, Saccharomyces servazii, and Saccharomyces castellii (Bon et al. 2000a,b; Casaregola et al. 2000; Cliften et al. 2003, 2006; Kellis et al. 2003), and several strains of S. cerevisiae have been entirely sequenced (Wei et al. 2007; Borneman et al. 2008; http://www.broad.mit.edu/annotation/genome/saccharomyces_cerevisiae/). This offers a unique view on the population structure and reproductive preferences of a group of yeasts playing an important role in fermentation processes, and a powerful tool for quantitative trait analyses (Demogines et al. 2008). Recently, wild populations of S. cerevisiae and S. paradoxus have also been characterized by hybridization to genome tiling oligonucleotide arrays and by partial genome sequencing (Liti et al. 2009; Schacherer and Kruglyak 2009).
The recognition of a whole-genome duplication in the ancestry of S. cerevisiae (Wolfe and Shields 1997) prompted many studies on the evolutionary and functional consequences of such events in yeasts (Wong et al. 2002; Byrnes et al. 2006) and in other eukaryotes such as plants (Arabidopsis Genome Initiative 2000; Yu et al. 2005; Jaillon et al. 2007), fishes (Jaillon et al. 2004), or ciliates (Aury et al. 2006). The subsequent loss of redundant gene copies, which may lead to speciation bursts (Scannell et al. 2006), destroys original gene neighborhood relationships and reshapes the genetic maps of post-duplication species, leaving typical dual synteny patterns (Dietrich et al. 2004; Jaillon et al. 2004; Kellis et al. 2004). Inference of the original gene order has been attempted by analyzing duplicated genomes (Byrne and Wolfe 2005; Byrnes et al. 2006; Scannell et al. 2007b; Conant and Wolfe 2008), but an authentic image of the original genome organization of this important group of yeasts is desirable.
We report the complete sequence and manual annotation of the genomes of three novel yeast species, Zygosaccharomyces rouxii, a member of the Zygosaccharomyces clade, Saccharomyces (Lanchancea) kluyveri, and Kluyveromyces (Lachancea) thermotolerans, two members of the Lachancea clade. We have compared these genomes with those of Kluyveromyces lactis and Ashbya (Eremothecium) gossypii (Dietrich et al. 2004; Dujon et al. 2004), providing a multispecies comparison among nonduplicated Saccharomycetaceae. We discovered that, far from being minimal genomes, these species contain many paralogous genes, which are often highly diverged in sequence and represent approximately a third of their total genes. We also found that, despite sharing significant conservation of synteny and a common protein repertoire of approximately 3300 families, these species span a large evolutionary range as evidenced from their important sequence divergence.
Results
General sequencing strategy and brief description of the genome sequences
Rationale of species selection
Our goal was to characterize and compare gene content and genome organization of the Saccharomycetaceae (as defined by Kurtzman and Robnett 2003) that did not experience the ancestral whole-genome duplication that sculpted the genomes of S. cerevisiae and its closest relatives. To this end, we determined the complete DNA sequence of three novel yeast genomes, Z. rouxii, K. thermotolerans, and S. kluyveri (see Table 1), and included in our analyses the previously published sequences of K. lactis (Dujon et al. 2004) and A. gossypii (Dietrich et al. 2004). These five yeast species represent four distinct clades of Saccharomycetaceae, respectively, designated as Zygosaccharomyces, Lachancea, Kluyveromyces, and Eremothecium (Fig. 1A). Given their complex phylogenetic relationship within the Saccharomycetacea, we designate them collectively as “protoploid” solely to distinguish them from the “duplicated” yeasts (clades 1–6) (Kurtzman 2003). Other protoploid genomes, such as Kluyveromyces waltii (Kellis et al. 2004) and Kluyveromyces marxianus (Llorente et al. 2000a), were not considered since our comparisons required complete genome sequences with a single contig per chromosome.
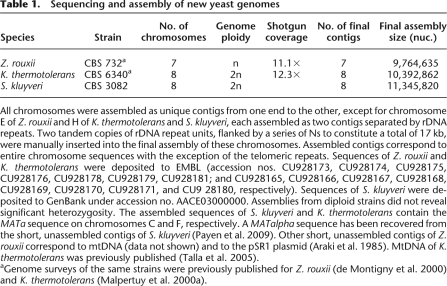
Table 1.
Sequencing and assembly of new yeast genomes
All chromosomes were assembled as unique contigs from one end to the other, except for chromosome E of Z. rouxii and H of K. thermotolerans and S. kluyveri, each assembled as two contigs separated by rDNA repeats. Two tandem copies of rDNA repeat units, flanked by a series of Ns to constitute a total of 17 kb, were manually inserted into the final assembly of these chromosomes. Assembled contigs correspond to entire chromosome sequences with the exception of the telomeric repeats. Sequences of Z. rouxii and K. thermotolerans were deposited to EMBL (accession nos. CU928173, CU928174, CU928175, CU928176, CU928178, CU928179, CU928181; and CU928165, CU928166, CU928167, CU928168, CU928169, CU928170, CU928171, and CU9 28180, respectively). Sequences of S. kluyveri were deposited to GenBank under accession no. AACE03000000. Assemblies from diploid strains did not reveal significant heterozygosity. The assembled sequences of S. kluyveri and K. thermotolerans contain the MATa sequence on chromosomes C and F, respectively. A MATalpha sequence has been recovered from the short, unassembled contigs of S. kluyveri (Payen et al. 2009). Other short, unassembled contigs of Z. rouxii correspond to mtDNA (data not shown) and to the pSR1 plasmid (Araki et al. 1985). MtDNA of K. thermotolerans was previously published (Talla et al. 2005).
aGenome surveys of the same strains were previously published for Z. rouxii (de Montigny et al. 2000) and K. thermotolerans (Malpertuy et al. 2000a).
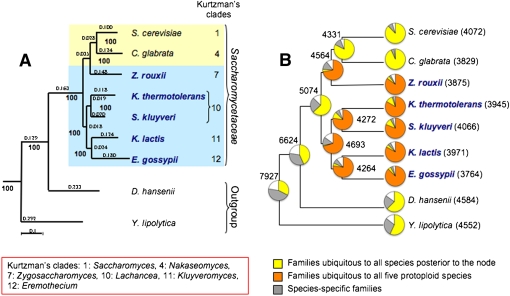
Figure 1.
Phylogeny and protein-coding repertoire of Saccharomycetaceae and outgroups. (A) The phylogenetic tree results from the alignment of 180 proteins (66,709 amino acids), selected from all universal one-member families having a homolog in S. pombe, using the MAFFT algorithm (Katoh et al. 2005), cleaned with Gblocks (Castresana 2000) before concatenation. Only families for which the ratio between the cleaned blocks and the initial alignment was higher than 75% were considered. The tree was constructed by maximum likelihood using PhyML (Guindon and Gascuel 2003) with a JTT substitution model corrected for heterogeneity among sites by a gamma-law distribution using four different categories of evolution rates. The proportion of invariable sites and the alpha-parameter of the gamma-law distribution were optimized according to the data. Branch length is indicated above or next to each branch, and bootstrap values (in bold) next to each node. S. pombe was used as an anchoring outgroup. Clades number and designation for Saccharomycetaceae are according to Kurtzman (2003) and Kurtzman and Robnett (2003). Protoploid species are highlighted (bold blue names). (B) Figures represent the total number of protein families in each species or node (pan-proteome, defined as the sum of all families present in all species posterior to the node). Pie charts indicate the proportion of families classified as “ubiquitous” (core-proteome, common to all species posterior to the node), “species-specific” (present in only one of the species posterior to the node), or other combinations. In orange, the proportion of families shared by all five protoploid species. Note the similar pie charts for the Saccharomycetaceae species, compared to the different pie charts of the outgroup.
The five yeast species studied here show diverse biological and metabolic properties. Z. rouxii is an osmo- and halotolerant species used in some fermentation processes (Solieri and Giudici 2007). It is able to grow on high concentrations of salt and/or sugar (Jansen et al. 2003) and is often considered as a food-spoiling agent. The type strain (CBS732) is haploid, but some wild isolates of Z. rouxii are diploid (Solieri et al. 2008). K. thermotolerans is usually associated with fruits or with insects feeding on plants, but one isolate was recently obtained from a wine re-fermentation process (Vilela-Moura et al. 2008). S. kluyveri is often used in protein production because it wastes less glucose by aerobic fermentation than does S. cerevisiae, and hence is better at biomass production (Møller et al. 2002, 2004). It has been developed as a model organism to study a variety of biological processes such as pyrimidine degradation (Beck et al. 2008), metabolic flux (Blank et al. 2005), or fatty acid desaturases (Oura and Kajiwara 2008). S. kluyveri appears widespread in the environment, with strains isolated from insect guts, soil, or trees in North America, Europe, and India. K. lactis and A. gossypii, are respectively, a haploid, lactose-utilizing yeast (Dujon et al. 2004), and a haploid phytopathogen able to form filaments (Dietrich et al. 2004).
Overall genome anatomy, composition, and duplications
We annotated the three novel yeast genomes sequenced here and the previously sequenced genome of K. lactis (see Methods and http://www.genolevures.org/). Annotations of A. gossypii were taken from AGD (http://agd.vital-it.ch/index.html). Table 2 lists major features and properties of these five genomes. For comparison, the genomes of S. cerevisiae (Goffeau et al. 1996) and Candida glabrata (Dujon et al. 2004) were included as representative of post-genome duplication species of Saccharomycetaceae, while the genomes of Debaryomyces hansenii and Yarrowia lipolytica (Dujon et al. 2004) served as outgroups. The five protoploid genomes vary in size (from 8.7 Mb to 11.3 Mb) and are smaller than the two post-duplication genomes and the two outgroup genomes. Genome size reduction in Z. rouxii and A. gossypii is accompanied by an increased gene density (76.1% and 79.6% compared to 69.2% to 72.3% for the three other protoploid yeasts).
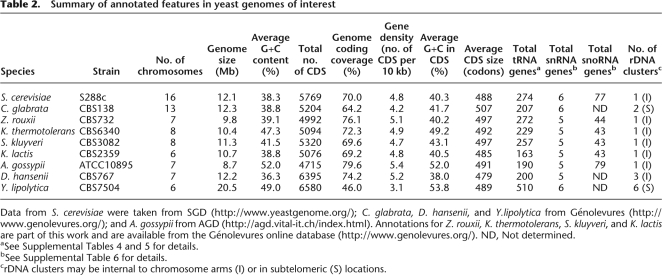
Table 2.
Summary of annotated features in yeast genomes of interest
Data from S. cerevisiae were taken from SGD (http://www.yeastgenome.org/); C. glabrata, D. hansenii, and Y.lipolytica from Génolevures (http://www.genolevures.org/); and A. gossypii from AGD (http://agd.vital-it.ch/index.html). Annotations for Z. rouxii, K. thermotolerans, S. kluyveri, and K. lactis are part of this work and are available from the Génolevures online database (http://www.genolevures.org/). ND, Not determined.
aSee Supplemental Tables 4 and 5 for details.
bSee Supplemental Table 6 for details.
crDNA clusters may be internal to chromosome arms (I) or in subtelomeric (S) locations.
Two species, K. thermotolerans and A. gossypii, show a markedly higher GC content than other yeasts, Y. lipolytica excepted (Table 2). In every species, the nucleotide composition is generally uniform across the entire genome, despite local fluctuations in some cases (Supplemental Fig. 1). S. kluyveri represents an astonishing exception to this rule. An ∼1-Mb-long region of chromosome C displays a GC content 12% higher that the rest of the genome (25% higher for the third codon position of CDS). The remaining 250-kb portion of this chromosome is identical in nucleotide composition to the rest of the genome. This heterogeneity is studied in detail in a separate work (Payen et al. 2009). In brief, it is found that the GC-rich chromosomal segment shows conserved synteny with K. thermotolerans, is devoid of transposable elements, and replicates later than other chromosomes during the S phase.
The protoploid yeast species have six, seven, or eight chromosomes. A single centromere composed of three short elements (CDEI, CDEII, and CDEIII) was identified for each chromosome of Z. rouxii, K. thermotolerans, and S. kluyveri (Supplemental Table 1; Supplemental Fig. 2B). Centromeres of K. lactis and A. gossypii were previously described (Dietrich et al. 2004; Dujon et al. 2004). Interestingly, the AT-rich CDE II spacers vary in length (Supplemental Fig. 2A). The correspondence between centromeres of the five protoploid Saccharomycetaceae genomes could be deduced from synteny conservation of flanking genes (Supplemental Table 2). In all likelihood, the protoploid ancestor was a species with eight chromosomes. The eight centromeres of K. thermotolerans and S. kluyveri show a simple one-to-one congruence. Nonambiguous correspondence with the former two species is also found for the seven centromeres of Z. rouxii and A. gossypii. The case of K. lactis is more complex with only six chromosomes and multiple rearrangements around several centromeres.
As anticipated from their phylogenetic position among the Saccharomycetaceae, the five yeast species studied here do not show traces of the whole-genome duplication that occurred in the ancestry of clades 1–6 (Kurtzman and Robnett 2003). Similarly, we can rule out the possible occurrence of another genome duplication in either of the four clades studied here since we could not find evidence of dual synteny in all pairwise map comparisons between our five species. Taking advantage of this fact, we have examined the possible presence of segmental duplications in these genomes (Supplemental Table 3). If one ignores known transposable elements and subtelomeric regions, only a few cases of segmental duplication are found in the genomes of K. thermotolerans, S. kluyveri, and K. lactis, and none in Z. rouxii or A. gossypii. Such a paucity of segmental duplications is surprising in view of the high frequency (10−7 per mitosis) of their spontaneous formation in S. cerevisiae (Payen et al. 2008), and contrasts with their abundance in mammalian genomes (for review, see Samonte and Eichler 2002) (see Discussion).
Genome content: Protein-coding genes and spliceosomal introns
Protein-coding genes were identified as described in Methods. The protoploid genomes contain from 4715 (A. gossypii) to 5320 (S. kluyveri) protein-coding genes (Table 2). This is 8%–18% less than S. cerevisiae, which, itself, contains ∼10% less genes than the two species used here as outgroups, D. hansenii and Y. lipolytica. With approximately 5000 protein-coding genes on average, protoploid yeasts have the smallest known gene set among hemiascomycetes. Protein size distributions and global amino acid compositions are similar for the nine yeasts considered here (mean: 492 ± 14 amino acids; median: 410 ± 11 amino acids). Spliceosomal introns are rare (3%–6% of protein-coding genes are interrupted by introns) in these five yeasts, and very few genes contain two introns (four in Z. rouxii, 11 in S. kluyveri). Splice sites and branch-point sequence motifs are similar to those or S. cerevisiae, and intron lengths are shorter, except for K. lactis. Additional information on yeast spliceosomal introns can be found at http://genome.jouy.inra.fr/genosplicing/.
tRNA-coding genes
The variety of tRNA species (identified by their anticodon), number of genes, and corresponding codon usage are given in Supplemental Table 4. The five protoploid yeasts follow the tRNA sparing rules previously described for other hemiascomycetes, enabling them to interpret the entire genetic code with only 43 or 44 tRNA species instead of the complete eukaryotic set of 46 (Marck et al. 2006). As expected for yeasts, a significant proportion (25%–35%) of tDNAs harbor an intron, all of which are short (except for the tDNA-Leu [CAG], where introns are 288, 150, and 134 nucleotides [nt] long for K. lactis, A. gossypii, and S. kluyveri, respectively).
In each yeast, most species of tRNA molecules are encoded by more than one gene (up to 17 copies), and the different paralogous gene copies are identical, or almost identical, in sequence, that is, there exists only one tRNA species for each anticodon. Two exceptions are notable here: (1) in Z. rouxii, the two copies of tDNA-Val (TAC) differ from each other at 26 positions; (2) in S. kluyveri, one of the three copies of tDNA-Glu (CTC) differs by 17 nt changes from the two other copies of tDNA-Glu (CTC). Interestingly, this tDNA, which is located on the GC-rich left arm of chromosome C, results from a silent T-to-C mutation in the anticodon but is otherwise identical in sequence to the 13 tDNA-Glus (TTC).
As in other yeasts, tDNAs are dispersed throughout the genomes. Distances between two successive tDNAs along chromosomes vary from a few hundred base pairs to >300 kb (median values 20–30 kb). Clusters of tDNAs are rare in yeasts and tandem arrays of identical tDNAs even rarer. Thus, it is worth mentioning the array of five tandemly repeated tDNA-Glus (CTC), each separated by ∼1 kb, that lies on chromosome B of K. thermotolerans. As previously described for other hemiascomycetes (Marck et al. 2006; Acker et al. 2008), probable di-cistronic tDNA pairs are also found in the yeast genomes we analyzed (Supplemental Table 5). In all cases, the intervals between the two successive tDNAs, that are always co-oriented, are very short and lack Pol III terminators. Some pairs, such as Arg (TCT)–Asp (GTC) or His (GTG)–Val (AAC), are common to several yeast species and probably result from ancestral fusions. Others appear specific to individual lineages.
Genes for other noncoding RNA molecules
A single rDNA locus is found in each protoploid species (Table 2). It is located within a chromosome arm, as in S. cerevisiae, not in a subtelomeric position as in some other yeasts. In each case, a gene encoding the 5S RNA molecule is part of the rDNA repeat, in opposite orientation of the 35S transcript (precursor of 18S, 5.8S, and 26S RNAs).
Genes for other major noncoding RNA molecules were annotated as described in Methods (Supplemental Table 6). The five spliceosomal RNAs (U1, U2, U4, U5, U6 snRNAs) are conserved in structure and size, and each is encoded by a single gene in the five yeast species. The same holds true for the U3 snoRNA, except for an interchromosomal duplication that created two identical genes in Z. rouxii. Similarly, the RNA moieties of the RNase P and SRP complexes are each encoded by a single gene in all yeast species, but their sequences and sizes differ owing to insertions of variable lengths around the conserved structural core. C/D snoRNA genes are highly conserved. (H/ACA snoRNAs were not systematically investigated because of their variable size and poor sequence conservation.) The five polycistronic clusters, which encode 17 snoRNA molecules, are conserved. Also conserved are the five snoRNA genes embedded in introns of protein-coding genes. Finally, nearly all of the 21 monocistronic C/D snoRNA genes were unambiguously identified in each yeast, including snR52 transcribed by RNA polymerase III. The three absent cases (snR4 in K. lactis and S. kluyveri, snR50 in K. thermotolerans, and snR62 in Z. rouxii) were missed, probably owing to extensive sequence divergence and lack of synteny. We did not precisely define telomerase RNAs (beside the two previously annotated ones) (McEachern and Blackburn 1995; Chappell and Lundblad 2004), owing to their extensive sequence variation.
Transposable elements
Very few transposable elements are present in the five yeast genomes we analyzed (Supplemental Table 7). Z. rouxii and A. gossypii have a few degenerate Ty3-like elements and no other class I elements (retro-elements). K. thermotolerans, S. kluyveri, and K. lactis have several solo-LTRs of Ty1-like elements, but only S. kluyveri has intact copies of that retro-element. In K. thermotolerans, there are only two degenerated Ty1 copies, and both are located in subtelomeric regions, as previously observed for K. lactis (Fairhead and Dujon 2006).
Our greatest surprise came from the presence of a novel class II element in the protoploid yeast genomes. Such elements have only been found so far in C. albicans (Goodwin et al. 2001, 2007) and Y. lipolytica (Neuvéglise et al. 2005). Now, we found sequences similar to the hAT DNA transposon of plants and fungi (Rubin et al. 2001) in K. thermotolerans, S. kluyveri, K. lactis, and A. gossypii (Supplemental Table 7). Four full-length elements of this family (called Rover) were recognized in total. All carry a single CDS (between 631 and 867 codons), possess Terminal Inverted Repeats (TIR), and create 8-bp target site duplications. The presence of this element in four yeasts only, suggests an invasive transfer (possibly in the common ancestor of clades 10, 11, and 12 of Saccharomycetaceae, as the element is not found in Z. rouxii). Judging from the limited number of Rover sequences or their remnants, this element had a limited evolutionary success.
Protein families and functional repertoire
Overall description of protein families and core protein repertoire
The predicted proteomes of the five protoploid yeast species (see Methods) were classified into families based on all-to-all sequence comparisons and consensus clustering, as previously described (Nikolski and Sherman 2007). Proteomes of S. cerevisiae, C. glabrata, D. hansenii, and Y. lipolytica were included in the comparisons, bringing the total to 48,889 yeast proteins. Among these, 98% could be partitioned into 7927 protein families (Table 3). The remaining 1015 proteins (2%) could not be unambiguously classified using our clustering parameters. The list of protein families and their content can be found at http://www.genolevures.org/. A third of the protein families are common to all nine yeasts. In each species, they are represented either by a single gene (1689 families) or by several paralogous genes (902 families). Another 45% of protein families are species-specific, and the remaining 22% are represented in various subsets of species. We examined the distribution of the three classes of families in the different yeast species and at each evolutionary node according to the phylogenetic tree (Fig. 1B). As expected, most species-specific families originate from the two outgroup species. Yet about a quarter of the 5074 families found in all Saccharomycetaceae is made up of species-specific proteins. The five protoploid proteomes are quite homogeneous, with only small proportions of species-specific families. They share additional protein families in addition to the ones common to all nine species, bringing the total of their common families to 3295. This “core protein repertoire” represents 81%–88% of the protein families present in each of the five protoploid species. As expected, most families of this core repertoire are represented in S. cerevisiae and C. glabrata, making it a characteristic feature of Saccharomycetaceae whose functions are worth elucidating.
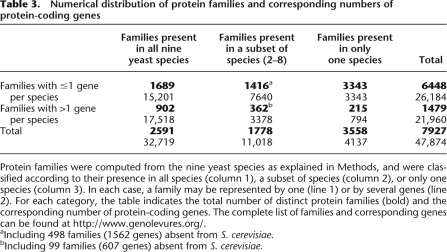
Table 3.
Numerical distribution of protein families and corresponding numbers of protein-coding genes
Protein families were computed from the nine yeast species as explained in Methods, and were classified according to their presence in all species (column 1), a subset of species (column 2), or only one species (column 3). In each case, a family may be represented by one (line 1) or by several genes (line 2). For each category, the table indicates the total number of distinct protein families (bold) and the corresponding number of protein-coding genes. The complete list of families and corresponding genes can be found at http://www.genolevures.org/.
aIncluding 498 families (1562 genes) absent from S. cerevisiae.
bIncluding 99 families (607 genes) absent from S. cerevisiae.
Functional categorization of the core protein repertoire
Because few functional data are available for yeasts other than S. cerevisiae, we restricted our analysis of the core repertoire to families having, among them, at least one S. cerevisiae representative with a functional annotation. A total of 4097 distinct S. cerevisiae proteins with an associated GO-term for biological processes were retrieved from GO resources at http://www.yeastgenome.org/. For each of the 32 informative GO-terms recovered, the proportion of families belonging to the core repertoire was computed (Supplemental Fig. 3). Genes involved in ribosome biogenesis, protein translation or modification, transport, RNA metabolism, or cellular respiration are highly represented among the core repertoire families. In contrast, processes such as sporulation, meiosis, or conjugation are more frequently based on species- or lineage-specific genes.
Paralogous genes, genome redundancy, and tandem arrays
Paralogous genes
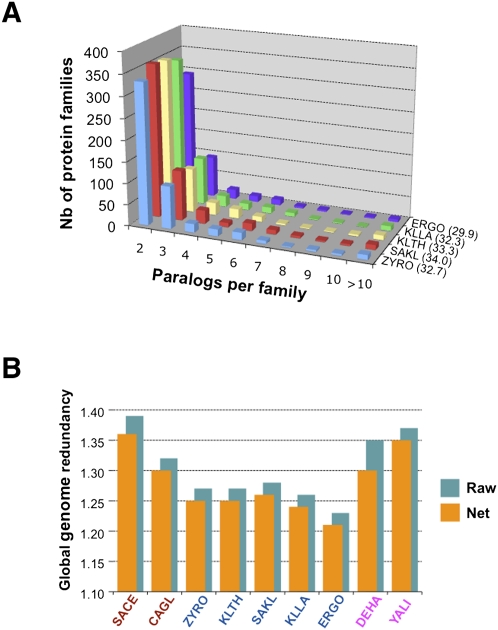
As noted above, a large number of protein families (1479) are represented by more than one protein in a given yeast species, revealing paralogs derived from ancestral duplications. Figure 2A shows that the distributions of protein family sizes for the five protoploid species are globally similar. With 300–350 families of two paralogous genes per species, 95–115 families of three paralogs, and a few larger families, a total of 30%–34% of protein-coding genes are members of multigene families in the protoploid genomes (compared to 44% for S. cerevisiae). Thus, ancestral duplications of various sorts (see below) contribute to a third of the genomes of Saccharomycetaceae, while the whole-genome duplication in the ancestry of S. cerevisiae contributes to only an additional 10% of paralogs. We also compared the global genome redundancy of all nine yeast species (Fig. 2B) (note that redundancy only refers here to sequence similarity, not to function). The five protoploid yeasts are similar in this regard (slightly lower for A. gossypii) and different from other yeasts. The higher genome redundancy of S. cerevisiae and C. glabrata reflects their ancestral whole-genome duplication. In contrast, the higher genome redundancy of D. hansenii and Y. lipolytica results from the expansion of specific gene families (if the 104 multigene families specific to Y. lipolytica were ignored, then its net redundancy would drop to the same figure as D. hansenii). In both cases, the total gene number increases without a corresponding increase in the protein repertoire.
Figure 2.
Genome redundancy. (A) Shown for each yeast species is the total number of protein families distributed according to their size (nb of paralogs per family; families of one member are not shown). The proportion (in percent) of the number of CDS belonging to multigene families to the total number of CDS is indicated in brackets next to the species abbreviation. (B) Compared genome redundancies for all nine yeast genomes. (SACE) S. cerevisiae; (CAGL) C. glabrata; (ZYRO) Z. rouxii; (KLTH) K. thermotolerans; (SAKL) S. kluyveri; (KLLA) K. lactis; (ERGO) A. gossypii; (DEHA) D. hansenii; (YALI) Y. lipolytica. Global genome redundancy is calculated as the ratio of total number of protein-coding genes in a genome vs. the total number of protein families in the same species. Raw redundancy counts all gene copies within tandem gene arrays (TGA); net redundancy considers only one gene-equivalent per TGA.
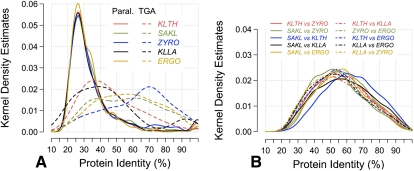
Ignoring TGAs (see below), paralogs appear randomly dispersed in the genomes of protoploid yeasts (Supplemental Table 8). This dispersion is compatible with interchromosomal as well as intrachromosomal duplication events that reshuffled the genomes over a long time. Global distributions of sequence identities between paralogous proteins are remarkably similar for the species studied (Fig. 3A). A major mode is observed at ∼27% amino acid identity, which corresponds to highly diverged proteins (probably very ancient gene duplications). Higher identity (∼35%–75%), representing more recent duplications or stronger functional constraints, is observed with slowly decreasing frequency. Note the relative excess of highly similar paralogs (>90% amino acid identity), representing even more recent gene duplications or stringent functional constraints (slightly more important in K. lactis and K. thermotolerans than for the three other species).
Figure 3.
Distribution of amino acid sequence identities between pairs of homologous proteins. Pairs of orthologous (as defined from SONS) or paralogous proteins (defined from protein families) were used to compute the distributions. (A) Paralogs; (B) orthologs. Amino acid identities were calculated from BLAST alignments with low complexity filter. Each distribution was computed from all pairwise alignments between two species for orthologs, and from pairwise alignments within families of two and three members for paralogs. (Solid lines) Dispersed paralogs; (dashed lines) TGAs; species abbreviations as in Figure 2.
Tandem gene arrays (TGAs)
The total number of TGAs, with either no (most cases) or a few intervening genes between the successive paralogous copies, ranges from 31 for A. gossypii to 51 for S. kluyveri, most of which consist of only two or three gene copies (Supplemental Table 9). The few TGAs of four and five genes may correspond to functional adaptations. Some TGAs consist of a gene and a pseudogene, indicating functional inactivation of a gene copy after the duplication. Interestingly, paralogs in TGAs are generally less diverged in sequence than dispersed paralogs (Fig. 3A), in agreement with the idea that they represent more recent gene duplication events, possibly of adaptive type. Accordingly, several TGAs appear as species-specific, as if the tandem duplications occurred independently in the various phylogenetic branches (Supplemental Table 10). They encompass a large variety of functions and, for the most part, protein families of limited sizes (one to three members per species). Other TGAs are conserved within the protoploid yeasts. Many consist of large multigene families (with more than 10 members per species). Using synteny conservation as an indication of common ancestry, a minimum of 18 TGAs were formed prior to species divergence and conserved (despite copy number variation; Supplemental Table 10). For example, genes encoding the TAF14 subunit of the polymerase II transcriptional machinery, protein kinases involved in mitotic exit network, and B-type cyclins comprise two-copy TGAs conserved in all five protoploid yeasts (the last one is duplicated in S. cerevisiae and C. glabrata). TGA expansion and conservation among all hemiascomycetous yeasts will be published elsewhere (L Despons, P Baret, L Frangeul, V Leh-Louis, P Durrens, and JL Souciet, in prep.).
Conservation of dispersed paralogous gene copies
We also examined the conservation of dispersed paralogs to pinpoint events of gene duplication or loss during the evolution of the five protoploid yeasts and to try to evaluate their functional consequences. Many families of two members (204) or three members (52) are conserved among the five species studied. Considering, as a conservative underestimate, only pairs of paralogs for which orthology of both members is clearly demonstrated for all five species by synteny conservation, a total of 114 cases could be retrieved (Supplemental Table 11). They are involved in a variety of functions, as deduced from their S. cerevisiae homologs, and have diverse degrees of sequence conservation. In addition to the genes encoding histones H4 and H2B, and mitochondrial, cytoplasmic, or peroxysomal protein isoforms that were expected, we found genes encoding a variety of protein kinases, translational elongation factors, thioredoxins, a nuclear pore component, an ATPase of the AAA family, the 1,3-beta-D-glucan synthase, factors involved in fatty acid metabolism, and specific components of the proteasome and ubiquitination pathway. Some conserved duplicated genes are of unknown function. Among the conserved paralogous proteins having undergone the greatest sequence variation (∼20% of amino acid identity) are a component of the SSU processosome containing the U3 snoRNA, a ubiquitin-specific protease, and a protein involved in structural maintenance of chromosomes.
Variation of dispersed paralogous gene copies
At the other extreme, events of gene duplication or loss that occurred during the evolution of protoploid yeasts are illustrated by several species-specific families of paralogs. Given the diversity of the resulting situations, we have examined three simple cases. If one considers (Supplemental Table 12) genes present in one copy in four species but in families of two or more paralogs in the fifth, the largest amplification is represented by five paralogs (including one tandem) in S. kluyveri that encode proteins with weak similarity to the Amn1 proteins of S. cerevisiae involved in daughter cell separation and chromosome stability. Other series of two or three paralogs are involved in a variety of functions. If one considers (Supplemental Table 13) genes present in families of two or more paralogs in one species but absent from all others, the most spectacular cases are two families of, respectively, 20 and 10 members observed in Z. rouxii, the first one related to the COS/DUP family of mostly subtelomeric genes in S. cerevisiae. Similarly, there are two families of seven members in K. lactis, two families of five members, and one family of four members in K. thermotolerans. Among this species-specific category, families of two or three paralogs are also found in all species, but, unfortunately, almost no functional inference is possible owing to lack of homologs. Finally, there are a variety of gene family expansions in some species that are probably related to functional adaptation (Supplemental Table 14). For example, K. lactis has 12 copies (compared to zero to four for other species) of a gene similar to the Mch2 protein of S. cerevisiae that may be involved in importing monocarboxylic acids. Similarly, S. kluyveri has 12 copies (compared to three to six for other species) of a gene encoding proteins weakly similar to cell surface proteins, and Z. rouxii has 10 copies (compared to zero to four for other species) of genes potentially encoding oxydoreductases involved in the formation of chiral alcohols.
Orthologous genes and synteny conservation
Definition of orthologs
The proper identification of orthologous genes is a major challenge (Kuzniar et al. 2008) because accumulation of gene-loss and duplication events tends to blur the recognition of true orthologs among the set of remaining homologs. Reciprocal best hits are often used to assign orthologs among a set of homologs (Rivera et al. 1998), but true orthologs are not necessarily those with the most similar sequences among all homologs (Lynch and Conery 2000). The present set of yeast species offered us a chance to define orthologous genes based on conservation of their genetic location. We started from a series of homologous genes identified from the corresponding protein families (see above), and examined their flanking chromosomal regions for the presence of other homologs (see Methods). In this way, we defined subsets of orthologous genes from each inspected family of homologs. The procedure was reiterated exhaustively until all protein-coding genes of all yeast species had been examined, defining SONS (Subset of Orthologs by Neighborhood and Similarity). In total, we identified 3896 SONS out of 3493 families (80% of families present in more than one species). A total of 27,926 genes (64% of the genes) are assigned to SONS, that is, belong to groups of orthologs confirmed by a shared neighborhood. Most of the remaining genes belong to Y. lipolytica and D. hansenii as expected from the very low conservation of synteny between those species and other yeasts. The complete list of genes in SONS can be found at http://www.genolevures.org/.
Conservation of orthologs
A total of 1609 SONS (41%) are common to all yeasts (i.e., they comprise a series of 1:1:1:1:1 orthologs confirmed by synteny). For the rest, some genes are missing in some species owing to gene loss during evolution. Using all alignments of orthologous proteins (as defined from cognate SONS), we computed the distributions of sequence identities for all pairwise comparisons of our five yeasts (Fig. 3B). Strikingly, their mean or median values (all distributions are monomodal, as expected for orthologs) underline the large evolutionary distances between protoploid species of Saccharomycetaceae. Even species belonging to the same clade, such as S. kluyveri and K. thermotolerans, show an average amino acid identity as low as 58.2%. Other pairwise comparisons show even lower similarity (48%–53%), consistent with their proposed phylogeny (see Fig. 1). Thus, even a homogeneous phylogenetic group such as the Saccharomycetaceae spans a very broad evolutionary range.
Synteny conservation
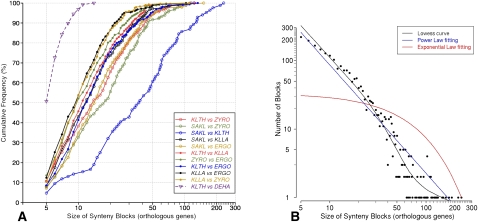
Using gene orthologs (as defined from SONS), we computed the maximal synteny conservation between the five protoploid yeast genomes as described in Methods. Pairwise comparisons between the five species (Supplemental Fig. 4) show the significant amount of map reshuffling that occurred during the evolution of this group of species, leaving numerous and relatively short synteny blocks scattered throughout these genomes. However, recognizable synteny blocks cover significant proportions of each genome (∼80%–90% of all protein-coding genes). The distributions of block size (14–26 genes) and number (180–291) vary only slightly between pairs of species, except for the S. kluyveri vs. K. thermotolerans comparison that shows larger (mean size 59 genes) and fewer (84) synteny blocks. An example of synteny block conservation is illustrated by Supplemental Figure 5. Note that, despite extensive map reshuffling, the five protoploid species clearly form a homogeneous group: When a more distantly related yeast such as D. hansenii is compared to any of them, the size of synteny blocks and their coverage of the genome diminish considerably (Supplemental Fig. 6). Distributions of size of synteny blocks in all pairwise species comparisons (Fig. 4) are consistent with the phylogenetic relationships between species. Interestingly, the frequency distribution of synteny blocks according to their size fits a power law rather than the expected exponential law implied by the random breakage model of Nadeau and Taylor (1984).
Figure 4.
Size of synteny blocks. (A) Distribution of sizes of synteny blocks (log scale abscissae) between all pairs of protoploid Saccharomycetaceae. The same distribution for synteny blocks between K. thermotolerans and an outgroup species (D. hansenii) is shown for comparison. Species abbreviations as in Figure 2. (B) Frequency distribution of synteny blocks (log scale ordinate) between all pairs of protoploid Saccharomycetaceae according to size (log scale abscissa).
Our definition of synteny blocks allows for the presence of “intervening” genes in them (see Methods). The number of such genes is roughly proportional to block sizes (Supplemental Fig. 6). Figures are similar for Z. rouxii, K. lactis, and A. gossypii (approximately seven intervening genes for 100 orthologs in syntenic blocks), but are significantly higher for S. kluyveri (approximately 12). An intermediate figure is observed for K. thermotolerans (approximately nine).
Relationship between sequence divergence and synteny conservation
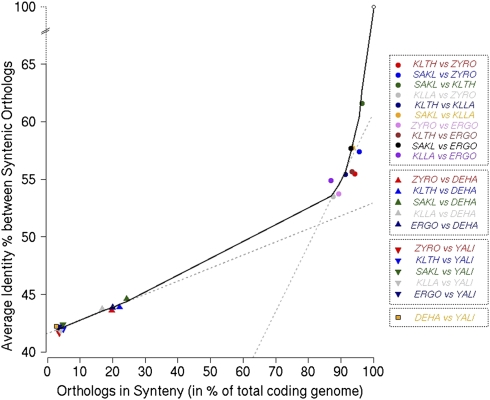
We examined the relationship between the rates of sequence divergence and chromosomal rearrangements during the evolution of hemiascomycetes (Fig. 5). There is a nonlinear relationship between average amino acid identity of orthologous proteins and the proportion of genes remaining in synteny. The five protoploid species are grouped in a sector of the curve in which sequence identity decreases rapidly while synteny conservation decreases slowly. Pairwise comparisons of the same species with those of the outgroup (D. hansenii or Y. lipolytica) extrapolate well with results of the most distant species among protoploids (Z. rouxii vs. K. lactis or A. gossypii). The nonlinear form of this relationship indicates that significant sequence divergence occurs before chromosomal maps become extensively rearranged. At larger evolutionary distances, chromosome reshuffling catches up to protein-sequence divergence, which becomes limited by saturation and functional constraints (see Discussion).
Figure 5.
Relationship between protein sequence divergence and synteny conservation. Average sequence identities between orthologous proteins (ordinate) in all pairwise species comparisons were calculated from Figure 3. The percent of orthologs remaining in synteny (abscissa) for the same pairs of yeast species is the ratio of the total number of orthologs within syntenic blocks over the total number of orthologs between the two species considered.
Genome rearrangements from inferred ancestors
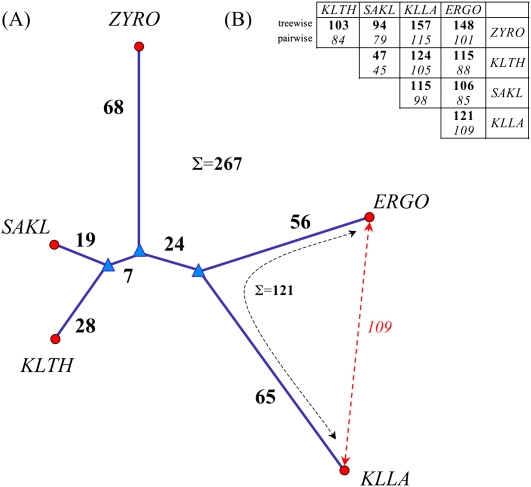
The slow pace of genome rearrangements enabled us to infer ancestral events from contemporary genomes based on conservation of synteny blocks. Such an inference is possible because, in protoploid yeasts, the probability of independent formation of such blocks in different lineages is likely to remain negligible compared to their inheritage from a common ancestor. Each genome was factored into a sequence of ordered synteny blocks common to all genomes, and super-blocks were computed using the method developed by G Jean, DJ Sherman, and M Nikolski (in prep.). From this, we computed median genomes for the five protoploid genomes, as well as rearrangement trees containing intermediate ancestral candidates for different branches (Goëffon et al. 2008). Figure 6 shows pairwise rearrangement distances and a rearrangement tree. The rearrangement tree separates Z. rouxii from the other four species, in agreement with the phylogenetic tree based on sequence alignments (see Fig. 1). Again, in agreement with phylogeny, S. kluyveri and K. thermotolerans are the closest in terms of rearrangements. However, in the rearrangement tree, A. gossypii and K. lactis are as diverged from each other as Z. rouxii is from the ancestor of all five species. This underlines the highly dynamic nature of yeast genomes, even within a related group like the Saccharomycetaceae. (The difference in rearrangement distance between the binary rearrangement tree and a simple star topology rooted at the median is only 10 events [3.5%].)
Figure 6.
Genome rearrangements. (A) Minimum rearrangement tree computed using the FAUCILS stochastic local search method (Goëffon et al. 2008); (●) contemporary genomes; (▲)inferred ancestral genomes; bold figures show the number of rearrangements in this minimum tree. (B) Contrast between rearrangement distances summing the branches in the tree (in bold, upper row), and pairwise distances (in italics, lower row) computed using the GRIMM method (Tesler 2002). Note that the requirement of going through a common ancestor usually increases treewise distances compared to pairwise distances owing to the triangle inequality, but decreases the sum of distances for the entire tree when all genomes are taken into account. This is illustrated in A for the pair ERGO and KLLA, Σ = 121 > 109, while the overall sum for the tree is 267. The rearrangement tree independantly corroborates the phylogenetic tree calculated using sequence similarity (see Discussion).
Discussion
With the complete genome sequencing and analysis of three novel yeast species, Z. rouxii, K. thermotolerans, and S. kluyveri, we have been able to perform the first comparative genomic study involving four distinct clades (and five species) of Saccharomycetaceae, that all separated from S. cerevisiae before its ancestral genome duplication. In two previous studies, yeasts of these clades, namely, A. gossypii (Dietrich et al. 2004) and K. waltii (Kellis et al. 2004), were individually compared to S. cerevisiae but not between themselves. In another study (Dujon et al. 2004), K. lactis was involved in multispecies comparative genomics with four other hemiascomycetes, but none was a protoploid Saccharomycetaceae. The present study was facilitated by the development of new methods that may be of general interest for other comparative genomic studies between species having sufficient conservation of synteny (see Methods). Among them is the method we used to define gene orthology (ML Seret and PV Baret, in prep.). Compared to YGOB, which implies manual curation of data (Byrne and Wolfe 2005), and to SYNERGY, which requires a priori definition of parameters (Wapinski et al. 2007), our method (IONS) simply subdivides precomputed sets of homologs (based on sequence similarity) using gene neighborhood evidence in a reiterative process with gradually decreasing neighborhood sizes until series of homologs with only one gene per species are obtained.
Genomes of the five species we examined reveal common signatures that confirm the monophyletic origin of Saccharomycetaceae and distinguish them from representatives of other subdivisions of hemiascomycetes, used here as outgoups for comparison. Among those signatures are short centromeres (Meraldi et al. 2006), triplication of mating cassettes (except for S. kluyveri) (Butler et al. 2004), a very limited number of spliceosomal introns (Bon et al. 2003), usage of the universal genetic code (Miranda et al. 2006), and a single rDNA locus containing the genes for the 5S RNA molecule (Acker et al. 2008). Additional discriminatory signatures are also found in their mitochondrial genomes: The genes encoding subunits of complex I of the respiratory chain are missing, while a gene for a variable ribosomal subunit and another one for an abnormal tRNA-Thr (UAG) altering the mitochondrial genetic code are present (http://www.ncbi.nlm.nih.gov/genomes/OrganelleResource.cgi?opt=organelle&taxid=4751).
The five protoploid genomes show only limited variation in the total number of protein-coding genes (<7% relative to the mean) and almost no variation for some noncoding RNA genes. But their chromosome number varies from six to eight, the number of tRNA genes varies from 163 to 272, and the average genome composition from ∼39% to 52% GC, indicating important lineage-specific evolution that may be correlated with the different lifestyles and properties of these five species. Such diversity is not surprising given the large evolutionary distances separating the five species, as revealed by the significant sequence divergence among orthologous proteins. This was not known before. Even the two species classified within the same Lachancea clade, S. kluyveri and K. thermotolerans, show average amino acid identity of only ∼58%. This is much less than between the two most distant species of the Saccharomyces sensu stricto clade (Bon et al. 2000a; Cliften et al. 2003; Kellis et al. 2003), which has nevertheless been more extensively studied, illustrating again previous emphasis on yeasts involved in biotechnological fermentation.
Evolutionary distances among protoploid yeasts are such that our sequence-based phylogeny (Fig. 1), which is in agreement with that of Fitzpatrick et al. (2006), is not entirely congruent with the phylogeny of Saccharomycetaceae published by Kurtzman (2003), where clade 12 (Eremothecium) is separated from clade 11 (Kluyveromyces). In both cases, however, Z. rouxii branches separately from the four other yeasts and shares a common origin with the duplicated yeasts of the Saccharomyces and Nakaseomyces clades. Results presented here definitely demonstrate that Z. rouxii separated from them before the whole-genome duplication. Given the tree topology of Kurtzman (2003) and the fact that the Vanderwaltozyma clade was recently demonstrated to have emerged after the duplication (Scannell et al. 2007b), Zygosaccharomyces represents the last clade having diverged from its relatives prior to the whole-genome duplication. Of all protoploid yeasts studied here, Z. rouxii is, therefore, the most closely related to the putative ancestral genome of S. cerevisiae.
Despite their long evolutionary branches, the five protoploid species we analyzed show a high degree of synteny conservation. Even if chromosome rearrangement breakages are relatively numerous (a few hundred, except between S. kluyveri and K. thermotolerans), they leave relatively long synteny blocks of one or two dozen genes on average, and large proportions of genomes (>80%) are present in still recognizable synteny blocks in all pairwise comparisons. Such a range of synteny conservation was not available for yeasts before. The Saccharomyces sensu stricto yeasts share almost completely conserved synteny with few breakpoints (Fischer et al. 2000, 2001), and other yeasts are too distantly separated to retain much synteny (Fischer et al. 2006). The nonlinear relationship between sequence divergence and synteny conservation is compatible with the idea that genetic maps are more robust than DNA sequences over evolutionary periods corresponding to entire families or even orders of the Linnean taxonomical hierarchy. The effects of genome rearrangements become significant over longer evolutionary times when sequence changes saturate. Similar ideas based on much more limited sets of data were already discussed (Langkjaer et al. 2000; Llorente et al. 2000b; Malpertuy et al. 2000b). The frequency distribution of synteny blocks according to their size is similar to results obtained from the 12 Drosophila genomes and other insects (Zdobnov and Bork 2007), suggesting that chromosome rearrangements are not random in either of these two groups of eukaryotes despite very distinct genome organizations. Accordingly, genome rearrangement trees built from inferred ancestors are not entirely coincident with phylogenetic trees built on sequence conservation of gene products. Differences in the rate of genome rearrangements between yeast lineages have been reported (Fischer et al. 2006). Note that conserved synteny blocks often contain short internal gene deletions and/or single gene insertions (“intervening” genes). Many “intervening” genes have homologs at ectopic locations in either the same or other genomes and may have arisen by single gene transfer (possibly by retroposition) or by dispersed short segmental duplications similar to the few that were recognized. A few “intervening” genes correspond to horizontally transferred genes, as previously illustrated by the presence of a bacterial transposase-type of sequence in A. gossypii (Hall et al. 2005). Gene transfer from bacteria to yeasts is also documented in S. kluyveri by the presence of six copies of a bacterial IS element (family IS607) on chromosomes B, G, and H (Rolland et al. 2009).
The genomes of protoploid yeasts contain remarkably few segmental duplications (except in subtelomeric locations), in contrast to genomes of higher eukaryotes, where they are a major source of genome rearrangements (Johnson et al. 2001; Samonte and Eichler 2002), and to the high frequency of their spontaneous formation in S. cerevisiae (Payen et al. 2008). The limited role of segmental duplications in the evolution of yeast genomes may be related to the important instability of duplicated copies of identical sequences. Compared to their paucity in recent duplications of nearly identical sequences, genomes of protoploid yeasts contain a surprisingly high number of paralogous genes: about a third of their gene content. Thus, we can say that there is no such thing as “nonduplicated” yeasts. Families of paralogs issue from ancient duplication events that leave diverged gene copies at dispersed genomic locations and from TGAs, in which gene copies are globally less diverged. Although some of the dispersed duplications appear ancestral and conserved in all protoploid species (some are also conserved in outgroup yeasts, suggesting they may be ancestral to all hemiascomycetes), we recognized a significant number of lineage-specific events of gene duplication and loss, some of which resulted in significant gene family expansions that may be correlated with specific biological properties of each yeast. But the remarkable point is that, despite such dynamics, the diverse protoploid species have similar levels of global genome redundancy (the figure for A. gossypii is slightly smaller, in agreement with a reductive and adaptive evolution [Dietrich et al. 2004]). The net redundancy, ∼1.25 on average, reflects an equilibrium between gene duplication and loss over long evolutionary periods, when this equilibrium is not affected by accidental events such as whole-genome duplication or adaptive events such as gene family expansions.
The genome sequences of the Saccharomycetaceae we described, which offer a more balanced representation of the different clades, provide the framework to understand the origin of the common properties and individual variations of these yeasts. Their core genetic repertoire (core proteome) consists of approximately 3300 protein families, within a pan-proteome of approximately 5000 families for all Saccharomycetaceae. The fact that the pan repertoire dramatically increases when outgroup species are considered underlines the major evolutionary gaps between the different subdivisions of hemiascomycetes and the need for additional genomic studies of this interesting fungal group.
Methods
Genome sequencing
Complete genome sequences of K. thermotolerans (CBS6340) and Z. rouxii (CBS732; determined at Genoscope) and of S. kluyveri (CBS3082; determined at the Washington University Genome Sequencing Center) were automatically assembled from shotgun Sanger sequencing reads using ARACHNE (Jaffe et al. 2003) and finished by dedicated sequencing for gaps and low-quality regions.
Genome annotation
CDS
Gene models for protein-coding genes were constructed using GeneMark (Borodovsky and McIninch 1993) trained with a conservative set of coding and noncoding sequences for each of the four genomes: Z. rouxii, K. thermotolerans, S. kluyveri, and K. lactis. All open reading frames (ORFs) greater than 80 codons were examined. Overlapping conflicts were solved by best GeneMark prediction and/or existence of BLASTP alignments with the UniProtKB database. Gene models were manually examined and simultaneously annotated for the four species after automated identification of homologs (DJ Sherman, T Martin, and P Durrens, in prep.).
Spliceosomal introns
Consensus splice sites and branch point sequences were defined for each yeast species from introns of ribosomal protein-coding genes (Bon et al. 2003). All combinations of the three motifs separated by appropriate distances (also defined for each yeast species) were examined for the possible formation of CDS greater than 80 codons after splicing. Note that introns in 5′-UTRs were, therefore, not systematically predicted. Overlapping intron predictions were manually curated.
Genes for noncoding RNAs
Genes for tRNAs were identified as described in Marck et al. (2006). Ribosomal DNA repeats were identified by comparison to other yeast genomes (Chindamporn et al. 1993; Rustchenko and Sherman 1994) and mapped to chromosomes using previous karyotype data (Sor and Fukuhara 1989; Neuvéglise et al. 2000; Sychrova et al. 2000). Genes for C/D snoRNAs and snRNAs were annotated by a combination of sequence similarity search with S. cerevisiae, secondary structure, and synteny conservation. H/ACA snoRNAs were not systematically identified.
Mobile genetic elements
Transposable elements were detected by TBLASTN on chromosome sequences using known fungal transposable elements as queries, and manually curated. Identified elements were then used to detect partial or degenerate elements and solo LTRs.
Protein families
Four complementary distance matrices were computed between the predicted translation products for the 47,874 protein-coding genes of the seven yeasts annotated by Génolevures (C. glabrata, Z. rouxii, K. thermotolerans, S. kluyveri, K. lactis, D. hansenii, and Y. lipolytica) (http://www.genolevures.org/), plus S. cerevisiae (http://www.yeastgenome.org/) and A. gossypii (http://agd.vital-it.ch/), combining BLAST and Smith-Waterman alignments with and without filtering for homeomorphy (Wu et al. 2004). Symmetric matrices derived from amino acid identities were constructed and submitted to MCL clustering (Enright et al. 2002) with a range of inflation parameters. These competing partitions were reconciled using the consensus method of Nikolski and Sherman (2007) and manually curated using literature search and systematic comparisons with COG (Tatusov et al. 2003) and PIRSF (Wu et al. 2004) classifications.
Orthologous genes
SONS were defined by the combination of two dimensions: sequence similarity of gene products (defined from families; see above), and conservation of gene neighborhood along chromosomes (Supplemental Fig. 7). Two genes of different yeast species whose translation products belong to the same family (homologs by similarity) will be members of the same SONS if they share at least one pair of neighbors that are also homologous to each other by similarity. The process is reiterated for all possible hetero-specific pairwise comparisons of homologs. At an initial step, neighboring is examined for 15 genes on each side of the two homologs considered, giving rise to (2 × 14)2/2 pairwise comparisons. Homologs that do not share a pair of homologous neighbors are separated in two distinct SONS. The process is reiterated (reducing the neighborhood size in order to decrease the probability of spurious connections) until each SONS contains ≤1 gene for each yeast species. Homologs in such a case are considered as orthologs confirmed by synteny. Details of the method and comparisons with other methods to infer orthology will be published separately (ML Seret and PV Baret, in prep.).
Syntenic block construction
Synteny blocks between two yeast genomes were constructed from the physical adjacency of orthologous genes (defined by SONS, above) along chromosomes (deduced from sequence-based map), using two parameters: A, the minimum number of ortholog pairs forming anchor points; and I, the maximum number of nonorthologous genes (“intervening”) between two successive anchor points. (A and I were set, respectively, to 5 and 10, following Fischer et al. [2006].) Note that TGAs (see text) were considered as equivalent to a single gene.
Acknowledgments
We thank E. Beyne and H. Soueidan for help in programming and data management, Y. Boulard for help in tRNA gene search, M. Santos for sharing unpublished results on tRNA genes, A.N. Nikolskaya for expertise in curating protein families, and M. Aigle and F. Tekaia for useful discussions. This work was supported in part by funding from the Consortium National de Recherche en Génomique (CNRG) to Génoscope, from CNRS (GDR 2354; Génolevures), ANR (ANR-05-BLAN-0331; GENARISE), ARC (grant 3738), Région Aquitaine (Pôle Recherche en Informatique; 20051306001AB), and Bureau des ressources génétiques (grant 347; Diversité fongique), and in part by grants awarded to R.W. by the National Human Genome Research Institute of the U.S. National Institutes of Health, and by funds provided to M.J. by the James S. McDonnell Foundation. B.D. is a member of Institut Universitaire de France.
Footnotes
[Supplemental material is available online at http://www.genome.org and at http://www.genolevures.org/. The sequence data for Zygosaccharomyces rouxii and Kluyveromyces thermotolerans have been submitted to EMBL-Bank (http://www.ebi.ac.uk/embl/) under accession nos. CU928173–CU928176, CU928178, CU928179, CU928181 and CU928165–CU928171, and CU928180, respectively. Saccharomyces kluyveri sequences were deposited to GenBank under accession no. AACE03000000.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.091546.109.
References
- Acker J, Ozanne C, Kachouri-Lafond R, Gaillardin C, Neuvéglise C, Marck C. Dicistronic tRNA-5S genes in Yarrowia lipolytica: An alternative TFIIIA-independent way for expression of 5S rRNA genes. Nucleic Acids Res. 2008;18:5832–5844. doi: 10.1093/nar/gkn549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Araki H, Jearnpipatkul A, Tatsumi H, Sakurai T, Ushio K, Muta T, Oshima Y. Molecular and functional organization of yeast plasmid pSR1. J Mol Biol. 1985;182:191–203. doi: 10.1016/0022-2836(85)90338-9. [DOI] [PubMed] [Google Scholar]
- Aslett M, Wood V. Gene ontology annotation status of the fission yeast genome: Preliminary coverage approaches 100% Yeast. 2006;23:913–919. doi: 10.1002/yea.1420. [DOI] [PubMed] [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplication revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Barnett JA. A history of research on yeasts 10: Foundations of yeast genetics. Yeast. 2007;24:799–845. doi: 10.1002/yea.1513. [DOI] [PubMed] [Google Scholar]
- Beck H, Dobritzsch D, Piskur J. Saccharomyces kluyveri as a model organism to study pyrimidine degradation. FEMS Yeast Res. 2008;8:1209–1213. doi: 10.1111/j.1567-1364.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- Blank LM, Lehmbeck F, Sauer U. Metabolic flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res. 2005;5:545–558. doi: 10.1016/j.femsyr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Boekhout T. Biodiversity: Gut feeling for yeasts. Nature. 2005;434:449–451. doi: 10.1038/434449a. [DOI] [PubMed] [Google Scholar]
- Bon E, Neuvéglise C, Casarégola S, Artiguenave F, Wincker P, Aigle M, Durrens P. Genomic exploration of the hemiascomycetous yeasts: 5. Saccharomyces bayanus var. uvarum. FEBS Lett. 2000a;487:37–41. doi: 10.1016/s0014-5793(00)02276-6. [DOI] [PubMed] [Google Scholar]
- Bon E, Neuvéglise C, Lépingle A, Wincker P, Artiguenave F, Gaillardin C, Casarégola S. Genomic exploration of the hemiascomycetous yeasts: 6. Saccharomyces exiguus. FEBS Lett. 2000b;487:42–46. doi: 10.1016/s0014-5793(00)02277-8. [DOI] [PubMed] [Google Scholar]
- Bon E, Casarégola S, Blandin G, Llorente B, Neuvéglise C, Munsterkotter M, Guldener U, Mewes HW, Van Helden J, Dujon B, et al. Molecular evolution of eukaryotic genomes: Hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Forgan AH, Pretorius IS, Chambers PJ. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 2008;8:1185–1195. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- Borodovsky M, McIninch J. Recognition of genes in DNA sequence with ambiguities. Biosystems. 1993;30:161–171. doi: 10.1016/0303-2647(93)90068-n. [DOI] [PubMed] [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. Evolution of the MAT locus and its HO endonuclease in yeast species. Proc Natl Acad Sci. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes JK, Morris GP, Li WH. Reorganization of adjacent gene relationships in yeast genomes by whole-genome duplication and gene deletion. Mol Biol Evol. 2006;23:1136–1143. doi: 10.1093/molbev/msj121. [DOI] [PubMed] [Google Scholar]
- Casaregola S, Lépingle A, Bon E, Neuvéglise C, Huu-Vang N, Artiguenave F, Wincker P, Gaillardin C. Genomic exploration of the Hemiascomycetous Yeasts: 7. Saccharomyces servazzii. FEBS Lett. 2000;487:47–51. doi: 10.1016/s0014-5793(00)02278-x. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chappell AS, Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol Cell Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chindamporn A, Iwaguchi S, Nakagawa Y, Homma M, Tanaka K. Clonal size-variation of rDNA cluster region on chromosome XII of Saccharomyces cerevisiae. J Gen Microbiol. 1993;139:1409–1413. doi: 10.1099/00221287-139-7-1409. [DOI] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterson R, Cohen BA, Johnston M. Finding functional features in Saccharomyces cerevisiae by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Cliften PF, Fulton RS, Wilson RK, Johnston M. After the duplication: Gene loss and adaptation in Saccharomyces genomes. Genetics. 2006;172:863–872. doi: 10.1534/genetics.105.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Probabilistic cross-species inference of orthologous genomic regions created by whole-genome duplication in yeast. Genetics. 2008;179:1681–1692. doi: 10.1534/genetics.107.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A, Smith E, Kruglyak L, Alani E. Identification and dissection of a complex DNA repair sensitivity phenotype in Baker's yeast. PLoS Genet. 2008;4:e1000123. doi: 10.1371/journal.pgen.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Montigny J, Straub ML, Potier S, Tekaia F, Dujon B, Wincker P, Artiguenave F, Souciet JL. Genomic exporation of the hemiascomycetous yeasts: 8. Zygosaccharomyces rouxii. FEBS Lett. 2000;487:52–55. doi: 10.1016/s0014-5793(00)02279-1. [DOI] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Puhlmann R, Luedi P, Choi S, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Dujon B. Hemiascomycetous yeasts at the forefront of comparative genomics. Curr Opin Genet Dev. 2005;6:614–620. doi: 10.1016/j.gde.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casarégola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead C, Dujon B. Structure of Kluyveromyces lactis subtelomeres: Duplications and gene content. FEMS Yeast Res. 2006;6:428–441. doi: 10.1111/j.1567-1364.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405:415–454. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- Fischer G, Neuvéglise C, Durrens P, Gaillardin C, Dujon B. Evolution of gene order in the genomes of two related yeasts species. Genome Res. 2001;11:2009–2019. doi: 10.1101/gr.212701. [DOI] [PubMed] [Google Scholar]
- Fischer G, Rocha EP, Brunet F, Vergassola M, Dujon B. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet. 2006;2:e32. doi: 10.1371/journal.pgen.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goëffon A, Nikolski M, Sherman DJ.An efficient probabilistic population-based descent for the median genome problem. Proceedings of the 2008 GECCO Conference Companion on Genetic and Evolutionary Computation; New York: ACM; 2008. pp. 315–322. [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Ormandy JE, Poulter RT. L1-like non-LTR retrotransposons in the yeast Candida albicans. Curr Genet. 2001;39:83–91. doi: 10.1007/s002940000181. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Busby JN, Poulter RT. A yeast model for target-primed (non-LTR) retrotransposition. BMC Genomics. 2007;8:263. doi: 10.1186/1471-2164-8-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall C, Brachat S, Dietrich FS. Contribution of horizontal transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann G, McIntyre M, Nielsen J. Fungal genomics beyond Saccharomyces cerevisiae. Curr Opin Biotechnol. 2003;14:226–231. doi: 10.1016/s0958-1669(03)00020-x. [DOI] [PubMed] [Google Scholar]
- Jaffe DB, Butler J, Gnerre S, Mauceli E, Lindblad-Toh K, Mesirov JP, Zody MC, Lander ES. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–476. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jansen M, Veurink JH, Euverink GJ, Dijkhuizen L. Growth of the salt-tolerant yeast Zygosaccharomyces rouxii in microtiter plates: Effects of NaCl, pH and temperature on growth and fusel alcohol production from branched-chain amino acids. FEMS Yeast Res. 2003;3:313–318. doi: 10.1111/j.1567-1364.2003.tb00172.x. [DOI] [PubMed] [Google Scholar]
- Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Fell JW. The yeast handbook. Springer-Verlag; New York: 2006. Yeast systematics and phylogeny—implications of molecular identification methods for studies in ecology; pp. 11–30. [Google Scholar]
- Kurtzman C, Piskur J. The yeast handbook. Springer-Verlag; New York: 2006. Taxonomy and phylogenetic diversity among the yeasts; pp. 29–46. [Google Scholar]
- Kurtzman CP, Robnett CJ. Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: Finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Langkjaer RB, Nielsen ML, Daugaard PR, Li W, Piskur J. Yeast chromosomes have been significantly reshaped during their evolutionary history. J Mol Biol. 2000;304:271–288. doi: 10.1006/jmbi.2000.4209. [DOI] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts I, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Malpertuy A, Blandin G, Artiguenave F, Wincker P, Dujon B. Genomic exploration of the hemiascomycetous yeasts: 12. Kluyveromyces marxianus var. marxianus. FEBS Lett. 2000a;487:71–75. doi: 10.1016/s0014-5793(00)02283-3. [DOI] [PubMed] [Google Scholar]
- Llorente B, Malpertuy A, Neuvéglise C, de Montigny J, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, et al. Genomic exploration of the hemiascomycetous yeasts: 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 2000b;487:101–112. doi: 10.1016/s0014-5793(00)02289-4. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Magee BB, Sanchez MD, Saunders D, Harris D, Berriman M, Magee PP. Extensive chromosome rearrangements distinguish the karyotype of the hypovirulent species Candida dubliniensis from the virulent Candida albicans. Fungal Genet Biol. 2008;45:338–350. doi: 10.1016/j.fgb.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpertuy A, Llorente B, Blandin G, Artiguenave F, Wincker P, Dujon B. Genomic exporation of the hemiascomycetous yeasts: 10. Kluyveromyces thermotolerans. FEBS Lett. 2000a;487:61–65. doi: 10.1016/s0014-5793(00)02281-x. [DOI] [PubMed] [Google Scholar]
- Malpertuy A, Tekaia F, Casarégola S, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, de Montigny J, et al. Genomic exploration of the hemiascomycetous yeasts: 19. Ascomycete-specific genes. FEBS Lett. 2000b;487:113–121. doi: 10.1016/s0014-5793(00)02290-0. [DOI] [PubMed] [Google Scholar]
- Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H. The RNA polymerase III-dependent family of genes in hemiascomycetes: Comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 2006;34:1816–1835. doi: 10.1093/nar/gkl085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda I, Silva R, Santos MA. Evolution of the genetic code in yeasts. Yeast. 2006;23:203–213. doi: 10.1002/yea.1350. [DOI] [PubMed] [Google Scholar]
- Møller K, Bro C, Piskur J, Nielsen J, Olsson L. Steady-state and transient-state analyses of aerobic fermentation in Sacchromyces kluyveri. FEMS Yeast Res. 2002;2:233–244. doi: 10.1111/j.1567-1364.2002.tb00088.x. [DOI] [PubMed] [Google Scholar]
- Møller K, Sharif MZ, Olsson L. Production of fungal alpha-amylase by Saccharomyces kluyveri in glucose-limited cultivations. J Biotechnol. 2004;111:311–318. doi: 10.1016/j.jbiotec.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Taylor BA. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvéglise C, Bon E, Lépingle A, Wincker P, Artiguenave F, Gaillardin C, Casarégola S. Genomic exploration of the hemiascomycetous yeasts: 9. Saccharomyces kluyveri. FEBS Lett. 2000;487:56–60. doi: 10.1016/s0014-5793(00)02280-8. [DOI] [PubMed] [Google Scholar]
- Neuvéglise C, Chalvet F, Wincker P, Gaillardin C, Casarégola S. Mutator-like element in the yeast Yarrowia lipolytica displays multiple alternative splicings. Eukaryot Cell. 2005;4:615–624. doi: 10.1128/EC.4.3.615-624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolski M, Sherman DJ. Family relationships: Should consensus reign? Consensus clustering for protein families. Bioinformatics. 2007;23:e71–e76. doi: 10.1093/bioinformatics/btl314. [DOI] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Genetics of Candida albicans, a diploid human fungal pathogen. Annu Rev Genet. 2007;41:193–211. doi: 10.1146/annurev.genet.41.042007.170146. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Pan X, Peyser BD, Ye P, Meluh PB, Yuan DS, Irizarry RA, Bader JS, Spencer FA, Boeke JD. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Oura T, Kajiwara S. Substrate specificity and regioselectivity of delta12 and omega3 fatty acid desaturases from Saccharomyces kluyveri. Biosci Biotechnol Biochem. 2008;72:3174–3179. doi: 10.1271/bbb.80361. [DOI] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Fischer G, Marck C, Proux C, Sherman DJ, Coppée J-Y, Johnston M, Dujon B, Neuvéglise C. Unusual composition of a yeast chromosome arm is associated with its delayed replication. Genome Res. 2009 doi: 10.1101/gr.090605.108. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23:494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, Neuvéglise C, Sacerdot C, Dujon B. Insertion of horizontally transferred genes within conserved syntenic regions of yeast genomes. PLoS ONE. 2009 doi: 10.1371/journal.pone.0006515. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E, Lithwick G, Levy AA. Structure and evolution of the hAT transposon superfamily. Genetics. 2001;158:949–957. doi: 10.1093/genetics/158.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko EP, Sherman F. Physical constitution of ribosomal genes in common strains of Saccharomyces cerevisiae. Yeast. 1994;10:1157–1171. doi: 10.1002/yea.320100904. [DOI] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE. Segmental duplications and the evolution of the primate genome. Nat Rev Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Butler G, Wolfe KH. Yeast genome evolution—the orgin of the species. Yeast. 2007a;24:929–942. doi: 10.1002/yea.1515. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci. 2007b;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of S. cerevisiae. Nature. 2009;458:342–346. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solieri L, Giudici P. Yeasts associated to traditional balsamic vinegar: Ecological and technological features. Int J Food Microbiol. 2007;125:36–45. doi: 10.1016/j.ijfoodmicro.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Solieri L, Cassanelli S, Croce MA, Giudici P. Genome size and ploidy level: New insights for elucidating relationships in Zygosaccharomyces species. Fungal Genet Biol. 2008;45:1582–1590. doi: 10.1016/j.fgb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Sor F, Fukuhara H. Analysis of chromosomal DNA patterns of the genus Kluyveromyces. Yeast. 1989;5:1–10. doi: 10.1002/yea.320050103. [DOI] [PubMed] [Google Scholar]
- Souciet JL, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, de Montigny J, Dujon B, et al. Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett. 2000;487:3–12. doi: 10.1016/s0014-5793(00)02272-9. [DOI] [PubMed] [Google Scholar]
- Sychrova H, Braun V, Potier S, Souciet JL. Organization of specific genomic regions of Zygosaccharomyces rouxii and Pichia sorbitophila: Comparison with Saccharomyces cerevisiae. Yeast. 2000;16:1377–1385. doi: 10.1002/1097-0061(200011)16:15<1377::AID-YEA637>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Talla E, Anthouard V, Bouchier C, Frangeul L, Dujon B. The complete mitochondrial genome of the yeast Kluyveromyces thermotolerans. FEBS Lett. 2005;579:30–40. doi: 10.1016/j.febslet.2004.10.106. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesler G. GRIMM: Genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- Vilela-Moura A, Schuller D, Mendes-Faia A, Côrte-Real M. Reduction of volatile acidity of wines by selected yeast strains. Appl Microbiol Biotechnol. 2008;80:881–890. doi: 10.1007/s00253-008-1616-x. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene tree. Bioinformatics. 2007;23:549–558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, Cao Z, Gu Z, Bruno D, Miranda M, Nguyen M, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci. 2007;104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wong S, Butler G, Wolfe KH. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc Natl Acad Sci. 2002;99:9272–9277. doi: 10.1073/pnas.142101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P, et al. PIRSF: Family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, Kronstad JW, DeAngelis YM, Reeder NL, Johnstone KR, et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci. 2007;104:18730–18735. doi: 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]