Abstract
Piwi proteins, a subclass of Argonaute-family proteins, carry ∼24–30-nt Piwi-interacting RNAs (piRNAs) that mediate gonadal defense against transposable elements (TEs). We analyzed the Drosophila ovary somatic sheet (OSS) cell line and found that it expresses miRNAs, endogenous small interfering RNAs (endo-siRNAs), and piRNAs in abundance. In contrast to intact gonads, which contain mixtures of germline and somatic cell types that express different Piwi-class proteins, OSS cells are a homogenous somatic cell population that expresses only PIWI and primary piRNAs. Detailed examination of its TE-derived piRNAs and endo-siRNAs revealed aspects of TE defense that do not rely upon ping-pong amplification. In particular, we provide evidence that a subset of piRNA master clusters, including flamenco, are specifically expressed in OSS and ovarian follicle cells. These data indicate that the restriction of certain TEs in somatic gonadal cells is largely mediated by a primary piRNA pathway.
While the diversity of recognized small RNAs continues to grow, the majority of regulatory processes involving <30-nucleotide (nt) RNAs is governed by three fundamental classes: ∼21–24-nt microRNAs (miRNAs), 21-nt small interfering RNAs (siRNAs), and ∼24–30-nt Piwi-interacting RNAs (piRNAs). The function of these RNA families is mediated via Argonaute (Ago) effector proteins, with miRNAs and siRNAs housed in Ago-subclass effectors, and piRNAs resident in Piwi-subclass effectors (Kim et al. 2009). Broadly speaking, miRNAs repress host gene expression, siRNAs repress host genes, transposable elements (TEs), and viruses, and piRNAs repress TEs.
A long history of genetic analyses of D. melanogaster oogenesis (Gillespie and Berg 1995; Cox et al. 1998; Harris and Macdonald 2001; Cook et al. 2004; Pane et al. 2007) and TE control (Rubin et al. 1982; Bucheton et al. 1984; Pelisson et al. 1994; Prud'homme et al. 1995) proved to uncover many of the key features and factors involved in the piRNA pathway. Recent cloning of small RNAs associated with Piwi-subclass proteins provided mechanistic insight into piRNA biogenesis in Drosophila (Saito et al. 2006; Vagin et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007; Nishida et al. 2007) and vertebrates (Aravin et al. 2006, 2007; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006). In addition, Piwi complexes could cleave complementary targets (Lau et al. 2006; Saito et al. 2006; Gunawardane et al. 2007), establishing their biochemical activity similar to certain Ago proteins (Kim et al. 2009).
In Drosophila ovaries, PIWI and Aubergine (AUB) predominantly carry antisense TE-piRNAs that exhibit strong 5′ uridine (5′ U) bias, while Argonaute 3 (AGO3) is the predominant carrier of sense TE-piRNAs with bias for adenine at the tenth position (A10) (Brennecke et al. 2007; Gunawardane et al. 2007). These properties of sense and antisense piRNAs reflect an amplification cycle known as “piRNA ping-pong.” In brief, antisense TEs embedded in noncoding “master piRNA transcripts” generate antisense piRNAs that load AUB (or PIWI). These piRNA complexes not only cleave active TE transcripts, but in so doing can generate sense piRNAs that load AGO3. Since target cleavage occurs opposite nucleotides 10 and 11 of the guide RNA 5′ end, this defines a piRNA pair that overlaps by 10 nt, for which the guide strand begins with U and its target strand bears A10. Conversely, AGO3-mediated cleavage of master piRNA transcripts can regenerate antisense piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). Altogether, this cycle constitutes an adaptive immune response that senses active TE transcripts and selectively amplifies piRNAs capable of directing their destruction, not only in Drosophila, but also in diverse metazoans (Aravin et al. 2007; Houwing et al. 2007; Grimson et al. 2008).
Despite recent progress, studies of the piRNA pathway in Drosophila are challenged by the tedious nature of gonad dissection. Larger masses of mammalian gonads are easily obtained, but there are fewer mutants available and only limited strategies to manipulate them genetically. A limitation of both systems is that gonads are composed of multiple cell types that express different complements of Piwi proteins, thus obscuring the cells-of-origin of piRNAs cloned from whole-gonad homogenates. For these reasons, identification of homogenous cell lines that express piRNAs would be desirable.
Niki et al. (2006) described D. melanogaster ovarian cell cultures, including a mixed female germline stem cell/ovarian somatic sheet line (fGS/OSS) and an apparently homogenous derivative culture lacking the germline stem cells termed OSS. We analyzed the OSS line in detail and found that in addition to miRNAs, it expresses high levels of endo-siRNAs and primary piRNAs. Detailed analysis of short RNAs mapping to TE sequences provided unique insight into the provenance and function of TE-siRNAs and TE-piRNAs, including the existence of an efficient PIWI-associated primary piRNA pathway in somatic ovarian cells.
Results
A simple strategy to deplete uncomplexed RNAs from total RNA
We recently used cation-exchange chromatography to enrich for piRNA complexes (Lau et al. 2006; Lau 2008). Naked nucleic acids are retained on the ionic matrix, while active regulatory RNAs contained in protein complexes are eluted with mild salt. This procedure is useful since 5′-phosphate-dependent cloning strategies do not exclude degradation fragments, due to the activity of RNA kinases such as CLP1 (Weitzer and Martinez 2007). In addition, this one-step procedure simultaneously enriches for RNAs in diverse complexes without the need for specific immunoprecipitations.
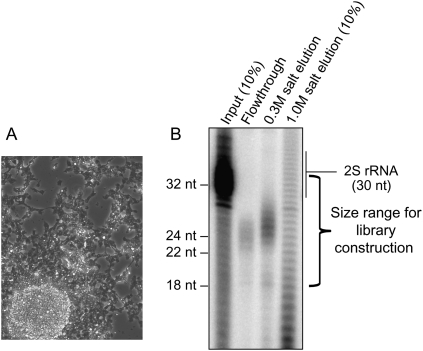
We fractionated extracts of OSS cells (Fig. 1A) on a HiTrap Q column, then radioactively labeled and analyzed the purified RNAs. Following this procedure, only ∼20–28-nt RNAs were present in the flowthrough and 300 mM salt eluate (Fig. 1B), with a slightly larger population of RNAs present in the latter fraction; 2S rRNA was effectively depleted. A ladder of RNAs with single-nucleotide resolution eluted with 1 M salt, which we inferred to comprise heterogeneously sized degradation fragments.
Figure 1.
Cation-exchange chromatography enriches for Argonaute- and Piwi-enclosed RNAs. (A) Phase contrast image of OSS cells illustrating their sheet-like morphology; under high density they also tend to form clumps. (B) pCp labeling of RNAs from total OSS RNA and various elutions from a HiTrap Q column. The mobility of the RNAs in the extract and elutions is slightly different from that of the synthetic markers owing to different salt concentrations of the loaded samples. Highly abundant 2S rRNA is visible in the input and ran slightly larger than its 30-nt sequence. In the flowthrough, ∼21–24-nt RNAs are visible, inferred to include miRNAs and siRNAs. In the 0.3 M salt elution, ∼24–28-nt RNAs are visible, inferred to represent the piRNA fraction. A heterogenous ladder of RNA fragments elutes with 1.0 M salt. Libraries were constructed from the flowthrough and 0.3 M salt elution.
In addition to separating noncomplexed RNAs, these fractionations provided the first indication that OSS cells express piRNAs. We then cloned the ∼18–34-nt RNA fraction from the flowthrough and low-salt eluate, and generated four lanes of sequence on two Illumina GA-II flow cells. In total, we mapped ∼14 million reads to the genome, and the data from the four sequencing reactions were indeed highly concordant (Supplemental Fig. S1).
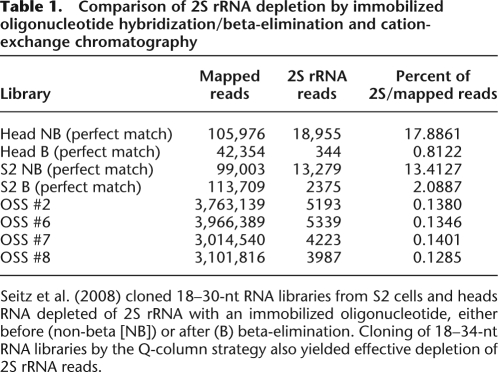
The efficacy of cation-exchange chromatography was evident in the depletion of 2S rRNA. For comparison, Seitz et al. (2008) showed that immobilized oligonucleotide hybridization followed by beta-elimination of 18–30-nt total RNA could quite effectively deplete 2S rRNA six- to 22-fold (Table 1), resulting in 2% and 0.8% 2S rRNA reads in S2 cell and head libraries, respectively. Although we cloned a wider size range encompassing most of the 2S rRNA population (which runs slightly larger than expected from its 30-nt sequence) (Fig. 1B), our Q-column-fractionated libraries yielded only 0.13% 2S rRNA reads. We inferred that these libraries should reciprocally be enriched in bona fide regulatory RNAs bound in protein complexes, including miRNAs, siRNAs, and piRNAs.
Table 1.
Comparison of 2S rRNA depletion by immobilized oligonucleotide hybridization/beta-elimination and cation-exchange chromatography
Seitz et al. (2008) cloned 18–30-nt RNA libraries from S2 cells and heads RNA depleted of 2S rRNA with an immobilized oligonucleotide, either before (non-beta [NB]) or after (B) beta-elimination. Cloning of 18–34-nt RNA libraries by the Q-column strategy also yielded effective depletion of 2S rRNA reads.
miRNA expression in OSS cells
We first identified ∼3.5 million reads that matched the currently annotated set of D. melanogaster miRNAs, encompassing 121 of 140 distinct mature miRNAs (i.e., not including precisely identical paralogs) previously identified by deep sequencing (Ruby et al. 2007b; Sandmann and Cohen 2007; Stark et al. 2007). While their read numbers were spread over five orders of magnitude, the relative abundance of miRNAs in the different sequencing reactions was nearly identical (Supplemental Table S1). The top 25 most abundant miRNAs, each contributing >1% of the total reads, accounted for 97.94% of miRNA reads and defined a core OSS profile. On the other hand, 106 distinct miRBase loci were cloned four or more times and were confidently annotated from our data without prior knowledge. Another 15 loci were sequenced one to three times, and presumably could have been confidently annotated with slightly deeper datasets (Fig. 2A; Supplemental Table S1).
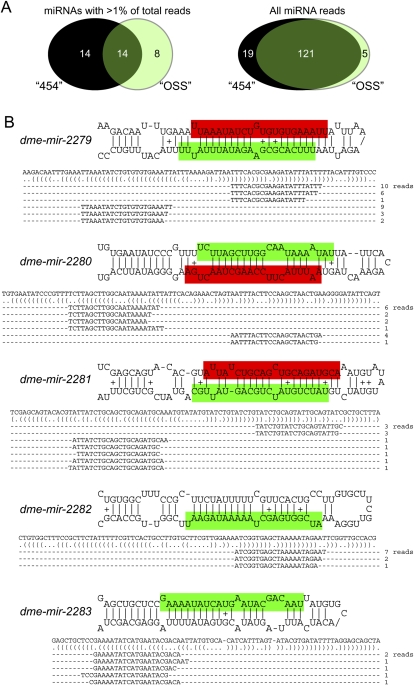
Figure 2.
miRNA expression in OSS cells. (A) OSS cells predominantly express a characteristic population of miRNAs (left) that overlaps only partially with the most abundant miRNAs annotated from across Drosophila development using 454 Life Sciences (Roche) sequencing (Ruby et al. 2007a). However, analysis of all reads indicates that the 454-annotated miRNAs and OSS miRNAs are very highly overlapping (right). Therefore, numerous ostensibly tissue-specific Drosophila miRNAs were captured at low levels in OSS cells by deep sequencing. (B) Five novel miRNA loci, expressed at a level of more than five reads/14 million library reads, were identified in OSS cells. Mature products were highlighted in green, and star strands in red; for some hairpins, the two small RNA products were cloned nearly equivalently.
It was therefore possible to identify >75%–85% of Drosophila miRNAs found by deep sequencing of heterogeneous and diverse tissues—including embryos and larvae of various stages, purified imaginal discs, adult heads, female and male bodies, dissected ovaries and testes, and S2 cells (Ruby et al. 2007b; Sandmann and Cohen 2007; Stark et al. 2007)—simply by massively sequencing this single cell type. We also identified some novel miRNAs, but these were surprisingly few, thus failing to fulfill the plausible expectation that many cell-type-specific miRNAs might be revealed by such deep sequencing. Only five new miRNAs (miR-2279, miR-2280, miR-2281, miR-2282, and miR-2283) were confidently annotated on the basis of more than four reads (i.e., expressed at >0.3 reads/million); three of these had star strand reads (Fig. 2B; Supplemental Table S1). All five miRNAs were poorly conserved and present only within the closest relatives of D. melanogaster (data not shown). Altogether, these data suggest that despite tens of thousands of genomic hairpins with qualities similar to miRNA precursors, a relatively limited set of bona fide miRNA genes exists in Drosophila.
TE-siRNA expression in OSS cells
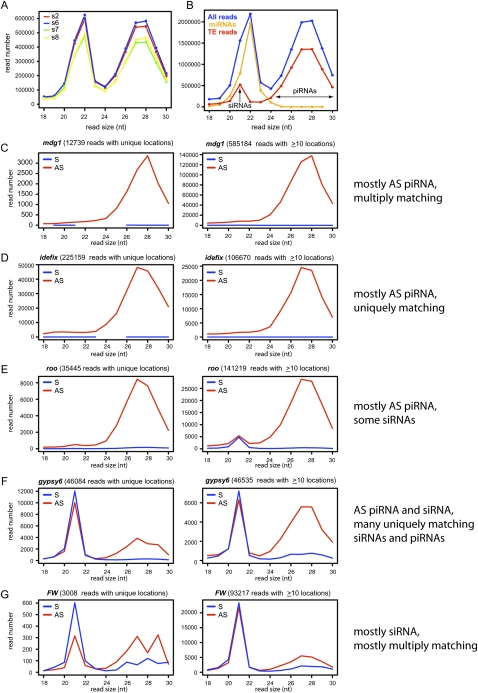
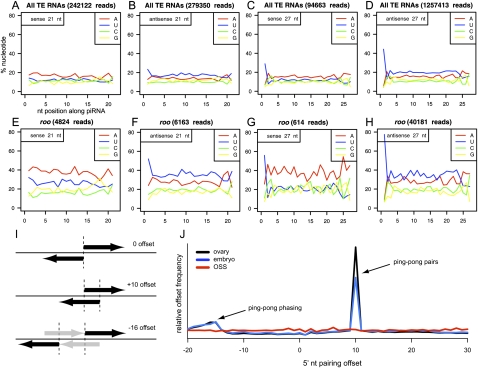
TEs were recently recognized as major sources of endo-siRNAs in Drosophila germline and somatic cells (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008). After subtracting the miRNA content of OSS libraries, a prominent class of precisely 21-nt reads derived from TEs was apparent, indicating that OSS cells have a robust endo-siRNA pathway (Fig. 3B). As is the case in Drosophila tissues and cells, TE-siRNAs were fairly equivalent between sense and antisense orientations, but exhibited a mild antisense bias (Fig. 4A,B). We systematically analyzed RNAs derived from 139 classes of TEs (RepeatMasker, UCSC Genome Browser), and observed that different TEs generated different levels of siRNAs relative to production of piRNAs. For example, mdg1 and idefix produced almost no siRNAs, and roo generated only a minor fraction of siRNAs (Fig. 3C–E). On the other hand, some TEs preferentially generated siRNAs relative to other size classes, with gypsy, gypsy6, FW, Doc, and Rover each generating >50,000 siRNA reads (Fig. 3F,G; Supplemental Fig. S2).
Figure 3.
TE-piRNAs and TE-siRNAs in OSS cells. (A) Reproducibility of overall read distribution in the four OSS sequencing reactions. Analysis of raw reads is shown; the normalized data are completely overlapping (Supplemental Fig. S1). (B) Combined OSS library data. Blue depicts the size distribution across all reads, orange depicts miRNA reads, and red depicts reads that mapped to transposons; the latter clearly segment into a siRNA population (peaking at 21 nt) and a piRNA population (∼24–30 nt). (C–G) Combined OSS read distribution mapped to various families of TEs (as annotated by RepeatMasker). (Red) Antisense reads; (blue) sense reads. Two graphs are shown for each TE family, depicting the size distribution of reads that map uniquely to the genome and the distribution of reads that map to the genome 10 or more times. Various patterns are observed across the TEs with respect to piRNAs vs. siRNAs, or the fraction of unique vs. multiply-matching reads. The complete analysis of all read bins across all TE families is available in Supplemental Figure S2.
Figure 4.
Sequence properties of TE-siRNAs and TE-piRNAs in OSS cells. (A–D) Aggregate nucleotide composition of all TE-derived reads of length 21 nt (siRNA) or 27 nt (as a representative piRNA size); in all graphs, the x-axis represents nucleotide position along the piRNA, while the y-axis represents the percent nucleotide composition and each position. 5′ U bias is observed for bulk AS-TE-siRNAs as well as S- and AS-TE-piRNAs, but is lacking in bulk S-TE-siRNAs. Note also that S-TE-piRNAs lack any bias for adenine at position 10 (above the adenine bias of neighboring nucleotide positions), as would be expected for ping-pong pairs. Analysis of other TE read sizes are available in Supplemental Figure S3. (E–H) Specific analysis of roo reads shows similar trends. Analysis of all other individual TEs are available in Supplemental Figure S4. (I) Overlap analysis of piRNAs with three hypothetical configurations of distance between 5′ ends of a piRNA and its nearest neighbor on the opposite strand; +10 offset is typical of piRNA ping-pong, while −16 offset is consistent with a phased arrangement of ping-pong pairs (gray arrows depict “missing” piRNAs). (J) Analysis of ovary (black) and 0–2-h embryo (blue) piRNAs reveals strong ping-pong (+10 offset pairs) and modest evidence for a phased ping-pong pairs. No ping-pong is observed for OSS piRNAs (red).
TEs could also be classified on the basis of whether they preferentially generated uniquely mapping reads, multiply mapping reads, or both (e.g., Fig. 3C–G). Bulk siRNAs mapped to many (>10) locations, consistent with their putative biogenesis from dispersed copies of actively transcribed TEs. However, specific TE classes such as gypsy and gypsy6 generated a high proportion of uniquely mapped siRNAs (Fig. 3F). The complete analysis of read lengths associated with all 139 TE families is provided in Supplemental Figure S2. Collectively, these observations indicated the distinct entry of different TEs into the piRNA and endo-siRNA pathways.
We observed a mild 5′ U bias in the ∼280,000 antisense reads (∼40%) but no 5′ U bias for the ∼240,000 sense siRNA reads (Fig. 4A,B; Supplemental Fig. S3). This was not an artifact of other TE reads that contaminated the siRNA fraction (e.g., incidental 21-nt degradation fragments of TE-piRNAs), as longer TE reads of both polarities exhibited 5′ U bias (Fig. 4C,D). Analyses of nucleotide composition across individual TE families showed that most exhibited 5′ U bias in antisense 21-nt reads, but that almost none were biased in their sense 21-nt reads (Fig. 4E–H; Supplemental Fig. S4). These data indicate that 5′ U bias represents a genuine, although otherwise subtle, characteristic of antisense TE-siRNAs.
TE-piRNA expression in OSS cells
TEs are also the major source of piRNAs in the Drosophila germline (Aravin et al. 2003; Vagin et al. 2006; Brennecke et al. 2007). The size distribution of reads mapped to TEs provided clear evidence for abundant 24–30-nt TE-piRNAs in OSS cells; indeed, we cloned about twice as many TE-piRNAs as miRNAs (Fig. 3B). Consistent with observations from ovaries, the TE-piRNA content was highly biased for antisense reads and for 5′U (Fig. 4D). TEs with exceptionally high antisense piRNA production (>500,000 reads) included 412, mdg1, GTWIN, and idefix (Fig. 3C,D; Supplemental Fig. S2). A small number of TEs generated dominantly sense piRNAs, including copia, DM-CR1A, and DNAREP1 (also known as DINE-1) (Supplemental Fig. S2). DM-CR1A was notable in that it also exhibited 5′ U bias in sense but not antisense siRNA reads, suggesting a polarity switch for both classes of small RNAs emanating from this TE class.
Even when cloning from PIWI/AGO3/AUB immunoprecipitates, the prepared RNAs must still avoid the 29–30-nt fraction due to the persistence of contaminating 2S rRNA (Brennecke et al. 2007). The efficient depletion of 2S rRNA reads from our 18–34-nt libraries (Fig. 1) allowed us to resolve a sizable population of 29–30-nt piRNAs; thus, the distribution of piRNAs is slightly larger than previously recognized (Fig. 3). We independently verified piRNA expression in OSS cells using Northern blotting and observed hybridization to ∼25–28-nt RNAs derived from roo, mdg1, and 297 (Supplemental Fig. S5). In summary, OSS cells are a homogenous Drosophila cultured cell line that expresses high levels of piRNAs.
Drosophila OSS cells express only PIWI amongst Piwi-class proteins
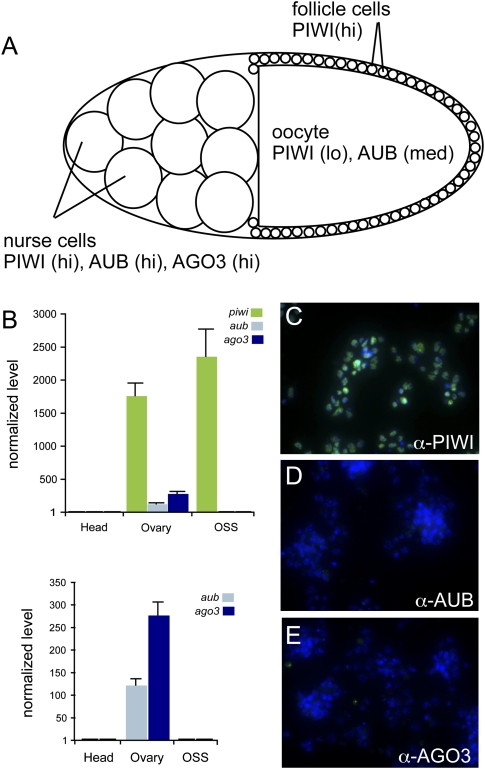
We next asked which Piwi proteins were involved in OSS piRNA expression. All three Drosophila Piwi genes—piwi, aub, and AGO3—are expressed by nurse cells and developing oocytes in the female germline (Williams and Rubin 2002). However, only piwi is additionally expressed in ovarian somatic stem cells and the follicle cells that line the oocyte (Cox et al. 2000; Brennecke et al. 2007; Gunawardane et al. 2007) (Fig. 5A).
Figure 5.
OSS cells are related to ovarian follicle cells. (A) Schematic of an individual mature germarium. The germline consists of 15 nurse cells and the developing oocyte; the latter is ensheathed by follicle cells of somatic origin. The accumulation of the three Piwi-class proteins—PIWI, AUB, and AGO3—in these celltypes is indicated. (B) Quantitative RT–PCR analysis of Piwi-class transcripts in OSS cells; ovaries were used as a positive control and female heads as a negative control. Transcript levels were normalized to RpL32 as a control, and expressed as the fold change above the level in heads. OSS cells express high levels of piwi, but not aub or AGO3. (C–E) Immunostaining of OSS cells with antibodies against the three Piwi-class proteins (green) verifies sole expression of PIWI; DNA was counterstained with DAPI (blue).
We performed real-time reverse-transcription PCR (qRT-PCR) analysis of piwi, aub, and AGO3 in OSS, using adult head RNA as a negative control and female ovary RNA as a positive control. As expected, ovaries contained 200- to 2000-fold the level of all three Piwi family genes relative to heads (Fig. 5B). With these gauges in place, we determined that OSS cells express high levels of piwi, comparable or higher than in ovaries, but they do not express aub or AGO3. This was also true at the protein level, since staining of OSS cells with antibodies against the three Piwi proteins (Brennecke et al. 2007) yielded reactivity only with anti-PIWI (Fig. 5C–E). We confirmed these results using an independent PIWI antisera (Saito et al. 2006) (data not shown). Taken together, these data support a follicular or prefollicular origin of OSS cells, as inferred earlier from their expression of FasIII (Niki et al. 2006).
Differential activity of piRNA master loci in somatic and germline cells
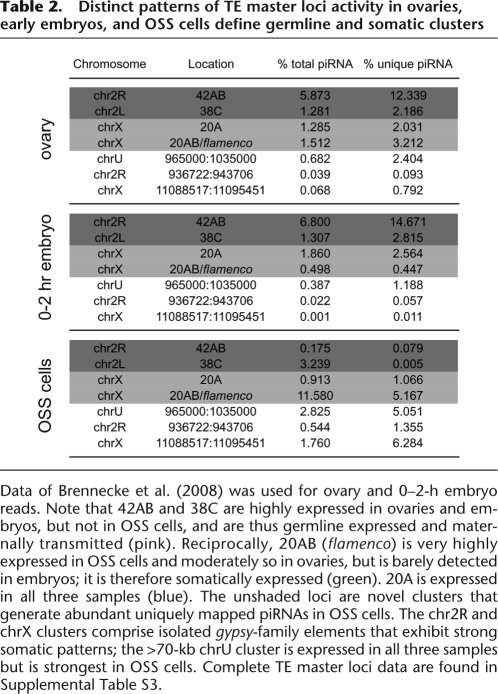
Analysis of ovary-derived piRNAs revealed the existence of master loci, defined as genomic intervals to which substantial numbers of piRNAs could be uniquely mapped (Brennecke et al. 2007). These were inferred to represent piRNAs that derived from the processing of much longer master transcripts, tens to hundreds of kilobases in length. Since this study analyzed whole ovaries, it was unclear whether there was tissue specificity of master transcript expression with respect to germline and soma.
We assessed this by comparing the distribution and frequency of uniquely mapping piRNAs between ovaries, early embryos, and OSS cells (Table 2; Supplemental Table S2; Supplemental Fig. S6). This analysis was aided by recently reported large-scale small RNA data from ovaries and 0–2-h embryos (Brennecke et al. 2008). As germline-expressed piRNAs are maternally deposited into the developing oocyte (Brennecke et al. 2008; Chung et al. 2008), piRNAs from these loci should be shared by ovaries and embryos. Conversely, there might exist piRNA loci solely active in somatic follicle cells; these might be shared only by ovaries and OSS cells. Finally, some master loci might be active in both soma and germline, and are thus present in all three populations.
Table 2.
Distinct patterns of TE master loci activity in ovaries, early embryos, and OSS cells define germline and somatic clusters
Data of Brennecke et al. (2008) was used for ovary and 0–2-h embryo reads. Note that 42AB and 38C are highly expressed in ovaries and embryos, but not in OSS cells, and are thus germline expressed and maternally transmitted (pink). Reciprocally, 20AB (flamenco) is very highly expressed in OSS cells and moderately so in ovaries, but is barely detected in embryos; it is therefore somatically expressed (green). 20A is expressed in all three samples (blue). The unshaded loci are novel clusters that generate abundant uniquely mapped piRNAs in OSS cells. The chr2R and chrX clusters comprise isolated gypsy-family elements that exhibit strong somatic patterns; the >70-kb chrU cluster is expressed in all three samples but is strongest in OSS cells. Complete TE master loci data are found in Supplemental Table S3.
This is indeed what we have found (Table 2; Supplemental Table 2). As expected, the previously described piRNA master loci generated a substantial fraction of uniquely mapping piRNAs. Notably, 42AB generated the highest percentage of unique piRNAs in both ovary and embryo populations (12%–15%). This is consistent with its known maternal transmission and role in hybrid dysgenesis (Brennecke et al. 2008). However, 42AB was barely active in OSS cells, contributing only 0.08% of unique piRNAs. We conclude that 42AB is predominantly active in the germline.
Reciprocally, 20AB/flamenco was the most active known piRNA cluster in OSS cells, generating 5.2% of unique piRNAs. However, it was only moderately abundant in ovaries, and barely detected in embryos. Since follicle cells comprise a minority of total ovary volume, OSS samples are comparatively enriched in follicle-specific transcripts. The only other known piRNA cluster expressed at appreciable levels in OSS cells was 20A. This locus was also well represented in ovaries and embryos, indicating that 20A is active in both soma and germline. Overall though, the other piRNA clusters defined previously (Brennecke et al. 2007) were barely active in OSS cells (Supplemental Table S2). Therefore, most of the previously described piRNA master loci are programmed for maternal transmission of piRNAs, but select master loci have specific functions in somatic cells of the gonad.
We wondered whether our OSS data might reveal novel follicle-cell master loci that were poorly represented in whole ovaries. We surveyed the genome in 5-kb intervals for loci that generated large numbers of TE-piRNAs, especially uniquely mapping piRNAs and collapsed adjacent regions where applicable. This showed the highest expressed piRNA loci in OSS cells to be 20AB/flamenco, and to a lesser extent 20A (Supplemental Table S3). However, we recovered several novel loci, including a cluster at chrU:965000:1035000 and isolated gypsy5- and gypsy-encoding regions of chromosomes X and 2R (Table 2; Supplemental Table S3; Supplemental Fig. S7). These putative soma-specific piRNA clusters share with 20AB/flamenco the properties of being much more highly expressed in OSS than in ovaries and of being less abundant in early embryos (in particular, the gypsy5 and gypsy clusters were nearly absent in embryos) (Table 2). The chrU cluster shared additional features with flamenco, including a long putative precursor (∼70 kb), nearly completely strand-specific piRNA production, a high degree of uniquely mapping piRNAs (∼100,000 reads, equivalent to the number of piRNAs uniquely mapped to flamenco), and a strong bias for antisense-oriented TEs, including multiple gypsy and idefix elements (Supplemental Fig. S7).
Lack of ping-pong signatures indicate that OSS cells express primary piRNAs
The ping-pong amplification model was proposed to explain the selective generation of abundant piRNAs from active transposon transcripts (Brennecke et al. 2007; Gunawardane et al. 2007). Its key signatures are: (1) the existence of complementary piRNA pairs that predominantly exhibit 10-nt offsets between their 5′ ends, and (2) the existence of distinct piRNA populations with either 5′ uridine bias or heterogenous first nucleotide and predominant adenine at the 10th position. Although ping-pong was elucidated by separately sequencing the piRNAs resident in the three Drosophila Piwi proteins, its signatures are evident in libraries constructed from total RNA (Aravin et al. 2007; Houwing et al. 2007; Grimson et al. 2008).
When analyzing piRNAs mapped to individual and consensus TEs, we observed that both antisense piRNAs and sense piRNAs in OSS cells exhibit strong preference for 5′U (Fig. 4C,D). This remained true when analyzing piRNAs mapped to individual TEs. Moreover, no TE class exhibited A10 bias for sense piRNA reads. We sensitized our analysis by searching for A10 bias in sense piRNAs lacking 5′ U, or in cohorts of piRNAs binned by size (Supplemental Figs. S3, S4; data not shown); however, no evidence for ping-pong was obtained. As a final test, we checked for 10-nt offsets to complementary piRNA pairs (Fig. 4I). We identified a strong signal for ping-pong pairs using large-scale TE-piRNA data from ovaries and 0–2-h embryos (Fig. 4J; Supplemental Fig. S8) (Brennecke et al. 2008). These analyses also revealed a minor signal for offset relationships of −16 to −18 nt, which might derive from phased ping-pong activity. In contrast, no particular dominant offset was observed for OSS TE-piRNAs (Fig. 4J; Supplemental Fig. S8).
When considered alongside our empirical observation that OSS cells contain little or no AGO3 or AUB (Fig. 5), we conclude that OSS sense and antisense piRNAs are produced by a primary pathway, as opposed to a ping-pong mechanism with segregated production of sense and antisense piRNAs. Since TE-piRNAs comprise the largest class of short regulatory RNA in OSS cells, collectively much more numerous than miRNAs and siRNAs combined (Fig. 3B), these data support the unexpected conclusion that abundant piRNA production is possible in the absence of ping-pong amplification. It remains formally possible that OSS cells acquired atypical behavior during the course of their derivation, but the close similarities in protein and piRNA expression between OSS cells and follicle cells strongly suggest that an efficient PIWI-associated primary piRNA pathway is a genuine property of somatic ovarian cells.
Novel subnuclear localization of PIWI in Drosophila OSS cells
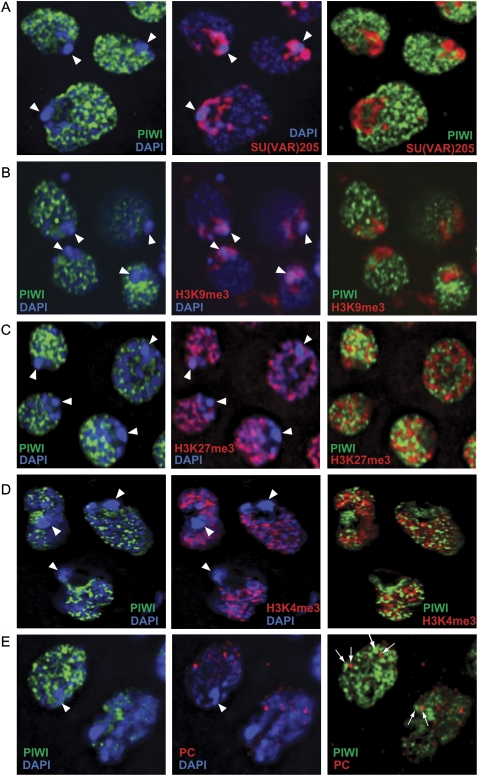
We reasoned that the subcellular localization of PIWI might be informative with respect to piRNA pathway activity in somatic ovarian cells. Similar to its endogenous localization in the germarium and embryo, PIWI was predominantly nuclear in OSS cells, as marked by the overlap with DAPI staining of DNA (Fig. 5C). However, high-magnification deconvolution microscopy revealed that PIWI was largely excluded from the highly condensed chromocenter (Fig. 6). The localization of PIWI in OSS cells contrasted with previous observations that PIWI was enriched at the chromocenter of polytene DNA of larval salivary glands (Brower-Toland et al. 2007). In that setting, PIWI colocalized with Suppressor of variegation 205 [SU(VAR)205, also known as heterochromatin protein 1a (HP1a)] and overlapped substantially with histone H3 trimethylated at lysine 9 (H3K9me3), a marker of transcriptionally repressed chromatin (Brower-Toland et al. 2007). In addition, PIWI was earlier reported to colocalize with Polyhomeotic (PH), a component of the Polycomb repressive complex 1 (PRC1), in nuclei of larval imaginal discs (Grimaud et al. 2006).
Figure 6.
PIWI protein localization in OSS cells appears nucleoplasmic and not specific for any chromatin state. Triple staining of PIWI and DAPI with SU(VAR)205 (also known as HP1a) (A), H3K9me3 (B), H3K27me3 (C), H3K4me3 (D), and Polycomb (PC) (E). Arrowheads mark the location of the chromocenter, a DAPI-dense congregation of heterochromatin. PIWI is specifically absent from the chromocenter and does not overlap appreciably with either markers of silent or active chromatin. Some PIWI foci are near Polycomb foci, but these appear to be incidental and are never overlapping (arrows, E).
Therefore, we examined PIWI with respect to additional chromatin markers. Triple staining of PIWI and DAPI with SU(VAR)205 (i.e., HP1a), H3K9me3, or H3K27me3 indicated that the bulk of PIWI in OSS cells was excluded from both constitutive and facultative heterochromatin (Fig. 6A–C). PIWI also did not overtly colocalize with Polycomb foci in OSS nuclei (Fig. 6E). Curiously, PIWI did not exhibit substantial overlap with H3K4me3 (Fig. 6D), a general marker of transcriptionally active domains. Taken together, these data demonstrate that bulk PIWI proteins in OSS cells are nucleoplasmic, but not fixed to any particular chromatin state.
Conclusions
A cultured cell system for studying primary piRNA production
We showed that OSS cells express the three major classes of small regulatory RNAs described in Drosophila: miRNAs, endo-siRNAs, and piRNAs. Moreover, the fact that OSS cells express only primary piRNAs makes them amenable to dissecting a key aspect of piRNA production that has been challenging to analyze in the intact animal. We found PIWI-associated production of primary piRNAs to be quite efficient and greater than miRNA and endo-siRNA output combined in these cells. Future studies are needed to understand how such massive piRNA generation is achieved. This may yield insight into the production of abundant pachytene piRNAs in mammalian testis. As with primary Drosophila piRNAs, pachytene piRNAs are presumably generated without ping-pong, but the mechanism of their genesis is otherwise mysterious.
Our study of siRNAs and piRNAs from this homogenous cell population provided unique insights into the generation and provenance of TE-derived small RNAs. Previous studies of ovaries were confounded by uncertainty in which cell types expressed various populations of small RNAs. Using OSS cells, we discerned follicular-specific piRNA master clusters, including 20AB/flamenco. We also identified novel somatic clusters, including a 70-kb cluster on chrU that exhibits many of the key features of flamenco, such as a high production of uniquely mapped piRNAs from a single-stranded precursor, and a strong bias for antisense-oriented TEs. The recognition of somatic piRNA clusters allowed us (1) to distinguish populations of maternally transmitted and ovary-restricted piRNAs in the animal, and (2) to demonstrate that somatic TE master loci such as flamenco are processed by a primary piRNA biogenesis mechanism that does not rely upon ping-pong exchange. These findings correlate well with genetic evidence that flamenco restricts gypsy, ZAM, and idefix specifically in somatic follicle cells (Prud'homme et al. 1995; Mevel-Ninio et al. 2007), and that gypsy and ZAM have indeed adopted a specific habitat in follicle cells (Pelisson et al. 1994; Leblanc et al. 2000). Taken together, these observations suggest that the primary piRNA pathway and specialized master clusters evolved in concert with the colonization of a follicular niche by specific TEs.
While this work was under review, Li et al. (2009) and Malone et al. (2009) obtained complementary results regarding the existence of a primary piRNA pathway; they analyzed small RNA libraries from various mutant ovaries and compared maternal transmission of piRNA populations to reach a similar conclusion that a PIWI-mediated primary pathway operates on flamenco for TE defense in somatic ovarian cells. We further note that the production of gypsy-family endo-siRNAs is also especially active in OSS cells (Fig. 3; Supplemental Fig. S2), suggesting that both piRNA- and siRNA-mediated defense are necessary in these specialized somatic gonadal cells.
Our immunohistochemical analysis of PIWI permitted a high-resolution view of the nuclear subcompartment of PIWI localization. In contrast with its reported colocalization with various markers of repressed chromatin in larval salivary glands and imaginal discs (Grimaud et al. 2006; Brower-Toland et al. 2007), PIWI was excluded from the chromocenter and from SU(VAR)205/HP1a- and PRC1-marked domains in OSS cells. Moreover, costaining with a panel of histone methylation marks revealed that PIWI was not preferentially associated with either transcriptionally repressed or active chromatin. Since OSS cells exhibit other molecular and genetic properties of follicle cells, where PIWI plays one of its most prominent roles, OSS cells may represent a relevant system for understanding the unique cell biology of PIWI, the only Drosophila Argonaute protein with predominant nuclear localization.
Methods
OSS cell methods
Culture media for OSS cells was prepared as described (Niki et al. 2006). For immunostaining, cells were cultured on glass chamber slides coated with Concanavalin-A (Sigma) and fixed in 1× PBS with 4% paraformaldehyde. After blocking for 1 h in abs-buffer (5% milk, 5% BSA, 1× PBS), cells were probed overnight with a 1:500–1:1000 dilution of primary antibodies: rabbit anti-PIWI, rabbit anti-AGO3, and rabbit anti-AUB (from Greg Hannon, Cold Spring Harbor Laboratory, NY), mouse anti-PIWI (from H. Siomi and M. Siomi, Keio University School of Medicine, Tokyo, Japan), rabbit anti-H3K9Me3 and rabbit anti-H3K27Me3 (Upstate/Millipore), rabbit anti-H3K4Me3 (Abcam), mouse anti-SU(VAR)205/HP1a (Developmental Studies Hybridoma Bank), or mouse anti-PC antibody (Saurin et al. 2001), followed by Alexa-488 or -Cy3 conjugated secondary antibodies (Molecular Probes).
Small RNA library construction
Two 10-cm dishes of confluent OSS cells were lysed in 1 mL of binding buffer (20 mM HEPES-KOH at pH 7.9, 10% glycerol, 100 mM KOAc, 0.2 mM EDTA, 1.5 mM MgCl2, 0.2 mM PMSF, 1.0 mM DTT, 1× Roche Complete EDTA-free protease inhibitor cocktail) by freeze-thaw fracture, and total cell lysate was clarified by 30-min centrifugation at full speed. A 1-mL HiTrap Q HP column (GE Healthcare) was equilibrated with binding buffer, and OSS whole-cell extract was applied and washed with 1 column volume (CV) of binding buffer to yield flowthrough. Two CV of binding buffer supplemented to 300 mM KOAc was applied to yield mild salt eluate. Flowthrough and eluate fractions were combined, extracted with phenol/chloroform, and RNAs were precipitated with ethanol. Small RNAs were subjected to library construction (NC Lau, T Ohsumi, M Borowsky, VV Kapitonov, RE Kingston, and MD Blower, in prep.) and sequenced using the Illumina GA-II platform.
Analysis of gene expression
We analyzed mRNA levels using quantitative reverse transcription–polymerase chain reaction (qPCR) and SYBR Green (Okamura et al. 2008) and the following primer sets:
piwi-Forward 5′-CTTCCCGAGGTAGTGGTGATGG-3′
piwi-Reverse 5′-GCTCTGTCGGGCCACCTTCACG-3′
aub-Forward 5′-GGAAATTCCGGATGACCGCAAC-3′
aub-Reverse 5′-ACCTGCGATGGCACCGGTCTG-3′
AGO3-Forward 5′-AAGCAGTTTGATCCTTCGCGTC-3′
AGO3-Reverse 5′-AGATCAACCAGCATTTCTAGAAC-3′
RpL32-A 5′-ATCGGTTACGGATCGAACA0-3′
RpL32-B 5′-ACAATCTCCTTGCGCTTCTT-3′
Small RNA Northern blotting was performed as described (Okamura et al. 2007) using LNA probes to monitor mdg1 and 297 (Chung et al. 2008) and DNA probes to monitor roo and miR-2 (Vagin et al. 2006); the miR-2 probe can likely hybridize to the products of several miRNA genes including mir-2a-1, mir-2a-2, mir-2b-1, and mir-2b-2).
Computational analyses of small RNA sequences
Reads were clipped of their 3′ linkers and mapped to the Drosophila Genome Release 5.3. These data were deposited at NCBI Gene Expression Omnibus under series GSE15378, with the four library datasets under GSM385744, GSM385748, GSM385821, and GSM385822. We extracted reads that matched perfectly to the genome using Novocraft (http://www.novocraft.com), with no internal mismatches permitted. Depending on the analysis, we either considered the total number of reads mapped to a given genomic location or considered the normalized read number obtained by dividing by the total number of mapped locations for a given sequence.
To analyze TE-piRNAs and TE-siRNAs, we mapped all reads to RepeatMasker annotations (extracted from http://www.repeatmasker.org/genomes/dm3/dm3.fa.out.gz) and considered as TE-reads the reads included entirely in the RepeatMasker coordinates. For the sequence composition analysis, we recorded the frequencies of each nucleotide for each position across the read (with read counts normalized against genomic locations). For the pairing analysis, as illustrated in Figure 4I, we counted occurrences of reads on one strand separated from reads on the opposite strand by an offset in 5′ nucleotides varying from −20 to +30. The numbers were then normalized (divided by their mean) for each data set.
For the master cluster analysis, we considered 21-nt reads and 24–30-nt reads that mapped to the previously defined master loci (Brennecke et al. 2007). In addition to OSS reads, we also analyzed previously published small RNA datasets of Hannon and colleagues (Brennecke et al. 2007, 2008). To define novel master loci, we analyzed 5-kb windows across the genome for the number of 24–30-nt reads, tabulating both the normalized number of reads and the number of uniquely matching reads. We considered those windows that contained annotated TEs and yielded >5000 uniquely mapped piRNAs, or at least 1% of the most abundant 5-kb window (resident in 20AB/flamenco).
Acknowledgments
We thank Martha Reed and Liz Perkins for help with fly extract; Mark Borowsky for assistance with the Illumina GA-II system; Vladimir Kapitonov for helpful discussions; and Gregory Hannon, Mikiko Siomi, and Haruhiko Siomi for generous gifts of antibodies. We thank Robert Kingston for his support during this project. N.C.L. was funded by the National Institute of Child Health and Human Development (K99-HD057298-01). Work in E.C.L.'s group was supported by the Sidney Kimmel Cancer Foundation, the Alfred Bressler Scholars Fund, and the US National Institutes of Health (R01-GM083300 and U01-HG004261).
Footnotes
[Supplemental material is available online at http://www.genome.org. The small RNA data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE15378.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094896.109.
Note added in proof
While this paper was in review, Kawaoka et al. (2009) described an ovary-derived cell line from Bombyx mori with endogenous expression of piRNAs and Piwi-class proteins.
References
- Aravin A, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: Identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. An endogenous siRNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes & Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;7:1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Lau NC. Analysis of small endogenous RNAs. Curr Protoc Mol Biol. 2008;Chapter 26 doi: 10.1002/0471142727.mb2607s81. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Leblanc P, Desset S, Giorgi F, Taddei AR, Fausto AM, Mazzini M, Dastugue B, Vaury C. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J Virol. 2000;74:10658–10669. doi: 10.1128/jvi.74.22.10658-10669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y, Yamaguchi T, Mahowald AP. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: The nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Cohen SM. Identification of novel Drosophila melanogaster microRNAs. PLoS One. 2007;2:e1265. doi: 10.1271/journal.pone.0001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]