Abstract
Magnetotactic bacteria are ubiquitous microorganisms that synthesize intracellular magnetite particles (magnetosomes) by accumulating Fe ions from aquatic environments. Recent molecular studies, including comprehensive proteomic, transcriptomic, and genomic analyses, have considerably improved our hypotheses of the magnetosome-formation mechanism. However, most of these studies have been conducted using pure-cultured bacterial strains of α-proteobacteria. Here, we report the whole-genome sequence of Desulfovibrio magneticus strain RS-1, the only isolate of magnetotactic microorganisms classified under δ-proteobacteria. Comparative genomics of the RS-1 and four α-proteobacterial strains revealed the presence of three separate gene regions (nuo and mamAB-like gene clusters, and gene region of a cryptic plasmid) conserved in all magnetotactic bacteria. The nuo gene cluster, encoding NADH dehydrogenase (complex I), was also common to the genomes of three iron-reducing bacteria exhibiting uncontrolled extracellular and/or intracellular magnetite synthesis. A cryptic plasmid, pDMC1, encodes three homologous genes that exhibit high similarities with those of other magnetotactic bacterial strains. In addition, the mamAB-like gene cluster, encoding the key components for magnetosome formation such as iron transport and magnetosome alignment, was conserved only in the genomes of magnetotactic bacteria as a similar genomic island-like structure. Our findings suggest the presence of core genetic components for magnetosome biosynthesis; these genes may have been acquired into the magnetotactic bacterial genomes by multiple gene-transfer events during proteobacterial evolution.
Magnetotactic bacteria are phylogenetically diverse microorganisms that produce well-ordered intracellular magnetic particles, also known as magnetosomes (Bazylinski et al. 1994). The various crystal morphologies and compositions observed are species or strain dependent, implying a high degree of biological control (Spring and Schleifer 1995). Recent comprehensive molecular studies, including proteomic (Okamura et al. 2000; Grünberg et al. 2001, 2004; Arakaki et al. 2003; Tanaka et al. 2006), transcriptomic (Schübbe et al. 2006; Suzuki et al. 2006; Würdemann et al. 2006), and whole-genomic analyses (Matsunaga et al. 2005; Schübbe et al. 2009), have revealed that the proteins associated with these particles play key roles in magnetosome biomineralization (Nakamura et al. 1995; Arakaki et al. 2003; Komeili et al. 2006; Scheffel et al. 2006). On the basis of the results of these studies, it has been described that the mechanism underlying magnetosome formation involves multiple processes including vesicle formation and alignment (Komeili et al. 2006; Scheffel et al. 2006), vesicular iron accumulation (Nakamura et al. 1995), and iron-oxide crystallization (Arakaki et al. 2003). Moreover, genome sequence analyses have revealed that the genes encoding magnetosome membrane proteins are conserved in the closely related magnetotactic bacteria in the form of a genomic island, also referred to as magnetosome island (MAI) (Grünberg et al. 2001; Ullrich et al. 2005; Fukuda et al. 2006; Jogler et al. 2009). However, most intricacies involved in the complete processes of magnetosome formation remain unclear. In addition, most of these studies have been conducted using α-proteobacteria.
On the basis of 16S rDNA sequence analyses of environmental samples, magnetotactic bacteria have also been categorized in other groups such as the subphyla δ-proteobacteria (DeLong et al. 1993; Kawaguchi et al. 1995; Simmons et al. 2006) and γ-proteobacteria (Simmons et al. 2004) and the phylum Nitrospirae (Spring et al. 1993). Various unique characteristics of bacterial morphotypes and magnetosome compositions have specifically been identified among the δ-proteobacteria. Large spherical magnetotactic bacteria, also called multicellular magnetotactic prokaryotes, collected from sulfidic environments, produce iron-sulfide magnetosomes; further, these bacteria can biomineralize iron oxide (Bazylinski et al. 1995). Barbell-shaped magnetotactic bacteria forming chains of two to five cocci have recently been reported (Simmons et al. 2006). These organisms are phylogenetically affiliated to the genus Desulforhopalus. Moreover, the δ-proteobacteria include several iron-reducing microorganisms that also produce iron-oxide minerals extracellularly (Lovley et al. 1987; Roden and Lovley 1993). Production of iron oxides is highly dependent on the growth conditions and is considered to be associated with the anaerobic respiration pathway (Fredrickson and Romine 2005). Despite the availability of the genomes of two Geobacter strains (JGI Microbial Genomics) (Methe et al. 2003), no comparative study has been conducted between magnetotactic bacteria and these iron-reducing microorganisms thus far.
Desulfovibrio magneticus strain RS-1 is the only isolate of magnetotactic bacteria classified under the δ-proteobacteria (Sakaguchi et al. 1993). Except for its magnetosome formation ability, strain RS-1 has the typical physiological and biochemical characteristics observed in other members of the Desulfovibrio spp., as it reduces sulfate and/or fumarate. Strain RS-1 therefore represents a bacterium with a novel metabolic feature among the magnetotactic bacteria (Sakaguchi et al. 2002). Moreover, this bacterium synthesizes irregular bullet-shaped magnetite crystals; the crystals are morphologically irregular, but share distinct crystallographic characteristics (Posfai et al. 2006), suggesting the presence of a unique biological regulation system of crystal morphology (Sakaguchi et al. 2002).
In the present study, we examined the whole-genome sequence of D. magneticus strain RS-1 and performed comparative genomic analysis with four magnetotactic bacteria classified under the α-proteobacteria. Further, we compared the genome with those of other Desulfovibrio members and dissimilatory iron-reducing bacteria, which exhibit uncontrolled synthesis of extracellular magnetite. By systematic analyses, we clarified common genome features and genes in the magnetotactic bacterial genomes.
Results and Discussion
General genome features
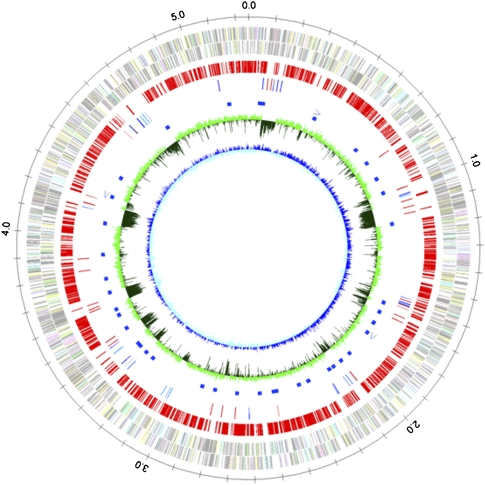
We used the whole-genome shotgun strategy to analyze the complete nucleotide sequence of the RS-1 genome. The genome consists of a circular chromosome of 5,248,049 base pairs (bp) and two circular plasmids, pDMC1 and pDMC2, of 58,704 and 8867 bp, respectively (Supplemental Table S1). The chromosome contains 4629 ORFs, and pDMC1 and pDMC2 contain 65 and 10 ORFs, respectively. Both plasmids are not similar to those identified in the other Desulfovibrio spp. and magnetotactic bacteria or bacterial strains. It is noteworthy that the genome size of strain RS-1 is ∼1.5-Mbp larger than the genome sizes of the other Desulfovibrio strains (Heidelberg et al. 2004), because of which the RS-1 genome contains 854 to 1688 more ORFs than those genomes. Although the average G+C content and ORF length in the RS-1 chromosome are similar to those of the chromosomes of other Desulfovibrio strains (Heidelberg et al. 2004), strain RS-1 is distinguishable by the nonuniform distribution of the G+C content along the chromosome (Fig. 1, eighth circle). The RS-1 chromosome contains numerous long genomic segments, as long as 115 kb, whose local G+C contents are considerably lower than the average content (Fig. 1, represented by dark-green “troughs”). The total length of the regions with the local G+C content below 50% (calculated with a window size of 100 bp) is ∼700 kb, corresponding to 12% of the total genome size. This percentage is significantly higher than that obtained from the genome of D. vulgaris Hildenborough (2%). The chromosomal segments with low G+C content often contain insertion sequence (IS) elements and their remnants. We found 55 copies of the IS elements, of which 18 encode imperfect transposase genes, almost exclusively within the chromosomal segments having low G+C content and in pDMC1. Furthermore, dot-plot comparison of the RS-1 genome revealed no colinearity with the D. vulgaris Hildenborough genome (Supplemental Fig. S1). The significantly large chromosome size, nonuniform distribution of the G+C content, presence of a large number of IS elements and their remnants, and lack of orthology in the chromosomal segments with low G+C content suggest that the RS-1 genome has acquired large stretches of genomic DNA through horizontal DNA transfer from distantly related organisms. The high plasticity of the genome is a unique characteristic of strain RS-1 in the genus Desulfovibrio.
Figure 1.
Representation of the circular chromosome of D. magneticus strain RS-1 in the form of concentric circles. The circles (from the periphery toward the center) indicate the following: first, scale in Mbp; second, ORFs predicted on the plus strand; third, ORFs predicted on the minus strand; fourth, ORFs orthologous to those in D. vulgaris Hildenborough; fifth, putative transposases (blue) and recombinases (red); sixth, rRNA operons; seventh, tRNA genes; eighth, G+C content; and ninth, GC skew.
BLAST search and functional categories
We searched all of the 4704 genes identified in the RS-1 genome by referring to the Universal Protein Resource (UniProt) database. Following a blast local alignment search tool (BLAST) search of the deduced amino acid sequences, 1726, 215, and 200 genes revealed the best BLAST hits with the genes of the members of Desulfovibrio, Syntrophobacter, and Geobacter, respectively (Supplemental Table S2). These genes encode the major metabolic components for energy production, including oxidative phosphorylation, and for the sulfate and fumarate respiration pathways. Among all of the genes of the RS-1 genome, 31, 28, and 15 genes revealed top BLAST hits with the genes from Candidatus Magnetococcus sp. strain MC-1, Magnetospirillum magneticum strain AMB-1, and Magnetospirillum gryphiswaldense strain MSR-1, respectively (Supplemental Table S2). To identify previously reported magnetotactic bacterial core genes (Richter et al. 2007), we also performed analyses of reciprocal best matches between five magnetotactic bacterial genomes (strains RS-1, AMB-1, MSR-1, MS-1, and MC-1). The results revealed that there are 456 core genes that correspond to ∼9.6% of the genetic content of RS-1.
We compared the distribution patterns of the ORFs predicted in the functional categories of the Clusters of Orthologous Group (COG) database between the magnetotactic bacteria and the other Desulfovibrio strains. The number of magnetotactic bacterial genes categorized in “Transcription;” “Replication, recombination, and repair;” “Signal transduction;” “Cell motility;” “Energy production and conversion;” “Inorganic ion transport and metabolism;” “Unknown;” and “General function prediction only” was greater than that of the genes present in the genomes of the other Desulfovibrio strains (Supplemental Table S3). The extra number of ORFs identified in the RS-1 genome was due to the presence of these genes. Although the genes categorized in “Inorganic ion transport and metabolism” were determined in the RS-1 genome, they only included ubiquitous ferrous and ferric ion transporters, also observed in the genomes of the other Desulfovibrio strains (Supplemental Table S4). Interestingly, the numerous genes categorized in “Signal transduction” were commonly observed in the genomes of D. magneticus strain RS-1 (383 genes), Magnetospirillum magneticum strain AMB-1 (308 genes) (Matsunaga et al. 2005), and Candidatus Manetococcus sp. strain MC-1 (265 genes) (Schübbe et al. 2009). The presence of numerous signal transduction components in magnetotactic bacteria suggests that the organisms greatly regulate their cellular function in order to adapt to varying environmental conditions. Therefore, in the case of magnetotactic bacteria, the strict regulation mechanism is considered to enable them to switch between magnet/nonmagnet synthesis and magnetotaxis.
Energy metabolism
Strain RS-1 produces energy by oxidation of substrates, such as lactate, pyruvate, malate, and oxaloacetate, coupled with the reduction of sulfate, thiosulfate, and fumarate (Sakaguchi et al. 2002). The genome sequence revealed the presence of sets of major metabolic components for the sulfate and fumarate respiration, acetyl-coenzyme A (acetyl-CoA), and oxidative phosphorylation pathways as reported in the other Desulfovibrio. We determined the genes encoding enzymes for terminal electron transfer including two sulfate adenylyltransferases (DMR_02810, DMR_39470), a set of adenylylsulfate reductases (DMR_05390, DMR_05400), a set of sulfite reductases (DMR_03600, DMR_03610, DMR_15890), and two sets of fumarate reductases (DMR_34270-90, DMR_05760-80). Pyruvate-flavodoxin oxidoreductase (DMR_20070) and pyruvate-formate lyase (DMR_14270) catalyze the conversion of pyruvate to acetyl-CoA. However, unlike the other Desulfovibrio, we did not identify authentic forms of the NAD- or cytochrome-dependent lactate dehydrogenases in the RS-1 genome. We therefore considered the existence of alternative NAD- or NADP-dependent dehydrogenases substituting for this function.
We also determined the components for hydrogen cycling as reported in the other Desulfovibrio strains (Heidelberg et al. 2004; Matias et al. 2005). A cytoplasmic membrane-bound Ech hydrogenase (DMR_02730-02780) catalyzes hydrogen production from protons and electrons generated in the lactate and pyruvate oxidation. Hydrogen is then reoxidized by periplasmic hydrogenases, such as four Fe-only hydrogenases (DMR_02480, DMR_07830, DMR_12950-60, DMR_43510) and two NiFe hydrogenases (DMR_15600-15610, DMR_02730-02780). A generated proton gradient between cytoplasm and periplasm could be used for ATP synthesis through F1F0ATP synthase (DMR_04760-04830, DMR_42160-42170). Ten c-type cytochromes (DMR_02420, DMR_02560, DMR_12830, DMR_18010, DMR_21540, DMR_29160, DMR_33620, DMR_35740, DMR_35840, DMR_42490) are probably involved in electron delivery through the cytoplasmic membrane via membrane-bound redox complexes for reduction of the terminal electron acceptors (Heidelberg et al. 2004). Interestingly, the genome sequence of strain RS-1 predicted the presence of six transmembrane redox complexes: Dsr (DMR_03600-03630, DMR_15890), Hmc (DMR_12830-12880), TpII-c3 (DMR_42490-42510), Qmo (DMR_05410-05430), and two sets of NADH:quinone oxidoreductase (complex I). Dsr, Hmc, TpII-c3, and Qmo have commonly been observed in the genus Desulfovibrio, but NADH:quinone oxidoreductase (complex I) has not been identified in this group (Matias et al. 2005). We found a gene set encoding NADH:quinone oxidoreductase (complex I) as a single-gene operon, nuoABCDEFGHIJKLMN (DMR_13310-13420), and another set in two separate gene regions (DMR_02470, DMR_27770-27880). The presence of nuo genes in strain RS-1 is therefore a unique characteristic in the genus Desulfovibrio.
Membrane-bound NADH:quinone oxidoreductase (complex I) is a ubiquitous enzyme that catalyzes the electron transfer from NADH to ubiquinone (UQ) in facultative anaerobic and aerobic microorganisms (Schneider et al. 2008). In contrast, anaerobic microorganisms including Desulfovibrio spp. generally utilize menaquinone (MQ) as an electron carrier in the electron transport chain. MK-7(H2) have previously been determined as a major MQ in strain RS-1 (Sakaguchi et al. 2002). UQ and MQ biosynthesis and relative concentration in facultative anaerobic microorganisms is regulated by growth conditions and oxygen supply, with UQ and MQ being the primary quinone under aerobic and anaerobic conditions, respectively. Quinone biosynthesis is therefore considered essential for bacterial survival (Collins 1981). In addition to the nuo genes encoding NADH:quinone oxidoreductase (complex I), the genome sequence in this study revealed the presence of multiple genes that may account for ubiquinone biosynthesis (DMR_04390, DMR_06940, DMR_11640, DMR_11910, DMR_21400, DMR_33520, DMR_33540, DMR_33560, DMR_35770, DMR_39330). Strain RS-1 may use both MQ and UQ to adapt to various environmental conditions, as the organism is known to change to either magnetic or nonmagnetic forms by utilizing electron acceptors during growth (Sakaguchi et al. 2002).
Comparative genomic analysis of magnetosome genes reveals common gene clusters in magnetotactic bacteria
Strain RS-1 exhibits the typical physiological and biochemical properties reported in most of the Desulfovibrio spp., except for the magnetosome-formation ability (Sakaguchi et al. 2002). The large chromosomal size of strain RS-1 among the members of this genus is therefore considered to be due to the presence of additional gene components, including the genes required for magnetosome biomineralization. To extract the genes conserved in all magnetotactic bacteria, we compared the RS-1 genome with the whole genomes of three Desulfovibrio strains and four magnetotactic bacteria classified under α-proteobacteria. First, we excluded 1841 genes exhibiting prominent similarities (E-value < 1e-05) with the three Desulfovibrio strains (highlighted in gray in Supplemental Table S5). These genes cover all of the COG categories and are considered to share the same origin as those of Desulfovibrio spp. We then compared the remaining 2858 genes with the genes of the four magnetotactic bacterial strains, and extracted 314 genes revealing significant similarities (E-value <1e-05) with all of the members (highlighted in red in Supplemental Table S5). The numbers of the extracted genes appearing in the different COG categories were as follows: “Transcription,” 18; “Replication, recombination, and repair,” 23; “Signal transduction,” 87; “Cell motility,” 20; “Energy production and conversion,” 28; “Amino acid transport and metabolism,” 17; “Inorganic ion transport and metabolism,” 27; and “General function prediction only,” 27. The distribution patterns were similar to those described earlier.
Several copies of the same transposase gene sets were identified in the RS-1 chromosome (eight sets) and pDMC1 (three sets). The gene sets consisted of three transposase genes and revealed best BLAST hits with those of a δ-proteobacterium, strain MLMS-1. In most of the cases, two sets were colocated in the genomic region. The transposases may play vital roles in gene acquirement and translocation events occurring in the genome. Moreover, three conspicuous gene regions (the nuo gene cluster, mamAB-like gene cluster, and gene region in pDMC1), wherein the genes common to all magnetotactic bacteria were concentrated, were elucidated by the comparative analysis. These gene regions showed lower G+C contents than those of the flanking regions, suggesting that the genes were acquired from other organisms during evolution. One of the two nuo gene subsets encoding NADH:quinone oxidoreductase (complex I) in strain RS-1 (DMR_13310–420) was identified in all the five magnetotactic bacteria as a conserved single gene cluster in their genomes. The gene cluster was also found in the genomes of dissimilatory iron-reducing bacteria, namely, Geobacter metallireducens strain GS-15 (Lovley et al. 1987), G. sulfurreducens strain PCA (Caccavo et al. 1994), and Shewanella putrefaciens strain CN32 (Glasauer et al. 2002), which exhibit uncontrolled synthesis of extracellular magnetites. These genes seem to be common in magnetite-forming bacteria. Furthermore, we identified orthologous genes encoding putative magnetosome membrane proteins such as MamA, MamB, MamK, MamE, and MamM. The corresponding gene region was called the mamAB gene cluster in the α-proteobacterial group of magnetotactic bacteria (Grünberg et al. 2001). The cryptic plasmid pDMC1 encoding three genes (DMR_p1_00060, DMR_p1_00260, DMR_p1_00380) showed the best BLAST hits with those of strain MC-1, although its role in with magnetosome formation is unknown.
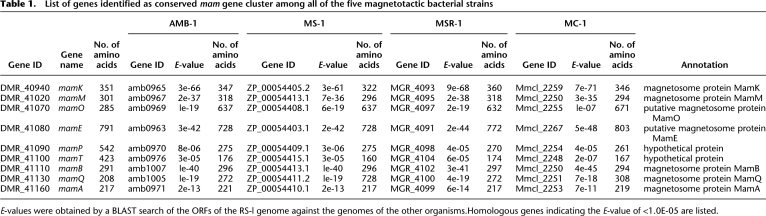
The mamAB-like gene cluster
We also identified a large gene region containing several orthologous gene pairs encoding magnetosome membrane proteins in α-proteobacterial strains in the RS-1 chromosome (Supplemental Table S5). Orthologs of mamA, mamB, mamE, mamK, mamM, mamO, and mamQ were identified as conserved genes in all of the magnetotactic bacteria. Furthermore, we found significantly similar genes for mamP and mamT in the adjacent region (Table 1). These nine genes are most probably candidates of the core genes essential for magnetosome formation. However, we could not clearly identify the genes and gene region in strain RS-1 as in the other four magnetotactic bacteria, because of their relatively low-sequence similarity and complex gene rearrangement (Fig. 2). In addition, no such gene cluster was determined in the other microorganisms, including other Desulfovibrio spp. and Geobacter spp., indicating that the gene region is unique to magnetotactic bacteria. The presence of the conserved gene region in strain RS-1 strongly suggests that the genetic components of the magnetosome formation apparatus have spread not only within the α-group but also to other bacterial classes by lateral gene transfer. We found direct repeats, including transposase genes, at both ends of the conserved gene region in the RS-1 genome, as well as in the genomes of the other magnetotactic bacteria. A gene encoding integrase (xerC), accounting for horizontal gene transfer of the MAIs in Magnetospirillum (Fukuda et al. 2006), was also observed in the gene region (DMR_40740). Therefore, these characteristics strongly suggest that the identified gene region is similar to currently identified MAIs. The MAI gene region in strain RS-1 was considered to be the region containing DMR_40710–DMR_41430, which corresponds to ∼71 kbp (Supplemental Table S5). The size accounts for ∼5% of the size difference between the genome of strain RS-1 and the genomes of other Desulfovibrio strains. The G+C content of the putative MAI of strain RS-1 was 62.1%, slightly lower than the chromosomal average (62.8%). G+C contents of the flanking 50-kbp gene regions at both ends of the MAI were 62.2% and 52.2%. We found unusually high numbers of transposases (18 genes) in the MAI. This gene region is considered to have the same origin as the other magnetotactic bacteria via the same gene transfer mechanism (Ullrich et al. 2005; Fukuda et al. 2006).
Table 1.
List of genes identified as conserved mam gene cluster among all of the five magnetotactic bacterial strains
E-values were obtained by a BLAST search of the ORFs of the RS-l genome against the genomes of the other organisms.Homologous genes indicating the E-value of <1.0E-05 are listed.
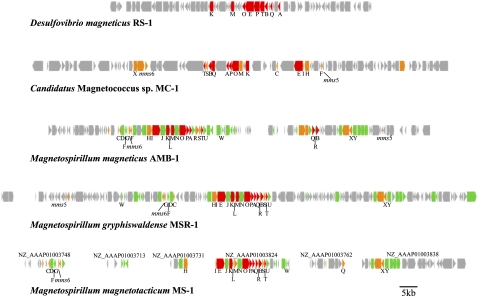
Figure 2.
Comparative analysis of the gene organization in five magnetotactic bacteria: D. magneticus strain RS-1, Magnetospirillum magneticum strain AMB-1, Candidatus Magnetococcus sp. strain MC-1, Magnetospirillum gryphiswaldense strain MSR-1, and Magnetospirillum magnetotacticum strain MS-1. The predicted ORFs are represented by boxes, with arrowheads indicating the direction of transcription. Red arrowheads indicate the ORFs that were identified to be common among all five magnetotactic bacteria: D. magneticus strain RS-1, Magnetospirillum magneticum strain AMB-1, Candidatus Magnetococcus sp. strain MC-1, Magnetospirillum gryphiswaldense strain MSR-1, and Magnetospirillum magnetotacticum strain MS-1. Orange and green arrowheads indicate the ORFs that were identified to be common in the α-proteobacterial magnetotactic bacteria and Magnetospirillum spp., respectively.
The nine conserved genes (mamA, mamB, mamE, mamK, mamQ, mamM, mamO, mamP, and mamT) in the RS-1 genome seem to constitute a single operon, as the reported mamAB operon in the other magnetotactic bacteria (Grünberg et al. 2001; Fukuda et al. 2006; Richter et al. 2007). However, the distinct characteristic of the MAI-like gene region of strain RS-1 within magnetotactic bacterial genomes is the absence of a number of genes including mms6, mms7 (mamD), mms13 (mamC), mamF, mamG, mamJ, mamX, and mamY. These genes were conserved in three Magnetospirillum strains and organized as mms6, mamGDFC, and mamXY operons with other genes. A set of identical gene components with rearrangements have also been identified in the genome of strain MC-1 (Richter et al. 2007). In addition, Richter et al. (2007) systematically extracted genes specific to the genomes of four α-proteobacterial strains by comparative genomic analysis and classified them into three gene groups: “Magnetospirillum-specific genes” (152 genes), “magnetotactic bacteria-specific genes” (11 genes), and “magnetotactic bacteria-related genes” (17 genes). All nine genes identified in the mamAB-like operon of strain RS-1 were categorized into “magnetotactic bacteria-related genes.” On the other hand, we did not find any homologous genes classifiable as “magnetotactic bacteria-specific genes,” and only seven gene homologs of “Magnetospirillum-specific genes” were found. Three of the seven genes showed remote similarities (E-value <1e-06) with function-known proteins, and the other four genes encoded hypothetical proteins. Therefore, the gene groups of “magnetotactic bacteria-related genes” and “Magnetospirillum-specific genes” do not seem to be specific to all of the magnetotactic bacteria. Multiple genes of hemerythrins that are known to bind and transport oxygen and iron have been reported within the MAI of Magnetospirillum spp. (Richter et al. 2007). In contrast, the MAI of strain RS-1 contains only a single copy of hemerythrin. These genes are considered to be acquired during α-proteobacterial evolution or eliminated from the RS-1 genome. Furthermore, the minimal gene set required for magnetosome formation is probably smaller than that proposed in the previous report (Richter et al. 2007).
The presence of common gene sets suggested that magnetosome formation in D. magneticus RS-1 basically involves a bioprocess similar to that described in other magnetotactic bacteria (Bazylinski and Frankel 2004; Matsunaga et al. 2007). MamB and MamM homologs may account for the transport of ferrous iron from the cytoplasm into the magnetosome vesicle (Grünberg et al. 2001, 2004). In addition, a homologous gene of magA (DMR_28020) encoding the proton-driving H+/Fe2+ antiporter protein in M. magneticum strain AMB-1 (Nakamura et al. 1995) was identified. The gene product might also be responsible for the iron transport into the magnetosome vesicles in strain RS-1. A filamentous structure formed by a bacterial actin-like protein, MamK (Komeili et al. 2006), played a role in magnetosome alignment. However, the mamGFDC cluster (Scheffel et al. 2008) and mms6 (Arakaki et al. 2003; Amemiya et al. 2007), playing important roles in controlling magnetosome size and/or morphology, were absent. The formation of irregular, bullet-shaped magnetosomes in strain RS-1 may be due to the deficiency of these genes in its genome. In place of these genes, we identified a number of RS-1-specific genes in the adjacent gene region. Alternatively, these gene products may have functional counterparts, which have only been identified in other magnetotactic bacteria in α-proteobacteria.
Conclusions
The whole-genome sequence of Desulfovibrio magneticus strain RS-1 revealed the unique characteristics of magnetotactic bacteria in their genomes. The genomes contain numbers of IS elements (55 genes) and integrases (28 genes), resulting in the acquirements of foreign genes and recombination events in the genome. The presence of numerous regulatory and signaling genes (383 genes) was also a common feature in the magnetotactic bacterial genomes. We identified the mamAB orthologs as a magnetotactic bacteria-specific gene cluster with the absence of other magnetosome formation-associated genes identified in the other magnetotactic bacteria. The gene region was located within MAI as reported for other magnetotactic bacteria. The nuo gene cluster was identified as a common gene subset among magnetite-forming microorganisms. Three genes revealing best BLAST hits with those of strain MC-1 were identified in a cryptic plasmid, pDMC1. The presence of the gene regions suggests that the genes were acquired by multiple gene-transfer events during the evolution of individual magnetotactic bacteria. Further, we found a number of novel genes, which may play specific roles in strain RS-1. Proteome analysis of pure-cultured magnetotactic bacteria and the comparison of genome sequences among various uncultured bacteria in environmental samples will clarify the complex biomineralization mechanism, origin, and transition pathway of magnetosome synthesis in magnetotactic bacteria.
Methods
Preparation of genomic DNA
Desulfovibrio magneticus strain RS-1 (NBRC 104933T = ATCC700980T = DSM 13731T) was grown anaerobically at 25°C for 7 d in the medium described by Sakaguchi et al. (2002). We extracted total DNA from the cells using the standard protocol.
Construction and sequencing of shotgun libraries
For constructing a plasmid library, we fractured total DNA by hydrodynamic shearing using Hydroshear (Genomic Solutions) to sizes between 1 and 3 kb, and then treated with BAL31 exonuclease. The DNA was further treated using Blunting High (Nippon Gene), according to the supplier's instructions, to clone it into the BAP-treated pUC118-HincII (Takara Bio). We subjected the DNA to electroporation into Escherichia coli DH10B-electrocompetent cells (Invitrogen) to create a shotgun library, and extracted plasmid DNA from the E. coli transformants by using the alkaline method. A cosmid library was also constructed by cloning the Sau3AI-digested DNA into the SuperCos-1 cosmid vector (Stratagene) cleaved by XhoI and BamHI. Approximately 120,000 plasmid clones and 5000 cosmid clones were sequenced from both ends of the inserts with dye-terminator chemistry by using an ABI 3700 DNA Analyzer (Applied Biosystems) or a BST-0100 DNA Fragment Analyzer (MJ Research). Raw sequence data corresponding to ∼12× coverage were assembled by using phred/phrap/consed software (http://www.phrap.org).
Gap closure
To close gaps, we selected cosmid clones bridging two neighboring contigs and determined their internal sequences by the primer-walking method using custom oligonucleotide primers or the transposon-mediated random insertion method with a TGS F-700 Template Generation System (Finnzyme). Alternatively, the gapped regions were amplified by PCR using an LA PCR Kit Ver. 2.1 (Takara Bio) and custom-made oligonucleotides, and the amplified DNA was sequenced. When high-quality sequence data could not be obtained under the standard reaction conditions, we adopted the following methods: (1) dye-terminator sequencing with dGTP as a substrate in place of dITP; (2) dye-terminator sequencing on the template PCR-amplified in the presence of 7-deaza-dGTP (Jung et al. 2002); or (3) transcriptional sequencing using the CUGA sequencing system (Nippon Genetech) on a MegaBace 1000 DNA Analysis System (GE Healthcare).
Gene identification and annotation
The Glimmer and GeneHacker programs were used to predict the positions of the ORFs. We further evaluated these predicted ORFs by using the Frameplot program. For predicting ORFs in the genomic regions where G+C contents were considerably lower or higher than the average values, followed by manual selection, we translated the genome sequence in six frames to obtain potential ORFs longer than 90 bp, considering ATG, GTG, and TTG as the potential initiation codons. Protein-function prediction was based on the following searches: (1) homology searches in the UniProt protein database; (2) profile searches in the HAMAP protein family database; and (3) domain or motif searches in the TIGRFAMs, Pfam, and PROSITE databases by using the InterPro program. We used the KEGG database for the reconstruction of metabolic pathways, and predicted signal peptides in proteins by using the SignalP program and transmembrane helices by using the TMHMM program. All of the information was integrated into an in-house database (OCSS; T Sekigawa and M Yamamoto, unpubl.) for semiautomatic assignments of protein functions and comprehensive manual curation. The genome sequence of strain RS-1 was deposited in the DDBJ database with the following accession numbers: AP010904 (chromosome), AP010905 (pDMC1), and AP010906 (pDMC2).
Comparative genomics
To compare the genome data sets, we conducted a BLAST search between all predicted ORFs in Magnetospirillum magneticum strain AMB-1 (Matsunaga et al. 2005), Magnetospirillum gryphiswaldense strain MSR-1 (Richter et al. 2007), Magnetospirillum magnetotacticum strain MS-1 (DDBJ/EMBL/GenBank), Candidatus Magnetococcus sp. strain MC-1 (JGI Microbial Genomics), Desulfovibrio vulgaris Hildenborough (Heidelberg et al. 2004), Desulfovibrio vulgaris strain DP4 (JGI Microbial Genomics), Desulfovibrio desulfuricans strain G20 (JGI Microbial Genomics), Geobacter metallireducens strain GS-15 (NCBI), Geobacter sulfurreducens strain PCA (Methe et al. 2003), and Shewanella putrefaciens strain CN-32 (JGI Microbial Genomics). In addition, the gene pairs for the nuo and mam gene clusters were manually inspected. A predicted probability score of <1e-05 was used as the standard cutoff to define a likely match. Reciprocal best matches were counted by a BLAST result with an E-value of <1e-05 and subject coverage of over 65%.
Acknowledgments
This work was partially funded by Grant-in-Aid for Scientific Research (A) No. 18206084 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence for Desulfovibrio magneticus strain RS-1 has been submitted to the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp) under accession nos. AP010904–AP010906.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.088906.108.
References
- Amemiya Y, Arakaki A, Staniland SS, Tanaka T, Matsunaga T. Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials. 2007;28:5381–5389. doi: 10.1016/j.biomaterials.2007.07.051. [DOI] [PubMed] [Google Scholar]
- Arakaki A, Webb J, Matsunaga T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J Biol Chem. 2003;278:8745–8750. doi: 10.1074/jbc.M211729200. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Garratt-Reed AJ, Frankel RB. Electron microscopic studies of magnetosomes in magnetotactic bacteria. Microsc Res Tech. 1994;27:389–401. doi: 10.1002/jemt.1070270505. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Frankel RB, Heywood BR, Mann S, King JW, Donaghay PL, Hanson AK. Controlled biomineralization of magnetite (Fe3O4) and greigite (Fe3S4) in a magnetotactic bacterium. Appl Environ Microbiol. 1995;61:3232–3239. doi: 10.1128/aem.61.9.3232-3239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccavo F, Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol. 1994;60:3752–3759. doi: 10.1128/aem.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MDJ. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Frankel RB, Bazylinski DA. Multiple evolutionary origins of magnetotaxis in bacteria. Science. 1993;259:803–806. doi: 10.1126/science.259.5096.803. [DOI] [PubMed] [Google Scholar]
- Fredrickson JK, Romine MF. Genome-assisted analysis of dissimilatory metal-reducing bacteria. Curr Opin Biotechnol. 2005;16:269–274. doi: 10.1016/j.copbio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Okamura Y, Takeyama H, Matsunaga T. Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB-1 reveals how magnetosome synthesis developed. FEBS Lett. 2006;580:801–812. doi: 10.1016/j.febslet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Glasauer S, Langley S, Beveridge TJ. Intracellular iron minerals in a dissimilatory iron-reducing bacterium. Science. 2002;295:117–119. doi: 10.1126/science.1066577. [DOI] [PubMed] [Google Scholar]
- Grünberg K, Wawer C, Tebo BM, Schüler D. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl Environ Microbiol. 2001;67:4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K, Muller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2004;70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM, et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. [DOI] [PubMed] [Google Scholar]
- Jogler C, Kube M, Schübbe S, Ullrich S, Teeling H, Bazylinski DA, Reinhardt R, Schüler D. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ Microbiol. 2009;11:1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- Jung A, Ruckert S, Frank P, Brabletz T, Kirchner T. 7-Deaza-2′-deoxyguanosine allows PCR and sequencing reactions from CpG islands. Mol Pathol. 2002;55:55–57. doi: 10.1136/mp.55.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Burgess JG, Sakaguchi T, Takeyama H, Thornhill RH, Matsunaga T. Phylogenetic analysis of a novel sulfate-reducing magnetic bacterium, RS-1, demonstrates its membership of the delta-Proteobacteria. FEMS Microbiol Lett. 1995;126:277–282. doi: 10.1111/j.1574-6968.1995.tb07430.x. [DOI] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Stolz JF, Nord GL, Phillips EJP. Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature. 1987;330:252–254. [Google Scholar]
- Matias PM, Pereira IAC, Soares CM, Carrondo MA. Sulphate respiration from hydrogen in Desulfovibrio bacteria: A structural biology overview. Prog Biophys Mol Biol. 2005;89:292–329. doi: 10.1016/j.pbiomolbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, Takeyama H. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 2005;12:157–166. doi: 10.1093/dnares/dsi002. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Suzuki T, Tanaka M, Arakaki A. Molecular analysis of magnetotactic bacteria and development of functional bacterial magnetic particles for nano-biotechnology. Trends Biotechnol. 2007;25:182–188. doi: 10.1016/j.tibtech.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, et al. Genome of Geobacter sulfurreducens: Metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- Nakamura C, Burgess JG, Sode K, Matsunaga T. An iron-regulated gene, magA, encoding an iron transport protein of Magnetospirillum sp. strain AMB-1. J Biol Chem. 1995;270:28392–28396. doi: 10.1074/jbc.270.47.28392. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Takeyama H, Matsunaga T. Two-dimensional analysis of proteins specific to the bacterial magnetic particle membrane from Magnetospirillum sp. AMB-1. Appl Biochem Biotechnol. 2000;84–86:441–446. doi: 10.1385/abab:84-86:1-9:441. [DOI] [PubMed] [Google Scholar]
- Posfai M, Moskowitz BM, Arato B, Schüler D, Flies C, Bazylinski DA, Frankel RB. Properties of intracellular magnetite crystals produced by Desulfovibrio magneticus strain RS-1. Earth Planet Sci Lett. 2006;249:444–455. [Google Scholar]
- Richter M, Kube M, Bazylinski DA, Lombardot T, Glockner FO, Reinhardt R, Schüler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J Bacteriol. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden EE, Lovley DR. Dissimilatory Fe(III) reduction by the marine microorganism Desulfuromonas acetoxidans. Appl Environ Microbiol. 1993;59:734–742. doi: 10.1128/aem.59.3.734-742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T, Burgess JG, Matsunaga T. Magnetite formation by a sulphate-reducing bacterium. Nature. 1993;365:47–49. [Google Scholar]
- Sakaguchi T, Arakaki A, Matsunaga T. Desulfovibrio magneticus sp. nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles. Int J Syst Evol Microbiol. 2002;52:215–221. doi: 10.1099/00207713-52-1-215. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gardes A, Grünberg K, Wanner G, Schüler D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol. 2008;190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Pohl T, Walter J, Dorner K, Kohlstadt M, Berger A, Spehr V, Friedrich T. Assembly of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochim Biophys Acta. 2008;1777:735–739. doi: 10.1016/j.bbabio.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Schübbe S, Würdemann C, Peplies J, Heyen U, Wawer C, Glockner FO, Schüler D. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2006;72:5757–5765. doi: 10.1128/AEM.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübbe S, Williams TJ, Xie G, Kiss HE, Brettin TS, Martinez D, Ross CA, Schüler D, Cox BL, Nealson KH, et al. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl Environ Microbiol. 2009 doi: 10.1128/AEM.02874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SL, Sievert SM, Frankel RB, Bazylinski DA, Edwards KJ. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl Environ Microbiol. 2004;70:6230–6239. doi: 10.1128/AEM.70.10.6230-6239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SL, Bazylinski DA, Edwards KJ. South-seeking magnetotactic bacteria in the Northern Hemisphere. Science. 2006;311:371–374. doi: 10.1126/science.1122843. [DOI] [PubMed] [Google Scholar]
- Spring S, Schleifer KH. Diversity of magnetotactic bacteria. Syst Appl Microbiol. 1995;18:147–153. [Google Scholar]
- Spring S, Amann R, Ludwig W, Schleifer KH, Vangemerden H, Petersen N. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a fresh-water sediment. Appl Environ Microbiol. 1993;59:2397–2403. doi: 10.1128/aem.59.8.2397-2403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Okamura Y, Calugay RJ, Takeyama H, Matsunaga T. Global gene expression analysis of iron-inducible genes in Magnetospirillum magneticum AMB-1. J Bacteriol. 2006;188:2275–2279. doi: 10.1128/JB.188.6.2275-2279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Okamura Y, Arakaki A, Tanaka T, Takeyama H, Matsunaga T. Origin of magnetosome membrane: Proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics. 2006;6:5234–5247. doi: 10.1002/pmic.200500887. [DOI] [PubMed] [Google Scholar]
- Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würdemann C, Peplies J, Schübbe S, Ellrott A, Schüler D, Glockner FO. Evaluation of gene expression analysis using RNA-targeted partial genome arrays. Syst Appl Microbiol. 2006;29:349–357. doi: 10.1016/j.syapm.2006.03.005. [DOI] [PubMed] [Google Scholar]