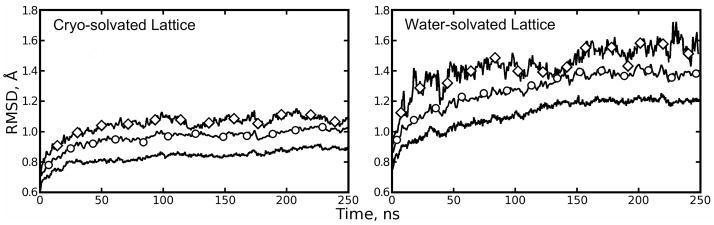
Figure 3.
Backbone root mean squared deviations (bkRMSDs) of lattices and lattice components relative to the 1MK5 structure. Plain line: average bkRMSD for each lattice’s eight asymmetric units (ASUs). Line with circles: average bkRMSD for four streptavidin tetramers (2 ASUs per tetramer). Line with diamonds: bkRMSD for the lattice as a whole. Quaternion alignment was used to determine RMSDs for ASUs and tetramers. A more restricted alignment procedure was used to compute the RMSD of each lattice as a whole (see Methods: Computation of atomic fluctuations and lattice disorder). The cryo-solvated lattice shows much lower overall RMSD than the water-solvated lattice; the differences may increase with further simulation.