Abstract
Eotaxin is an eosinophil-selective chemokine that is constitutively expressed in a variety of organs such as the intestine. Previous studies have demonstrated that the recruitment of eosinophils during inflammation is partially dependent on eotaxin, but the function of constitutive eotaxin during homeostasis has not been examined. To elucidate the biological role of this molecule, we now examine tissue levels of eosinophils in healthy states in wild-type and eotaxin-deficient mice. The lamina propria of the jejunum of wild-type mice is demonstrated to express eotaxin mRNA, but not mRNA for the related monocyte chemoattractant proteins. Wild-type mice contained readily detectable eosinophils in the lamina propria of the jejunum. In contrast, mice genetically deficient in eotaxin had a large selective reduction in the number of eosinophils residing in the jejunum. The reduction of tissue eosinophils was not limited to the jejunum, because a loss of thymic eosinophils was also observed in eotaxin-deficient mice. These studies demonstrate that eotaxin is a fundamental regulator of the physiological trafficking of eosinophils during healthy states. Because a variety of chemokines are constitutively expressed, their involvement in the baseline trafficking of leukocytes into nonhematopoietic tissue should now be considered.
In healthy subjects eosinophils account for only 1–3% of peripheral blood leukocytes, but in patients with allergic diseases, parasitic infections, and cancer, eosinophils can selectively accumulate in the circulation and a variety of tissues. Tissue accumulation of eosinophils can be detrimental because eosinophils are potent pro-inflammatory effector cells capable of causing severe host tissue damage (1, 2). In other instances, eosinophil accumulation may be beneficial because these cells can participate in host defense against parasites (3). Therefore, elucidating the molecular processes involved in eosinophil trafficking is of fundamental importance in understanding the role of this cell in immune surveillance and the pathogenesis of a variety of diseases. Numerous investigations have focused on understanding the processes involved in regulating the pathological accumulation of eosinophils (4, 5). Fewer studies have characterized the molecular signals controlling eosinophil tissue levels during steady-state healthy processes.
Chemokines are a large family of chemotactic cytokines that regulate diverse properties of leukocytes, including hematopoiesis at baseline and cell trafficking and activation during inflammation (6). Chemokine receptors, G-protein-linked seven-transmembrane molecules, not only mediate chemokine signaling but also are HIV coreceptors (7). Chemokines have been divided into four groups, designated CXC, CC, C, and CX3C, depending upon the spacing of conserved cysteines. The CXC and CC group, in contrast to the C and CX3C chemokines, contain many members. Of all the groups, the CC group is most active on eosinophils. Multiple CC chemokines target eosinophils, including monocyte chemoattractant protein (MCP)-2, MCP-3, MCP-4, RANTES, macrophage inflammatory protein (MIP)-1α, eotaxin, and the recently identified myeloid progenitor inhibitory factor 2 (eotaxin-2) (8). These have a conserved intron–exon structure and are located at a common chromosome locus.
Eotaxin is unique among the CC chemokines because in vivo protein administration studies have demonstrated that this molecule is chemoattractive primarily for eosinophils (9–11). Additionally, the eotaxin receptor is predominantly expressed by the hematopoietic cells involved in allergic responses: eosinophils, basophils, and T helper type 2 cells. Antigen challenge in rodents and humans markedly induces eotaxin. However, a combination of approaches, including eotaxin gene targeting, neutralization by antibodies, and receptor antagonism, have uniformly shown only a 2- to 3-fold reduction in eosinophil recruitment after antigen challenge (12–16). Eotaxin may therefore have other unappreciated functions in vivo. Eotaxin is constitutively produced by a variety of tissues such as the intestine in all species examined (10, 17–19), but the function of this constitutive protein has not been examined because the tissues previously studied (lung and skin) were devoid of detectable eosinophils at baseline. We now report that eosinophils normally reside in the jejunum and thymus of mice and that eotaxin has a critical role in regulating the level of tissue-dwelling eosinophils at baseline.
MATERIALS AND METHODS
Generation of Eotaxin-Deficient Mice.
Mice that were deficient in eotaxin were maintained inbred in a 129/SvEv background (13). Control 129/SvEv wild-type mice (Taconic Farms) were age- and sex-matched, and mated and maintained under identical pathogen-free conditions.
Histological Analysis of Tissue Eosinophils.
Serial 1.5-μm-thick glycomethacrylate-embedded sections of jejunum or thymus were stained with double-strength hematoxylin/azure II/aqueous eosin Y (HAE) and ethylene glycol monomethyl ether (20), or with Wright’s–Giemsa stain (21), or for cyanide-resistant endogenous peroxidase activity (20). Sections were examined for the number of eosinophils in a rectangular field, 135 μm high × 213 μm wide, with a 50× objective or 270 μm × 416 μm with a 25× objective, in the jejunum and thymus, respectively.
For immunohistochemical analysis, the jejunum (at 7 cm proximal to the cecum) or thymus was fixed with formalin and embedded in paraffin, and 5-μm sections were stained with an antiserum against eosinophil major basic protein (MBP) by using a modification of previous procedures (13, 22). Endogenous peroxidase was quenched with H2O2, and slides were blocked with normal goat serum, incubated with rabbit anti-murine MBP serum (gift of G. Gleich and J. Lee, Mayo Clinic, Scottsdale, AZ), followed by biotinylated goat anti-rabbit antibody and avidin-peroxidase complex (Vectastain ABC Peroxidase Elite kit; Vector Laboratories). They were then developed by nickel diaminobenzidine, enhanced by cobalt chloride to form a black precipitate, and counterstained with nuclear fast red. Controls included omission of the primary antibody to check for endogenous biotin and peroxidase activity as well as nonspecific binding of the secondary antibody. In the jejunum, the number of eosinophils associated with >15 randomly selected villi were counted for each mouse. Villi were selected from sections that demonstrated the longitudinal view of a villus from base to tip. The boundaries of the villus were the width of the villus and the length from the base of the submucosa to the tip of the villus. In the thymus, the number of eosinophils was enumerated in a square field, 120 μm × 120 μm, with a 40× objective. Statistical analysis was performed by Student’s t test.
In Situ Hybridization Analysis of Eotaxin.
In situ hybridization was performed as described (23). In brief, the full-length murine eotaxin cDNA in plasmid BlueScript (18) was linearized by BamHI or ApaI digestion, and antisense and sense RNA probes, respectively, were generated by T7 and T3 RNA polymerase (Riboprobe Gemini Core System II transcription kit; Promega). The radiolabeled ([α-[35S]thio]UTP) probes were reduced to an average length of 200 bases by controlled alkaline hydrolysis. The hybridized slides were washed under either high- or low-stringency conditions. For low stringency, the slides were washed for 60 min at 50°C in 50% formamide/5× SSC/20 mM DTT. This was followed by a 3-min RNase A digestion (20 μg/ml) at 37°C and 15-min washes in 2× SSC and 0.1× SSC (1× SSC = 0.15 M NaCl/0.015 M sodium citrate, pH 7.0). For high stringency, the slides were washed at 65°C for 30 min in 50% formamide/2× SSC/10 mM DTT; rinsed three times in 500 mM NaCl/10 mM Tris⋅HCl, pH 7.5/5 mM EDTA; digested with RNase A for 30 min at 37°C; and rinsed in fresh buffer. The high-stringency wash was repeated and then followed by two 15-min washes at room temperature, one in 2× SSC and one in 0.1× SSC/1 mM DTT. Autoradiography was performed for 7–40 days at 4°C. The specificity of the hybridization was established by using a sense probe, which did not hybridize above background levels observed with the antisense probe. Sections from both wild-type (n = 3) and eotaxin-deficient mice (n = 3) were hybridized and autoradiographed in triplicate under identical conditions.
RESULTS
Eotaxin mRNA Expression Pattern in the Jejunum.
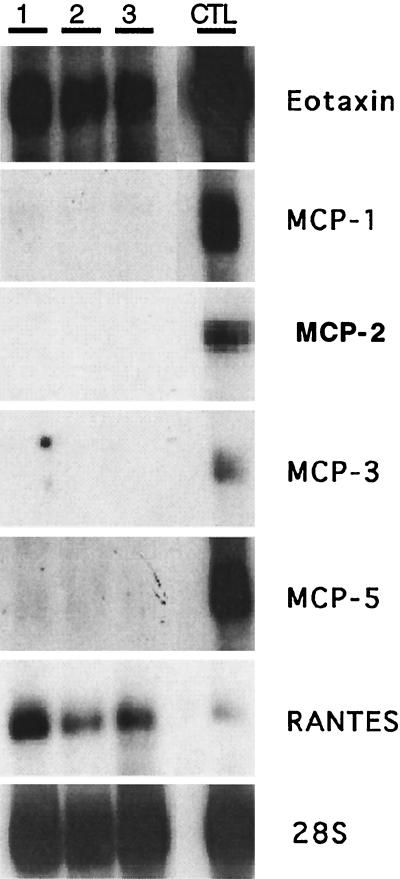
Previous studies have demonstrated that eotaxin is an inducible cytokine involved in the selective accumulation of pulmonary eosinophils (9, 17, 24). Few studies have directly addressed the role of eotaxin in other tissues or examined the physiological role of constitutive eotaxin. To begin to examine these issues, the constitutive expression pattern of several chemokines, including eotaxin, was examined in the jejunum, the longest gastrointestinal segment in mice. Only eotaxin and RANTES mRNA, and not that for MCP-1, MCP-2, MCP-3, or MCP-5 was uniformly detected in a Northern analysis of jejunum RNA (Fig. 1). A positive control for CC chemokine mRNA expression was lung RNA isolated from interleukin-4-overexpressing clara cell lung transgenic mice (25), because these mice have a widespread increase in chemokine expression (M.E.R. and F. Finkelman, unpublished results).
Figure 1.
Northern analysis of chemokine expression in the jejunum. Total RNA (20 μg) from the jejunum of wild-type mice was electrophoresed and transferred to a membrane that was hybridized under conditions of high stringency with cDNA probes for murine eotaxin, MCP-1, MCP-2, MCP-3, MCP-5, RANTES, and 28S ribosomal protein (37) by using methods previously described (17). Control RNA is derived from the lung of interleukin-4-overexpressing clara cell lung transgenic mice. Each numbered lane represents a different animal. Autoradiographs were exposed for 1–4 days. The chemokine cDNA probes were previously described (8), except for MCP-2, which has recently been cloned (M.N.S. and A.D.L., unpublished data).
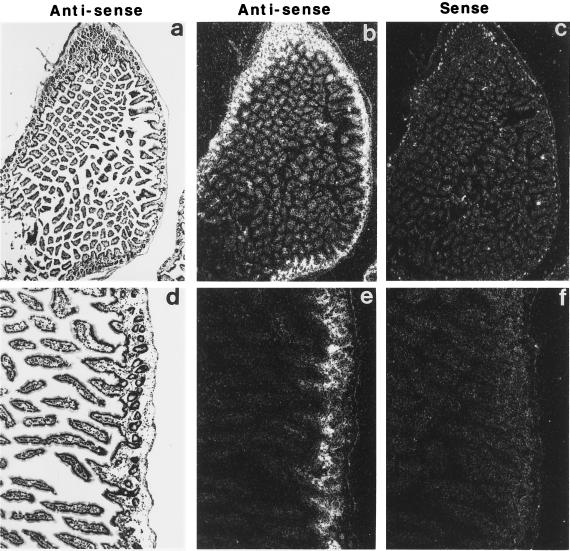
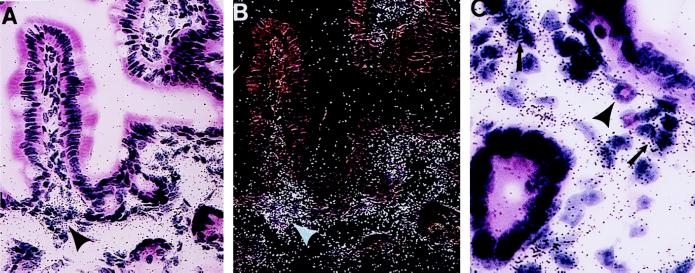
To further examine the expression of eotaxin in the jejunum, in situ hybridization analysis was performed. When an antisense riboprobe (Fig. 2 a, b, d, and e) derived from the murine eotaxin cDNA was used, dark-field microscopy (Fig. 2 b and e) showed eotaxin mRNA to be associated with a specific pattern that appeared to outline the interstitial tissue. Bright-field microscopy (Fig. 2 a and d) revealed that the staining was localized to the lamina propria of the mucosa layer and the submucosa. In contrast, no specific staining in any location was seen with the control sense probe (Fig. 2 c and f). The specific staining pattern with the antisense probe was eliminated in mice deficient in the eotaxin gene (data not shown). Likewise, the sense probe revealed no specific staining in the eotaxin-deficient mice (data not shown), indicating that the in situ hybridization conditions were specific for eotaxin. On higher power magnification (Fig. 3 A and B), it was apparent that eotaxin staining was primarily in the lamina propria and was of higher intensity at the necks of the intestinal crypts (arrowheads). No appreciable staining was seen in the epithelium or the lamina propria of the upper villi (Fig. 3 A and B). The enriched staining at the base of the villi was associated with aggregations of mononuclear cells (Fig. 3C). The frozen sections did not permit further identification of the mononuclear cells. Resident cells in the lamina propria have not been distinguished on the basis of location, but mononuclear cells known to exist in the lamina propria include macrophages, lymphocytes, plasma cells, dendritic cells, and fibroblasts. These results indicate that, in contrast to the respiratory tract (10, 14, 16, 24, 26), in the intestine eotaxin is not produced constitutively by epithelial cells but is produced by mononuclear cells in the lamina propria.
Figure 2.
In situ hybridization of eotaxin mRNA localization in the jejunum. Microscopy of frozen tissue sections hybridized with radiolabeled antisense (a, b, d, and e) or sense (c and f) eotaxin cRNA probes. The sections were washed under high-stringency conditions and autoradiographed for 40 days. Bright-field photomicrographs (a and d) of the same dark-field sections (b, c, e, and f) are shown. (Original magnification is ×36 in a–c and ×90 in d–f.)
Figure 3.
High magnification of eotaxin mRNA localization in the lamina propria of the jejunum. Bright-field (A and C) and dark-field (B) microscopy of jejunum hybridized with a radiolabeled eotaxin antisense cRNA probe. In A and B, taken in the same field, hybridization signals are located predominantly at the neck of the villus (arrowheads). In a higher-power magnification (C), hybridization signals are predominantly localized to an aggregation of mononuclear cells (arrows). An eosinophil is indicated (arrowhead) adjacent to the mononuclear cell aggregation. (Original magnification is ×460 in A and B and ×1360 in C.)
Detection of Tissue Eosinophils.
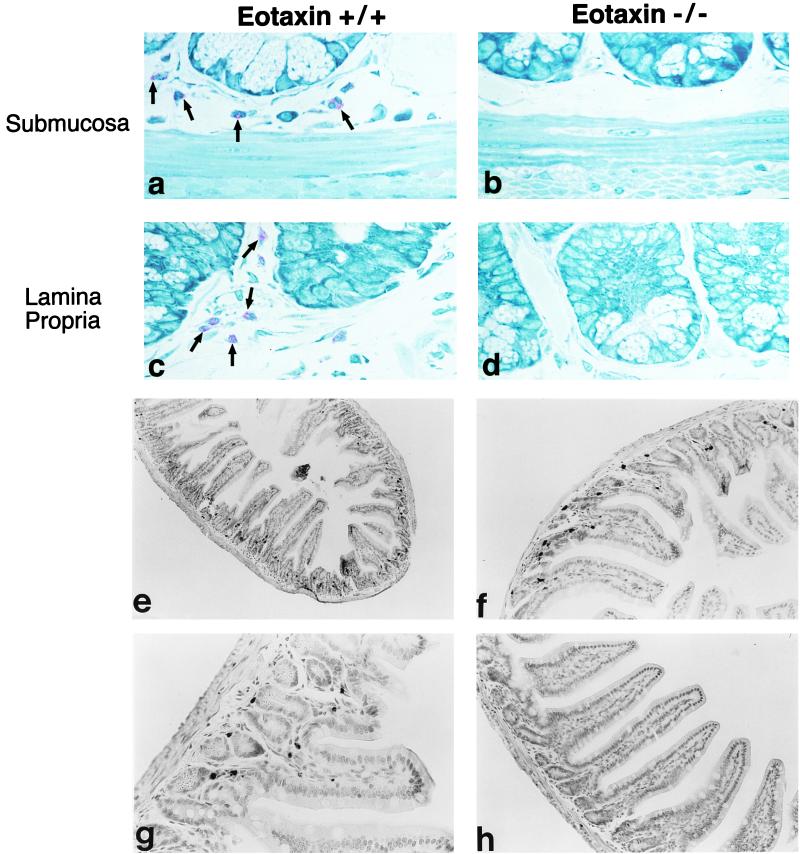
Constitutive eotaxin may be involved in the regulation of baseline numbers of eosinophils in various tissues (14, 17, 19, 27). We were previously unable to test this hypothesis, because we were focusing on tissues that contained no detectable levels of tissue eosinophils at baseline. Therefore, the identification of eosinophils normally residing in other tissues was undertaken. We continued to examine the jejunum because this tissue contained eotaxin mRNA and was known to contain diverse leukocyte populations. Serial 1.5-μm-thick glycomethacrylate-embedded sections were stained with Wright’s–Giemsa stain or HAE, and eosinophils were demonstrated to be prominent throughout the submucosa and the lamina propria of the mucosa. Photomicrographs of Wright–Giemsa-stained jejunum from wild-type mice are shown in Fig. 4 a and c, demonstrating the presence of easily detectable eosinophils. Most eosinophils resided in the lamina propria at the base of the villi and in the crypt area, although some eosinophils were also present throughout the length of the villi. An independent immunohistochemical approach was also used to detect tissue eosinophils. Using a rabbit polyclonal anti-murine MBP to specifically label eosinophils, we confirmed localization of eosinophils to similar regions of the jejunum (Fig. 4 e–g). The immunohistochemical identification of eosinophils with anti-MBP has the advantage that standard paraffin-embedded tissue samples can be examined for resident eosinophils. All three approaches demonstrated that eosinophils resided in various densities throughout the interstitial tissue of the jejunum.
Figure 4.
Eosinophil distribution in the jejunum of wild-type and eotaxin-deficient mice. The jejunum is stained with Wright’s–Giemsa stain (a–d) and the eosinophils are indicated in the submucosa and lamina propria by arrows. Wild-type (+/+) and eotaxin-deficient (−/−) mice are indicated. The jejunum in wild-type mice (e–g) and eotaxin-deficient mice (h) is stained with anti-MBP, and immunoreactivity is detected by nickel/cobalt enhancement of peroxidase activity. Eosinophils are indicated by arrows. (Original magnification is ×500 in a–d, ×30 in e, ×60 in f and h, and ×125 in g.)
Eosinophil Levels in the Jejunum in the Absence of Eotaxin.
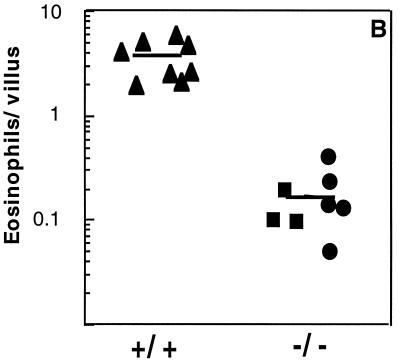
To test the hypothesis that constitutive eotaxin regulated the baseline level of tissue eosinophils, numbers of this leukocyte were examined in the jejunum of mice that were eotaxin-deficient. When Wright’s–Giemsa staining (Fig. 4 b and d) or anti-MBP immunohistochemistry (Fig. 4h) was used, eosinophils were rarely encountered. Whereas wild-type mice had 5.3 ± 0.4 (mean ± SD, n = 6) eosinophils per high-power field (hpf), eotaxin-deficient mice had 0.17 ± 0.26 eosinophils per hpf as measured by using the Wright’s–Giemsa staining (P < 0.001). Similarly, whereas wild-type mice had 3.8 ± 1.8 (mean ± SD, n = 8) eosinophils per villus, eotaxin-deficient mice had 0.15 ± 0.06 as measured by using the anti-MBP staining (Fig. 5) (P < 0.001). When the eotaxin-deficient mice were generated, two mouse lines were established from independent embryonic stem cells that had been transfected with the same targeting construct (13). When the second gene-targeted line was analyzed, a similar reduction in intestinal eosinophils was seen, indicating that the phenotype was not due to a nonspecific event (Fig. 5). In preliminary experiments, the total number of leukocytes in the lamina propria was modestly reduced in the eotaxin-deficient mice. Wild-type mice and eotaxin-deficient mice had 166 ± 19 and 103 ± 19 (mean ± SD, n = 6) leukocytes per hpf, respectively (P < 0.001).
Figure 5.
Quantitation of eosinophils in the jejunum. Eosinophils were enumerated from tissue sections that were stained with anti-MBP antiserum. The data are presented for wild-type (+/+) and eotaxin-deficient (−/−) mice. Two eotaxin-deficient mouse lines are indicated by the • and ▪ data points. The horizontal line represents the mean.
Tissue Eosinophils in the Thymus.
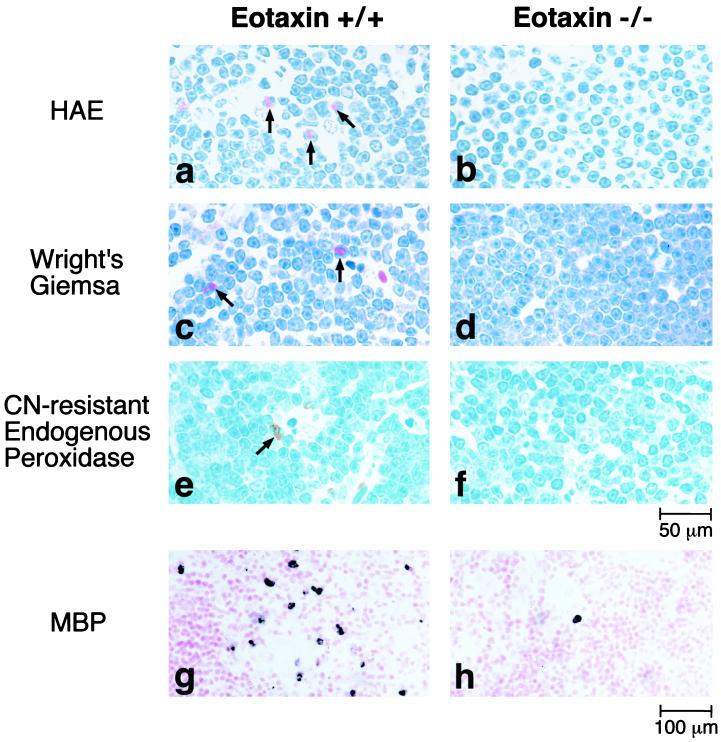
Next, studies were conducted to determine whether the results found in the jejunum were applicable in another tissue. Eotaxin mRNA is readily detectable in the thymus (17, 18, 24). Although eotaxin deficiency did not affect the level or phenotype of thymocytes, the concentration of thymic eosinophils has not been examined previously (13). Four independent methods were used to detect eosinophils in the thymus. By using HAE, Wright’s–Giemsa staining, cyanide-resistant endogenous peroxide staining, and anti-MBP staining, sections of the thymus from wild-type mice were shown to have detectable levels of eosinophils (Fig. 6). Most eosinophils were present in the thymic medulla. In contrast, the thymus from eotaxin-deficient mice had only rare eosinophils (Fig. 6). Wild-type mice and eotaxin-deficient mice had 24 ± 1 and 1.2 ± 0.8 eosinophils per high-power field, respectively (P < 0.001). These results indicated that the regulation of baseline concentrations of eosinophils by eotaxin was not restricted to the intestine.
Figure 6.
Photomicrographs of thymic eosinophils. Tissue sections were analyzed with HAE staining (a and b), Wright’s–Giemsa staining (c and d), cyanide-resistant endogenous peroxidase activity (e and f), or anti-MBP immunoreactivity (g and h). The data are presented for wild-type (+/+; Left) and eotaxin-deficient (−/−; Right) mice. The arrows point to eosinophils.
DISCUSSION
The marked and selective reduction in tissue eosinophils in healthy eotaxin-deficient mice indicates that eotaxin has a critical nonredundant role at baseline. This is particularly important because at least six chemokines signal through the eotaxin receptor in vitro and their relative importance has not been established (8). Additionally, a variety of approaches have shown that during inflammatory events, eotaxin’s role in the recruitment of eosinophils overlaps with the roles of other chemoattractants (12–16). The nonredundant role for eotaxin is not strictly related to its higher constitutive expression, because RANTES is also constitutively expressed in the intestine (Fig. 1). Furthermore, the targeted disruption of eotaxin does not affect the expression of CC chemokines genetically linked to eotaxin (M.E.R., unpublished results), indicating that the observed phenotype is indeed dependent upon eotaxin.
Three chemokine receptors (CXCR-2, CCR-1, and CCR-2) and two chemokines (MIP-1α and stromal-derived factor (SDF)-1α) have been gene targeted (28–33), but none of the resulting mice have been reported to have a defect in the baseline levels of leukocytes resident in nonhematopoietic tissues. These gene-targeted mice have variable levels of alterations in hematopoiesis, inflammation, or embryonic development. The unique role of constitutive eotaxin in regulating the baseline tissue level of a mature leukocyte in a nonhematopoietic tissue expands the role of chemokines. Several chemokines besides eotaxin are constitutively produced in different tissues. A role for these chemokines in regulating baseline leukocyte trafficking should therefore be examined.
While eotaxin is critical for regulating eosinophil tissue levels, other properties of the microenvironment, such as interleukin 5 levels or pH, may also contribute to eotaxin activity and/or eosinophil tissue levels (34, 35). It will be interesting to determine the mechanism by which eotaxin is active in regulating eosinophil tissue concentrations. For example, although eotaxin may promote eosinophil tissue homing, other mechanisms, such as alterations in apoptosis, may also be operational. The induction of eosinophil apoptosis may be delayed in the intestine, accounting for the higher number of eosinophils in this tissue. Eotaxin itself does not appear to be an eosinophil survival factor (11), but engagement of eosinophils by the extracellular matrix may induce the autocrine generation of survival factors (36). Eosinophils and their precursors are at normal levels in the bone marrow of eotaxin-deficient mice, so impaired eosinophil development is unlikely to account for reduced eosinophil tissue levels (13). Additionally, in the inbred strain of mice examined in this study, eosinophil levels in the circulation of eotaxin-deficient mice are equivalent to those in wild-type mice (M.E.R., unpublished results). This observation indicates that eotaxin is exerting its effects directly in the tissue rather than in the circulation or bone marrow.
Unequivocal evidence is presented showing that eosinophils are tissue-dwelling cells at baseline. The intestine and the thymus, under healthy conditions, are demonstrated to contain resident eosinophils. It is likely that eosinophils will also be detectable in tissues not yet analyzed but in which eotaxin is also expressed. This possibility has important implications in understanding the physiological properties of eosinophils because the normal life cycle of an eosinophil should include this tissue phase. It is hoped that further elucidation of the mechanisms of eosinophil physiological trafficking at steady state will provide knowledge concerning the poorly understood function of eosinophils. Tissue-dwelling eosinophils are likely to have a physiological function that is different from the function of recruited eosinophils during allergic inflammation. Tissue-dwelling eosinophils may have an immunomodulating role, because they are positioned in a physical location in the jejunum that allows them to have early contact with enteric antigens. Further analysis of eotaxin-deficient mice may uncover the role of tissue-dwelling eosinophils in a variety of physiological and pathological responses, including food allergy, eosinophilic gastroenteritis, and intestinal parasitic infections.
These studies underscore the critical role for eotaxin in regulating the tissue level of eosinophils and indicate that, in addition to regulating leukocyte recruitment during inflammation, eotaxin has a fundamental role under normal (i.e., healthy) conditions.
Acknowledgments
We are grateful to Dr. Phil Leder for helpful guidance, Dr. Paul Foster for review of the manuscript, and Dr. Anil Mishra for technical assistance.
ABBREVIATIONS
- MCP
monocyte chemoattractant protein
- HAE
hematoxylin, azure II, and eosin
- MBP
major basic protein
References
- 1.Gleich G J, Adolphson C R. Adv Immunol. 1986;39:177–253. doi: 10.1016/s0065-2776(08)60351-x. [DOI] [PubMed] [Google Scholar]
- 2.Weller P F. N Engl J Med. 1991;324:1110–1118. doi: 10.1056/NEJM199104183241607. [DOI] [PubMed] [Google Scholar]
- 3.Capron M. Ann Parasitol Hum Comp. 1991;66:41–45. [PubMed] [Google Scholar]
- 4.Bochner B S, Schleimer R P. J Allergy Clin Immunol. 1994;94:427–438. doi: 10.1016/0091-6749(94)90195-3. [DOI] [PubMed] [Google Scholar]
- 5.Hamelmann E, Oshiba A, Loader J, Larsen G L, Gleich G, Lee J, Gelfand E W. Am J Respir Crit Care Med. 1997;155:819–825. doi: 10.1164/ajrccm.155.3.9117011. [DOI] [PubMed] [Google Scholar]
- 6.Rollins B J. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- 7.Bates P. Cell. 1996;86:1–3. doi: 10.1016/s0092-8674(00)80070-7. [DOI] [PubMed] [Google Scholar]
- 8.Luster A D, Rothenberg M E. J Leukocyte Biol. 1997;62:620–633. doi: 10.1002/jlb.62.5.620. [DOI] [PubMed] [Google Scholar]
- 9.Jose P J, Griffiths-Johnson D A, Collins P D, Walsh D T, Moqbel R, Totty N F, Truong O, Hsuan J J, Williams T J. J Exp Med. 1994;179:881–887. doi: 10.1084/jem.179.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponath P D, Qin S X, Ringler D J, Clark-Lewis I, Wang J, Kassam N, Smith H, Shi X J, Gonzalo J A, Newman W, Gutierrez-Ramos J C, Mackay C R. J Clin Invest. 1996;97:604–612. doi: 10.1172/JCI118456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenberg M E, Ownbey R, Mehlhop P D, Loiselle P M, Vanderijn M, Bonventre J V, Oettgen H C, Leder P, Luster A D. Mol Med. 1996;2:334–348. [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalo J-A, Lloyd C M, Kremer L, Finger E, Martinez-A C, Siegelman M H, Cybulsky M, Guitierrez-Ramos J-C. J Clin Invest. 1996;98:2332–2345. doi: 10.1172/JCI119045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothenberg M E, MacLean J A, Pearlman E, Luster A D, Leder P. J Exp Med. 1997;185:785–790. doi: 10.1084/jem.185.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humbles A A, Conroy D M, Marleau S, Rankin S M, Palframan R T, Proudfoot A E, Wells T N, Li D, Jeffery P K, Griffiths-Johnson D A, Williams T J, Jose P J. J Exp Med. 1997;186:601–612. doi: 10.1084/jem.186.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teixeira M M, Wells T N C, Lukacs N W, Proudfoot A E I, Kunkel S L, Williams T J, Hellewell P G. J Clin Invest. 1997;100:1657–1666. doi: 10.1172/JCI119690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamkhioued B, Renzi P M, Abi-Younes S, Garcia-Zepeda E A, Allakhverdi Z, Ghaffar O, Rothenberg M E, Luster A D, Hamid Q. J Immunol. 1997;159:4593–4601. [PubMed] [Google Scholar]
- 17.Rothenberg M E, Luster A D, Lilly C M, Drazen J M, Leder P. J Exp Med. 1995;181:1211–1216. doi: 10.1084/jem.181.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothenberg M E, Luster A D, Leder P. Proc Natl Acad Sci USA. 1995;92:8960–8964. doi: 10.1073/pnas.92.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Zepeda E A, Rothenberg M E, Ownbey R T, Celestin J, Leder P, Luster A D. Nat Med. 1996;2:449–456. doi: 10.1038/nm0496-449. [DOI] [PubMed] [Google Scholar]
- 20.Beckstead J H, Halverson P S, Ries C A, Bainton D F. Blood. 1981;57:1088–1098. [PubMed] [Google Scholar]
- 21.Boyce J A, Friend D, Matsumoto R, Austen K F, Owen W F. J Exp Med. 1995;182:49–57. doi: 10.1084/jem.182.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain-Vora S, Wert S E, Temann U, Rankin J A, Whitsett J A. Am J Respir Cell Mol Biol. 1997;17:541–551. doi: 10.1165/ajrcmb.17.5.2883. [DOI] [PubMed] [Google Scholar]
- 23.Wert S E, Glasser S W, Korfhagen T R, Whitsett J A. Dev Biol. 1993;156:426–443. doi: 10.1006/dbio.1993.1090. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalo J-A, Jia G-Q, Aquirre V, Friend D, Coyle A J, Jenkins N A, Lin G-S, Katz H, Lichtman A, Copeland N, Kopf M, Gutierrez-Ramos J-C. Immunity. 1996;4:1–14. doi: 10.1016/s1074-7613(00)80293-9. [DOI] [PubMed] [Google Scholar]
- 25.Rankin J A, Picarella D E, Geba G P, Temann U-A, Prasad B, DiCosmo B, Tarallo A, Stripp B, Whitsett J, Flavell R A. Proc Natl Acad Sci USA. 1996;93:7821–7825. doi: 10.1073/pnas.93.15.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Wang D, Griffiths-Johnson D A, Wells T N, Williams T J, Jose P J, Jeffery P K. Eur Respir J. 1997;10:1946–1954. doi: 10.1183/09031936.97.10091946. [DOI] [PubMed] [Google Scholar]
- 27.Jose P J, Adcock I M, Griffiths-Johnson D A, Berkman N, Wells T N C, Williams T J, Power C A. Biochem Biophys Res Comm. 1994;205:788–794. doi: 10.1006/bbrc.1994.2734. [DOI] [PubMed] [Google Scholar]
- 28.Cacalano G, Lee J, Kikly K, Ryan A M, Pitts-Meek S, Hultgren B, Wood W I, Moore M W. Science. 1994;265:682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- 29.Cook D N, Beck M A, Coffman T M, Kirby S L, Sheridan J F, Pragnell I B, Smithies O. Science. 1995;269:1583–1585. doi: 10.1126/science.7667639. [DOI] [PubMed] [Google Scholar]
- 30.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Nature (London) 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 31.Gao J L, Wynn T A, Chang Y, Lee E J, Broxmeyer H E, Cooper S, Tiffany H L, Westphal H, Kwon-Chung J, Murphy P M. J Exp Med. 1997;185:1959–1968. doi: 10.1084/jem.185.11.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boring L, Gosling J, Chensue S W, Kunkel S L, Farese R V, Jr, Broxmeyer H E, Charo I F. J Clin Invest. 1997;100:2552–2561. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurihara T, Warr G, Loy J, Bravo R. J Exp Med. 1997;186:1757–1762. doi: 10.1084/jem.186.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mould A W, Matthaei K I, Young I G, Foster P S. J Clin Invest. 1997;99:1064–1071. doi: 10.1172/JCI119234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dairaghi D J, Oldham E R, Bacon K B, Schall T J. J Biol Chem. 1997;272:28206–28209. doi: 10.1074/jbc.272.45.28206. [DOI] [PubMed] [Google Scholar]
- 36.Kita H. J Allergy Clin Immunol. 1996;97:889–892. doi: 10.1016/s0091-6749(96)80061-3. [DOI] [PubMed] [Google Scholar]
- 37.Rich B E, Steitz J A. Mol Cell Biol. 1987;7:4065–4074. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]