Abstract
Here we report on a 3.0 Å crystal structure of a ternary complex of wild-type Thermus thermophilus argonaute bound to a 5′-phosphorylated 21-nucleotide guide DNA and a 20-nucleotide target RNA containing cleavage-preventing mismatches at the 10–11 step. The seed segment (positions 2 to 8) adopts an A-helical-like Watson–Crick paired duplex, with both ends of the guide strand anchored in the complex. An arginine, inserted between guide-strand bases 10 and 11 in the binary complex, locking it in an inactive conformation, is released on ternary complex formation. The nucleic-acid-binding channel between the PAZ- and PIWI-containing lobes of argonaute widens on formation of a more open ternary complex. The relationship of structure to function was established by determining cleavage activity of ternary complexes containing position-dependent base mismatch, bulge and 2′-O-methyl modifications. Consistent with the geometry of the ternary complex, bulges residing in the seed segments of the target, but not the guide strand, were better accommodated and their complexes were catalytically active.
RNA-induced silencing complex (RISC)-associated argonaute (Ago) proteins composed of PAZ- and PIWI-containing modules have a central role in mediating distinct assembly and cleavage steps of the RNA interference (RNAi) catalytic cycle1-4. The Ago protein, as the sole component of RISC exhibiting RNA ‘slicer’ activity5-7, is a critical player in the RNAi pathway8, effecting transcriptional and post-transcriptional gene regulation in plants and animals1-4. In this capacity, Agos have essential roles ranging from maintaining genomic integrity to heterochromatin formation. Some Ago proteins with active endonuclease domains contribute to the maturation of bound short interfering RNAs (siRNAs) by degradative cleavage of the passenger strand and subsequent guide-strand-mediated sequence-specific cleavage of target RNAs9-12. An improved understanding at the molecular level of guide-strand recognition in a binary complex with Ago and RNA target recognition and cleavage at the ternary Ago complex level could effect the application of RNAi approaches as a therapeutic modality against diverse human diseases13,14.
Bacteria also exhibit Ago-based cleavage activity of target RNAs and hence use the RNAi machinery, although details of the players and pathways at the cellular level are at present poorly understood. Most of the structural research on Ago proteins has initially focused on archaeal and eubacterial thermophilic Agos4,15-18. Crystal structures have been reported for archaeal Pyrococcus furiosus Ago5 and eubacterial Aquifex aeolicus Ago19, both in the free state, thereby defining the relative alignments of the PAZ-containing (N and PAZ domains and linkers L1 and L2) and PIWI-containing (Mid and PIWI) modules. Given that binding and cleavage studies have identified these eubacterial and archaeal Agos to be site-specific DNA-guided endoRNases19, our group has recently solved the crystal structure of eubacterial T. thermophilus Ago with 5′-phosphorylated 10- and 21-nucelotide guide DNAs20, thereby identifying the guide strand-binding channel between the basic PAZ- and PIWI-containing lobes of the bilobal19 Ago scaffold. This structure established that the 5′-phosphate and 3′-hydroxyl ends are positioned in their respective binding pockets in the Mid21,22 and PAZ23,24 domains of Ago, the 2–10 segment of the bound guide DNA forms a stacked array, such that the solvent-exposed Watson–Crick edges of bases 2 to 6 in the ‘seed’ segment are positioned for nucleation with target RNA20. In addition, the side chain of R548 was inserted between bases 10 and 11, thereby disrupting base stacking, and locking this step in a non-catalytically competent conformation. We now report on the structural characterization of the ternary T. thermophilus Ago complex containing target RNA, thereby defining in molecular detail the alignment and pairing of guide and target strands spanning the seed segment and their positioning within the expanded nucleic-acid-binding channel of the bilobal Ago scaffold.
Structure of ternary complex
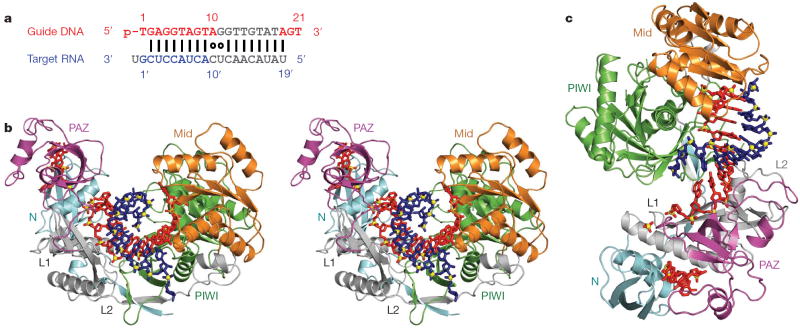
The 3.0 Å crystal structure of wild-type T. thermophilus Ago complexed with a 5′-phosphorylated (5′-phos-T1GAGG5TAGTA10GGTTG15TATAG20T) 21-nucleotide guide DNA and its essentially complementary 20-nucleotide target RNA, containing cleavage-preventing A-C and G-U mismatches at the 10 and 11 steps (Fig. 1a), is presented in a stereo view in Fig. 1b, with an alternative perspective shown in Fig. 1c. A movie of the ternary Ago complex is included in Supplementary Movie 1. The Ago protein, in a ribbon representation, is colour-coded by domains and linker elements, with the traceable segments of the guide DNA in red and the target RNA in blue.
Figure 1. Crystal structure of T. thermophilus Ago bound to 5′-phosphorylated 21-nucleotide guide DNA and 20-nucleotide target RNA.
a, Sequence of the guide DNA-target RNA duplex. The traceable segments of the bases of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. Disordered segments of the bases on both strands that cannot be traced are shown in grey. b, Stereo view of the 3.0 Å crystal structure of the Ago ternary complex. The Ago protein is colour-coded by domains (N in cyan, PAZ in magenta, Mid in orange and PIWI in green) and linkers (L1 and L2 in grey). The bound 21-nucleotide guide DNA is in red and traced for bases of the 1–10 and 19–21 segments, whereas the bound 20-nucleotide target RNA is in blue and traced for bases of the 1′ to 9′ segment. Backbone phosphorus atoms are in yellow. c, An alternate view of the complex.
The bases of the guide DNA can be traced for the 1–10 segment (electron density map, Supplementary Fig. 1a), in which the 5′-phosphate is inserted into its binding pocket in the Mid domain21,22 (Supplementary Fig. 2a), and can be traced for the 18/19 to 21 segment (electron density map, Supplementary Fig. 1b, c), in which nucleotides 20 and 21 at the 3′ end are inserted into their binding pocket in the PAZ domain (Supplementary Fig. 2b)23,24. The observed anchoring of both ends of the guide DNA in the ternary complex with RNA target2 was unexpected on topological grounds, although we anticipate the release of the 3′-end if pairing was to go beyond the seed segment observed in the present structure.
Guide-target duplex spanning seed segment
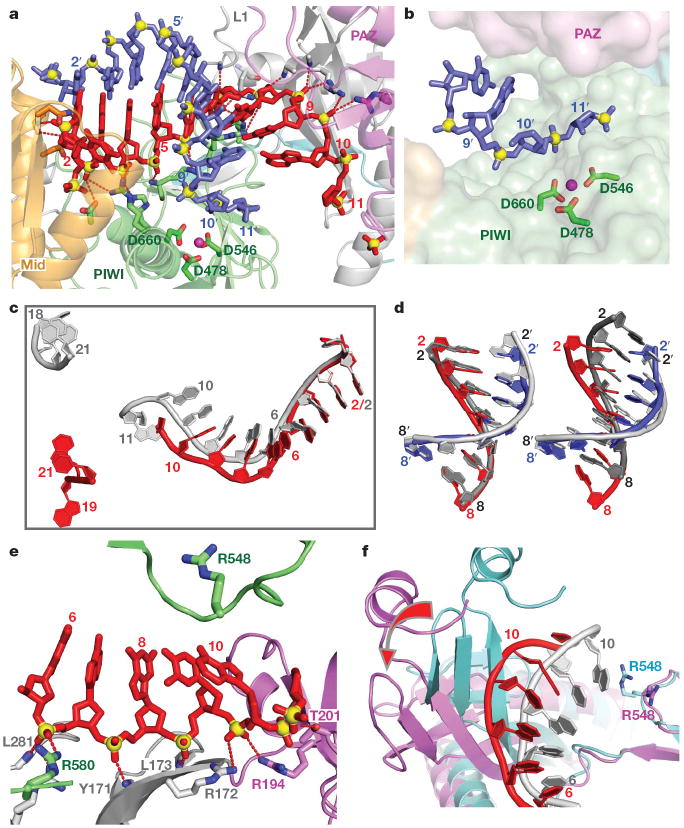
The bases of the target RNA can be traced for the 1′–9′ segment (electron density map, Supplementary Fig. 3), with Watson–Crick alignment of the target RNA with the guide DNA, spanning the entire 2–8 seed segment (Fig. 2a). In addition, we can trace the sugar-phosphate backbone of the guide DNA and target RNA strands spanning the mismatch-containing 10–11 step (electron density map, Supplementary Fig. 4), thereby defining the orientation of the mismatch-distorted 10–11 cleavage step on the target RNA relative to the Asp catalytic triad of the RNase H fold of the PIWI domain of Ago (Fig. 2b).
Figure 2. Comparison of structural details between the binary Ago complex with bound guide DNA and the ternary complex with added target RNA.
a, Expanded view of the ternary complex highlighting the guide DNA (1–10)-target RNA (1′–9′) duplex and Mg2+-coordinated catalytic residues (D478, D546 and D660) of the RNase H fold of the PIWI domain. Intermolecular hydrogen bonds between the Ago protein and the DNA guide strand in red are shown by dashed lines. b, Positioning of the sugar-phosphate backbone of the target RNA strand spanning the mismatch-containing 10–11-step relative to the catalytic residues of the PIWI domain. c, Comparison of the trajectory of traceable bound guide DNA in the binary (bases 1–11 and 18–21 in silver) and ternary (bases 1–10 and 19–21 in red) Ago complexes after superposition of their 5′-phosphate-binding pockets. d, Superposition of the guide DNA (red)-target RNA (blue) duplex spanning the 2–8 seed segment on A-form (left panel) and B-form (right panel) helices (silver) after best-fit superposition of the target RNA strand of the ternary Ago complex with one strand of the A/B-form helices. e, Positioning of stacked residues 6–10 of the DNA guide strand relative to R548, with emphasis on intermolecular interactions involving the sugar-phosphate backbone. f, Relative positioning of the 6 to 10/11 segment of the bound guide DNA strand and R548 in the binary (guide strand in silver, protein in cyan) and ternary (guide strand in red, protein in magenta) Ago complexes. The conformational change in the protein on proceeding from binary to ternary Ago complexes is indicated by a red arrow.
The relative positions of the guide DNA in the binary (1–11 and 18–21, in silver) and ternary (1–10 and 19–21, in red) Ago complexes after superposition of their 5′-phosphate-recognizing Mid binding pockets is shown in Fig. 2c. Although the guide DNA superpositions well for the 1–4 segment, there is an 8.9 Å displacement by position 10 and an 17.1 Å displacement by position 21, suggesting that a noticeable change occurs in the trajectory of the guide strand on proceeding from the binary to the ternary complex.
The 2–8 seed segment formed by the guide DNA-target RNA duplex in the ternary Ago complex superpositions quite well with an A-form helix (Fig. 2d, left panel) and less well with its B-form helical counterpart (Fig. 2d, right panel). The guide-target duplex adopts different trajectories in the Archaeoglobus fulgidus Piwi ternary complex reported previously21,22, which lacks the PAZ-containing module, and in the T. thermophilus Ago ternary complex (stereo view in Supplementary Fig. 5).
Conformational transitions
We had previously identified an orthogonal alignment of bases 10 and 11 of the guide DNA as a result of insertion of the R548 side chain into the 10–11 step in the binary Ago complex20 (Supplementary Fig. 6). In contrast, the side chain of R548 no longer inserts between bases 10 and 11 in the ternary complex (Fig. 2e), as a result of conformational changes on addition of target RNA. Thus, the non-helical conformation at the 10–11 step in the binary Ago complex (Supplementary Fig. 6)—an arrangement that would be detrimental for catalytic cleavage of target RNA by the RNase H fold of the PIWI domain—is released on ternary Ago complex formation (Fig. 2e).
A single Mg2+ cation can be identified bound to the catalytic Asp residues for one molecule (Fig. 2a and Supplementary Fig. 7a) but not for the other in the asymmetric unit of the ternary complex, with reasonable superposition of the Asp side chains between molecules (biscuit and green colours in Supplementary Fig. 7b). The relative alignments of these catalytic Asp residues changes on proceeding from the binary (in silver, Supplementary Fig. 7c) to the ternary (in green, Supplementary Fig. 7c) Ago complex with added target RNA.
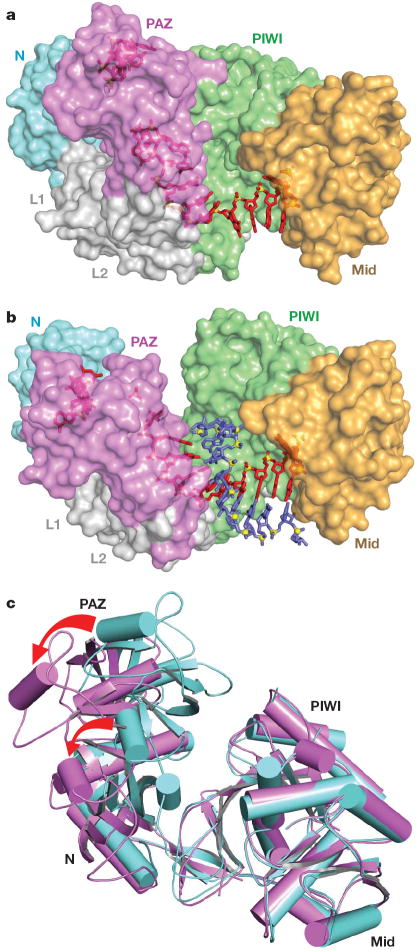
There is a marked conformational change in the relative alignments of the PAZ- and PIWI-containing lobes of the bilobal Ago scaffold on proceeding from the binary T. thermophilus Ago complex (with bound guide DNA) to the ternary complex (with added target RNA). This is readily observable on comparing the trajectory of the bound DNA guide strand (silver and red for the binary and ternary complexes, Fig. 2f) and adjacentprotein segments (cyan and magenta for the binary and ternary complexes, Fig. 2f; conformational change highlighted by a red arrow) between the two complexes. Specifically, the PAZ- and PIWI-containing lobes move further apart on proceeding from binary (Fig. 3a) to ternary (Fig. 3b) complex formation, resulting in an opening movement that widens the nucleic-acid-binding channel, thereby facilitating insertion, alignment and pairing of the target RNA.
Figure 3. Conformational changes within the bilobal Ago scaffold on proceeding from the binary (guide) complex to the ternary (guide + target) complex.
a, b, The nucleic acid binding canyon changes width on proceeding from the binary (a) to the ternary (b) complex. The Ago protein is shown in a space-filling representation with labelled domains and linkers colour-coded as in Fig. 1. The guide DNA (red) and target RNA (blue) are shown in stick representation with backbone phosphorus atoms in yellow. c, View of alignment of binary (cyan) and ternary (magenta) Ago complexes, after superpositioning of their PIWI-containing modules. The red arrows indicate the magnitude of the conformational changes on proceeding from binary to ternary complexes.
These movements can be tracked (Supplementary Movie 2) and are reinforced in a superpositioned view comparing the binary (in cyan) and ternary (in magenta) Ago complexes (Fig. 3c). The opening movement on ternary complex formation is reflected in a large conformational change in the N (rotation of 18.6° and translation of 0.6 Å) and PAZ (rotation of 23.1° and translation of 1.4 Å) domains (see red arrows in Fig. 3c) after superposition of their PIWI-containing modules (Mid and PIWI domains).
We also note that although bases 20 and 21 are similarly positioned in the PAZ binding pocket, the 18–19 segments of the binary and ternary complexes adopt different trajectories, associated with conformational changes in nearby protein segments (Supplementary Fig. 8).
Effect of mismatches and bulges on cleavage
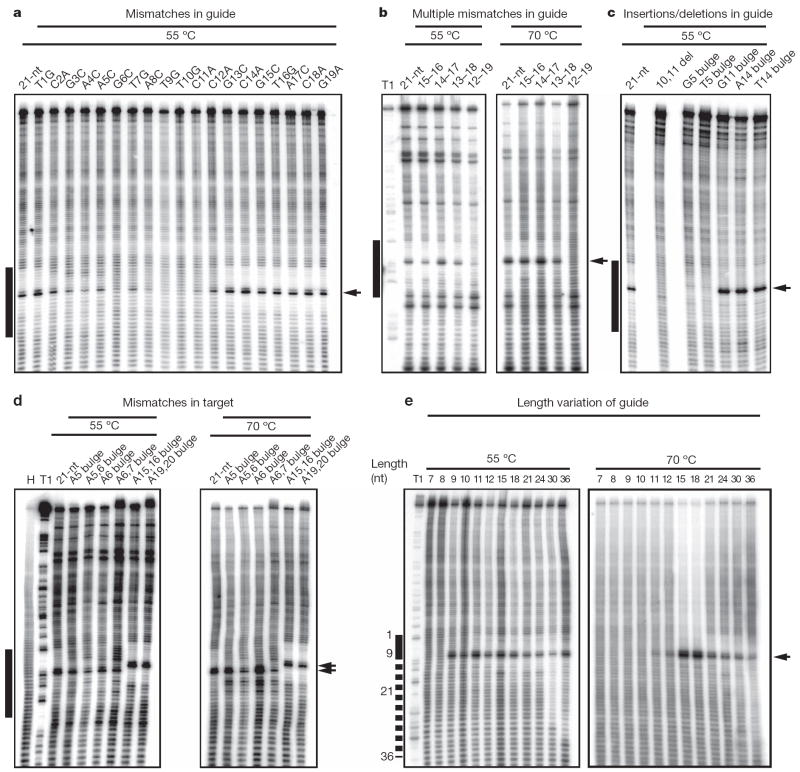
Previous biochemical studies showed RNase H catalytic activity of bacterial Ago proteins loaded with guide DNA19,20. To assess the importance of base pairing of the guide DNA to target RNA, we introduced single nucleotide mismatches at positions from 1 to 19 in the 21-nucleotide guide strand (Fig. 4a). Cleavage reactions were carried out by first preincubating the Ago protein with guide DNA strands at 55 °C, followed by the addition of target RNA and further incubation at either 55 or 70 °C; both enhanced cleavage and background hydrolysis were observed at the higher temperature. Single mismatches were fully tolerated in the 3′-guide DNA regions (positions 13–19), and at the very 5′ end (position 1), whereas cleavage was either abolished (position 9 and 10) or reduced (positions 11 and 12) by mismatches surrounding the cleavage site at 55 °C. Single mismatches in the seed region (position 2–8) showed reduced cleavage activity.
Figure 4. Target RNA cleavage activity of T. thermophilus Ago loaded with mismatched, bulge-containing or length-altered guide DNA strands.
Besides Ago-mediated target RNA cleavage, extensive chemical hydrolysis is observed over the entire RNA substrate length, except for a 12-nucleotide (nt) region across guide DNA positions 2 to 13, which was protected from hydrolysis by the interaction with Ago and its guide DNA. Mismatches or bulges in the 5′ region reduce protection, and frequently catalysis, because of reduced base-pairing stability and deviation from hydrolysis-protecting helical geometry. The black bar to the left of the images defines the region of the cleavage substrate complementary with the 21-nucleotide (nt) guide DNA (a–d) or the region complementary with the 7-nucleotide guide DNA with the dotted line indicating pairing for 3′-extended guide DNAs (e). Arrows indicate the cleavage site. Del, deletion; H, hydrolysis ladder of substrate RNA; T1, partial RNase T1 digest of substrate RNA. a, Single mismatches were placed in the 21-nucleotide guide, at positions 1 to 19. b, Double, quadruple, sextuple and octuple mismatches were introduced in the DNA guide 3′ region at indicated positions. c, Cleavage assay using DNA guides with a deletion or insertions of bulged nucleotides at indicated positions. d, Insertion of bulges in the target RNA. Insertion of nucleotides upstream of the cleavage site leads to shifts in the cleavage site by the number of inserted nucleotides. e, DNA guide length was increased from 7 to 36 nucleotides. The sequences of DNA guides and RNA targets are listed in Supplementary Table 2. As evident from increased hydrolysis of the target region recognized by the guide, insertion of bulges seems to destabilize guide-target interactions.
We next introduced several mismatches in the 3′-region and observed unaltered cleavage activity for up to six contiguous mismatches (positions 13–18) and reduced activity for eight contiguous mismatches (positions 12–19) at both 55 and 70 °C (Fig. 4b). These alterations of catalytic activity as a consequence of mismatches run parallel to earlier structure-function analysis in animals11,25, emphasizing the importance of contiguous base pairing for RNase H-mediated cleavage26 in the 5′-seed segment (positions 2–8), especially near the cleavage site (step 10–11), and minimal contribution of the 3′-half (positions 13–19).
The importance of the seed segment became more apparent when we introduced single nucleotide insertions or deletions into the guide DNA (Fig. 4c). Cleavage was abrogated by a single nucleotide insertion at position 5 in the seed segment, whereas they had little effect when introduced at position 11 adjacent to the cleavage site or at position 14 in the 3′ region of the guide strand. This suggests that the seed segment of the guide strand cannot accommodate a bulge nucleotide when paired to the target in the context of a helical duplex geometry, without disruption of the extensive intermolecular interactions that anchor the guide strand within the Ago scaffold (Fig. 2a; see schematic in Supplementary Fig. 9).
We next assessed the importance of single and double bulges within the target RNA. Unexpectedly, insertion of single bulges opposite positions 4–5 and 5–6 of the guide strand 5′-seed segment had minimal effect on cleavage, but cleavage was reduced after insertion of dual bulges at the same positions (Fig. 4d). In contrast to the guide strand, the sugar-phosphate backbone of the target strand complementary to the seed segment makes no contacts with the Ago scaffold (Supplementary Fig. 10 and Supplementary Movie 3), and seemingly accommodates single and to a lesser extent double bulges within the paired seed segment. Insertion of double bulges opposite positions 14–15 and 18–19 of the guide strand 3′ region did not impair target RNA cleavage (Fig. 4d), again reinforcing the conclusion that helical imperfections disrupting pairing interactions involving the 3′ region of the guide strand have a minimal affect on cleavage activity.
Effect of guide length and 2′-O-methyl on cleavage
To investigate the constraints on guide strand length for cleavage activity, we systematically investigated the cleavage activity of guide strands spanning from 7 to 36 nucleotides. At a reaction temperature of 55 °C, cleavage is observed for 5′-phosphorylated guide strand sequences that are as short as 9 nucleotides and as long as 36 nucleotides (Fig. 4e, left panel). Increasing the reaction temperature to 70 °C restricts cleavage to 12–36-nucelotides guide strands (Fig. 4e, right panel). Furthermore, cleavage activity is strictly dependent on the presence of a 5′-phosphate on the guide strand (Supplementary Fig. 11), a dependence not strictly observed for animal Ago complexes27,28. The observed cleavage for guide strands as short as 9-nucleotides at 55 °C (Fig. 4e, left panel), which lack residues proximal to the cleavage site that could not be mutated without loss of activity (Fig. 4a), was unanticipated. This suggests that mismatch accommodation introduces steric constraints that interfere with proper placement of the target strand cleavage residues, and that the geometry required for cleavage can also be adopted in the absence of residues 10 and 11 of the guide strand. The absence of cleavage for 9- to 11-nucleotides guide strands at 70 °C (Fig. 4e, right panel) probably reflects the thermodynamic instability of short duplex hybridization at higher temperatures. The observed cleavage activity by guide strands up to 36-nucleotides in length (Fig. 4e) could be rationalized by the guide strand adopting alternate trajectories for its 3′-segment leading into the PAZ pocket. This flexibility is presumably a consequence of fewer intermolecular contacts between the protein side chains and the sugar-phosphate backbone of the 3′-guide strand segment (residues 12–19; Supplementary Fig. 12), which is in contrast to the extensively sugar-phosphate backbone-anchored 5′-seed counterpart (Fig. 2a and Supplementary Fig. 9). In addition, the nucleic-acid-binding channel that encompasses the 3′-segment of the bound guide strand is accessible to the outside and permissive to looping-out longer 3′-segments (Supplementary Fig. 12).
In mammalian systems, altering the guide strand 2′-hydroxyl to 2′-O-methyl was shown to influence the cleavage activity depending on the extent and position of modification29. To probe the importance of the 2′-position in the eubacterial guide DNA, we introduced 2′-O-methyl modifications at various positions and monitored cleavage at 55 and 70 °C (Supplementary Fig. 13). We observed that fully modified 2′-O-methylated guide molecules, as reported for mammalian RNAi30, abrogated cleavage, whereas substitutions spanning the 5′-half (residues 2–9) or the 3′-half (residues 12–19) severely reduce activity. In contrast, pairwise replacement of DNA residues by 2′-O-methyl had minimal effects on catalytic activity, independent of the position (Supplementary Fig. 13). These results indicate that extensive 2′-O-methyl modifications of the guide strand may introduce cumulative steric constraints that inhibit the adoption of cleavage competent conformations or even interfere with guide strand accommodation into the Ago protein.
Our structural results on the ternary Ago complex have highlighted details of guide-strand-mediated target RNA recognition spanning the seed segment, and the conformational transitions that expand the nucleic-acid-binding channel within the bilobal Ago scaffold, thereby accommodating and aligning the target RNA for eventual site-specific cleavage. We have been unable in the present study to trace the electron density accurately for base 11 of the DNA guide and bases 10′ and 11′ of the RNA target strands of the bound duplex, with the disorder probably due to distortions associated with mismatch incorporation at the 10–11 step.
Functional implications
The biochemical and structural information helps us to understand how microRNA (miRNA)-target RNA duplexes can be accommodated in Ago complexes, and rationalizes how one class of siRNA off-targets31 requiring extensive base pairing across the cleavage site can be subjected to cleavage. We can now rationalize how single-site alterations in a target RNA within the sequence defined by the seed can be accommodated without interference with cleavage activity, provided that sufficient pairing is retained along the cleavage site. It is also conceivable that miRNA targets with more extensive base pairing extending outside the seed region, as observed for a subset of miRNA targets, may be subject to direct messenger RNA cleavage, possibly contributing to the general destabilization encountered in miRNA-mediated mRNA targeting.
Our results on eubacterial T. thermophilus Ago are similar to the finding that Drosophila Piwi proteins32 and human AGO proteins33 can accept different size guide strands to mediate target RNA cleavage. In essence, our studies emphasize that Piwi and Ago scaffolds have sufficient plasticity to accommodate different length guide strands, and that the size range of naturally accommodated small RNA classes are a direct consequence of their distinct biogenesis mechanisms.
It remains unclear at this time whether mammalian Agos will function in a manner suggested by the structure of the eubacterial Ago ternary complex outlined here. Therefore, the recent report on successful insect cell expression of a soluble form of human AGO2 (also known as EIF2C2 protein; ref. 34) opens opportunities and holds promise for future extension of continuing structural research to eukaryotic Ago complexes, thereby expanding on insights from chemical approaches into the molecular basis of target RNA recognition and cleavage by human RISC35,36.
METHODS SUMMARY
Wild-type T. thermophilus Ago was overexpressed from Escherichia coli and purified by chromatography. The ternary Ago complex was generated in a stepwise manner by initially mixing Ago with 21-nucleotide guide DNA, followed by addition of 20-nucleotide target RNA. Crystals were grown by hanging-drop vapour diffusion. The ternary Ago complex structure was determined by molecular replacement using the domains of the binary Ago complex structure20 as search models. Cleavage assays were undertaken using guide oligodeoxynucleotides and a longer target RNA. Details of all biochemical and crystallographic procedures are listed in Methods.
Supplementary Material
Acknowledgments
The research was supported by funds from the National Institutes of Health and the Starr Foundation to D.J.P. and T.T. We would like to thank the staff of NE-CAT beam line at the Advanced Photon Source, Argonne National Laboratory, supported by the US Department of Energy, for assistance with data collection.
Footnotes
Author Contributions Y.W. and G.S. expressed and purified T. thermophilus Ago, and grew crystals of the ternary complex. H.L. and Y.W. collected X-ray diffraction data on the micro-focus beam line, and Y.W. solved the structure of the ternary complex. The structural studies were undertaken with the supervision of D.J.P. S.J. was responsible for the cleavage assays on Ago with modified guide strands under the supervision of T.T. D.J.P. and T.T. were primarily responsible for writing the paper and all authors read and approved the submitted manuscript.
Author Information The structural coordinates of the ternary complex of T. thermophilus Ago bound to 5′-phosphorylated 21-nucleotide guide DNA and 20-nucleotide target RNA have been submitted to the Protein Data Bank under accession number 3F73. Reprints and permissions information is available at www.nature.com/reprints.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W. The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 4.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nature Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 5.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, et al. Argonaute2 is the catalytic engine of RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Parker JS, Roe S, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez J, Tuschl T. RISC is a 5′-phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 13.De Fougerolles A, Vornlocher H-P, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nature Rev Drug Discovery. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature Rev Genetics. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 15.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Hall TM. Structure and function of argonaute proteins. Structure. 2005;13:1403–1408. doi: 10.1016/j.str.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Parker JS, Barford D. Argonaute: a scaffold for the function of short regulatory RNAs. Trends Biochem Sci. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Patel DJ, et al. Structural biology of RNA silencing and its functional implications. Cold Spring Harb Symp Quant Biol. 2006;71:81–93. doi: 10.1101/sqb.2006.71.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Structure of the guide-strand-containing Argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma JB, et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the Paz domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 Paz domain. Nature Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 25.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nature Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 26.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 28.Chen PY, et al. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson AL, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nature Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 31.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 32.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′-end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 33.Martinez J, et al. Single-stranded anti-sense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 34.MacRae IJ, et al. In vitro reconstitution of human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 36.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature Rev Mol Cell Biol. 2007;8:23–26. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.