Abstract
Background
It was demonstrated previously that the three-locus RFLP haplotype, TaqI B-TaqI D-TaqI A (B-D-A), at the DRD2 locus constitutes a powerful genetic marker and probably reflects the most ancient dispersal of anatomically modern humans.
Results
We investigated TaqI B, BclI, MboI, TaqI D, and TaqI A RFLPs in 17 contemporary populations of the East European Plain and Siberia. Most of these populations belong to the Indo-European or Uralic language families. We identified three common haplotypes, which occurred in more than 90% of chromosomes investigated. The frequencies of the haplotypes differed according to linguistic and geographical affiliation.
Conclusion
Populations in the northwestern (Byelorussians from Mjadel'), northern (Russians from Mezen' and Oshevensk), and eastern (Russians from Puchezh) parts of the East European Plain had relatively high frequencies of haplotype B2-D2-A2, which may reflect admixture with Uralic-speaking populations that inhabited all of these regions in the Early Middle Ages.
Background
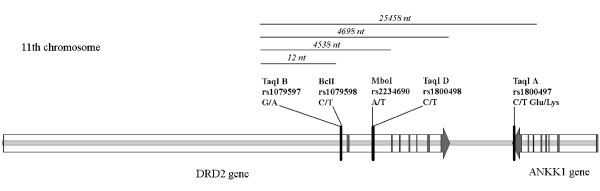
The DRD2 gene is located on chromosome 11 and encodes the neuronal dopamine receptor D2, which plays a role in movement, emotional memory, and appetitive behavior [1]. The DRD2 locus was an object of numerous genetic association studies [2-5], and the most extensively studied polymorphism is a TaqI A RFLP (rs1800497; in the vicinity of the DRD2 gene), which has been associated with the pathology of psychoses (schizophrenia and manic-depressive disorder), Parkinson's disease, and various substance abuse syndromes. It has been proposed that TaqI A might be in linkage disequilibrium with some unidentified polymorphisms within the exons or regulatory regions of the DRD2 gene, but recently it has been mapped to the last exon of the ANKK1 (ankyrin repeat and kinase domain containing 1) gene, and it results in a Glu-Lys substitution [6]. Other frequently studied RFLPs, for example, TaqI B and D (rs1079597 and rs1800498, respectively) are located in the introns of the DRD2 gene and, most probably, have no functional significance.
TaqI B, TaqI D, and TaqI A polymorphisms have also been studied on a worldwide scale [[7-11]; the ALFRED database, http://alfred.med.yale.edu/alfred/index.asp], and centers of dispersal, which probably reflect the most ancient dispersal of anatomically modern humans, have been proposed for their three-locus haplotypes [7]. It has been shown that the B2, D2, and A1 alleles are ancestral alleles common to other hominoids [12-14]. Kidd et al. [7] proposed the following evolutionary sequence for the most common haplotypes: evolution of B2-D2-A1 to B2-D2-A2 and B1-D2-A1 and evolution of B2-D2-A2 to B2-D1-A2. The other less frequent haplotypes probably arose by recombination. Three-locus haplotypes exhibit pronounced geographical differentiation. With the exception of some tribal populations of India [11], the ancestral haplotype B2-D2-A1 is mainly confined to African groups. The singly derived haplotype B1-D2-A1 is most widespread among people of East Asian descent, including Native Americans. Haplotype B2-D2-A2 is present among all populations but is least prevalent in Western European and American populations. The doubly derived haplotype B2-D1-A2 is common in Europe and rare in East Asia. The other haplotypes are extremely rare and have sporadic distribution.
Here we provide data on the variability of DRD2 haplotypes in a previously uninvestigated region, the East European Plain. We investigated 14 contemporary populations of the East European Plain and Siberia that belong to the Indo-European and Uralic language families. In addition, two populations of the Altaic language family (Yakuts and Kalmyks) and a population of the North Caucasian language family (Adygeis) were included as reference groups. We also performed an updated global analysis of B-D-A haplotype frequencies using our data and data from the ALFRED database.
Results
We studied five RFLP loci, TaqI B, BclI, MboI, TaqI D, and TaqI A. The locations of these polymorphisms are shown on the gene map (Figure 1), and allele frequencies in the populations studied are presented in Table 1. Allele 1 (the restriction site was absent) at the TaqI B locus was always present with allele 1 at the BclI locus; allele 2 (the restriction site was present) at the former locus was always found with allele 2 at the latter locus. The same tight linkage was observed for the MboI and TaqI D loci, and there were no exceptions among the 3198 chromosomes studied. In most populations, all loci exhibited Hardy-Weinberg equilibrium (assessed by the exact test using a Markov chain, P > 0.05). Significant deviations from Hardy-Weinberg equilibrium were observed only for the MboI and TaqI D loci in Khants (P = 0.044) and the TaqI A locus in Yakuts (P = 0.023).
Figure 1.
Map of polymorphisms in the DRD2 and ANKK1 genes. Restriction polymorphisms, corresponding SNPs, and distances between them are shown. Exons are indicated by gray vertical bars.
Table 1.
Geographical and linguistic affiliations of populations sampled in this study, and allele frequencies in the studied populations
| SNP No. | rs1079597 | rs1079598 | rs2234690 | rs1800498 | rs1800497 | |||
| SNP allele | Aa | Tb | Tc | Cd | Te | |||
| RFLP | TaqI B | BclI | MboI | TaqI D | TaqI A | |||
| Language family | Population | Geographical region | Number of individuals | allele 1 | allele 1 | allele 1 | allele 2 | allele 1 |
| Northern Caucasian | Adygeis (Shapsugs)f | Russia, Krasnodar region | 98 | 0.117 | 0.117 | 0.367 | 0.367 | 0.153 |
| Indo-European | Byelorussians 1g | Belarus, Brest region, Pinsk | 70 | 0.129 | 0.129 | 0.450 | 0.450 | 0.143 |
| Byelorussians 2g | Belarus, Minsk region, Mjadel' | 75 | 0.113 | 0.113 | 0.487 | 0.487 | 0.120 | |
| Byelorussians 3g | Belarus, Mogilev region, Klimovichi | 85 | 0.153 | 0.153 | 0.400 | 0.400 | 0.165 | |
| Russians 1 | Russia, Tver region, Andreapol' | 109 | 0.252 | 0.252 | 0.463 | 0.463 | 0.261 | |
| Russians 2g | Russia, Smolensk region, Sychevka | 117 | 0.145 | 0.145 | 0.440 | 0.440 | 0.154 | |
| Russians 3g | Russia, Kursk region, Ponyri | 65 | 0.131 | 0.131 | 0.446 | 0.446 | 0.154 | |
| Russians 4h | Russia, Ivanovo region, Puchezh | 95 | 0.158 | 0.158 | 0.526 | 0.526 | 0.189 | |
| Russians 5g | Russia, Archangelsk region, Oshevensk | 71 | 0.211 | 0.211 | 0.577 | 0.577 | 0.232 | |
| Russians 6 | Russia, Archangelsk region, Mezen' | 147 | 0.153 | 0.153 | 0.554 | 0.554 | 0.156 | |
| Uralic | Veps | Russia, Vologda region, Babaevo | 97 | 0.196 | 0.196 | 0.500 | 0.500 | 0.180 |
| Komi 1 (Izhemski)i | Russia, Komi Republic, Izhma | 112 | 0.138 | 0.138 | 0.411 | 0.411 | 0.152 | |
| Komi 2 (Priluzski)i | Russia, Komi Republic, Obyachevo | 109 | 0.206 | 0.206 | 0.482 | 0.482 | 0.225 | |
| Khantsj | Russia, Khanty-Mansi autonomous region | 62 | 0.234 | 0.234 | 0.750 | 0.750 | 0.258 | |
| Nenets | Russia, Yamalo-Nenetsky autonomous region | 65 | 0.223 | 0.223 | 0.762 | 0.762 | 0.262 | |
| Altaic | Yakutsg | Russia, Saha Republic, Tiungiuliu | 118 | 0.269 | 0.269 | 0.932 | 0.932 | 0.214 |
| Kalmyksg | Russia, Republic Kalmykiya, Elista | 104 | 0.356 | 0.356 | 0.856 | 0.856 | 0.346 | |
SNP accession numbers and corresponding RFLPs names are presented for the polymorphisms studied. Alleles without restriction sites are named "allele 1", alleles with restriction sites "allele 2".
a only A and G were detected at this SNP locus in a sample of 2450 chromosomes from 17 populations http://www.hapmap.org, http://www.ncbi.nlm.nih.gov/SNP
b only T and C were detected at this SNP locus in a sample of 1204 chromosomes from 14 populations http://www.ncbi.nlm.nih.gov/SNP
c only A and T were detected at this SNP locus in a sample of 142 chromosomes from three populations http://www.ncbi.nlm.nih.gov/SNP
d only T and C were detected at this SNP locus in a sample of 946 chromosomes from 12 populations http://www.ncbi.nlm.nih.gov/SNP
e only T and C were detected at this SNP locus in a sample of 2628 chromosomes from 21 populations http://www.hapmap.org, http://www.ncbi.nlm.nih.gov/SNP
f population initially described in [62]
g population initially described in [38]
h population initially described in [37]
i population initially described in [53]
j population initially described in [39]
Alleles 1 at the TaqI B, BclI, and TaqI A loci were more frequent in the populations of Russians 1 and 5, Veps, Komi 2, Khants, Nenets, Yakuts, and especially Kalmyks, than in the other populations (Table 1). Allele 1 at the MboI locus and allele 2 at the TaqI D locus were the most frequent alleles in Asian populations, i.e. Khants, Nenets, Yakuts, and Kalmyks. It is notable that combinations of allele 2 at the TaqI B locus with allele 1 at the BclI locus and allele 2 at the MboI locus with allele 2 at the TaqI D locus, found in the sequenced chimpanzee genome [15], were absent in human populations (Table 1).
Pairwise linkage disequilibrium (D') was strong in most cases (Table 2). However, disequilibrium values were lower for the MboI-TaqI A and TaqI D-TaqI A pairs in some populations, which is similar to the findings of Kidd et al. [7]. In Yakuts, disequilibrium was very low, except for the TaqI B-TaqI A and BclI-TaqI A pairs. Only two of the SNPs studied have been involved in the international HapMap project http://www.hapmap.org: TaqI A (rs1800497) and TaqI B (rs1079597). Linkage disequilibrium between these SNPs is high and significant in all 11 populations included in the HapMap project phase 3. The MboI and TaqI D loci are located between SNPs rs2587548 and rs2734836 investigated in the HapMap project. Linkage disequilibrium between SNP rs2587548 and the TaqI A locus is low in the Utah population of European ancestry and high but nonsignificant in the Chinese and Japanese populations (HapMap project phase 2), which is in agreement with our results.
Table 2.
Pairwise linkage disequilibrium values for the studied populations
| Population | TaqIB/BclI - MboI/TaqID | TaqIB/BclI - TaqIA | MboI/TaqID - TaqIA |
| Adygeis | 1.000 * | 1.000 * | 0.458 * |
| Byelorussians 1 | 1.000 * | 1.000 * | 0.870 * |
| Byelorussians 2 | 1.000 * | 1.000 * | 0.824 * |
| Byelorussians 3 | 1.000 * | 0.953 * | 0.835 * |
| Russians 1 | 1.000 * | 1.000 * | 0.956 * |
| Russians 2 | 1.000 * | 1.000 * | 1.000 * |
| Russians 3 | 1.000 * | 1.000 * | 0.855 * |
| Russians 4 | 1.000 * | 1.000 * | 0.507 * |
| Russians 5 | 1.000 * | 1.000 * | 0.774 * |
| Russians 6 | 1.000 * | 1.000 * | 0.919 * |
| Veps | 1.000 * | 1.000 * | 1.000 * |
| Komi 1 | 1.000 * | 1.000 * | 0.930 * |
| Komi 2 | 1.000 * | 1.000 * | 0.833 * |
| Khants | 1.000 * | 1.000 * | 1.000 * |
| Nenets | 1.000 * | 1.000 * | 0.498 P = 0.155 |
| Yakuts | 0.061 P = 0.827 |
1.000 * | 0.020 P = 0.927 |
| Kalmyks | 1.000 * | 0.978 * | 1.000 * |
The modulus of the standardized linkage disequilibrium coefficient (D') of Lewontin [57] is shown. An asterisk indicates significance level P < 0.05; greater P values are written in full.
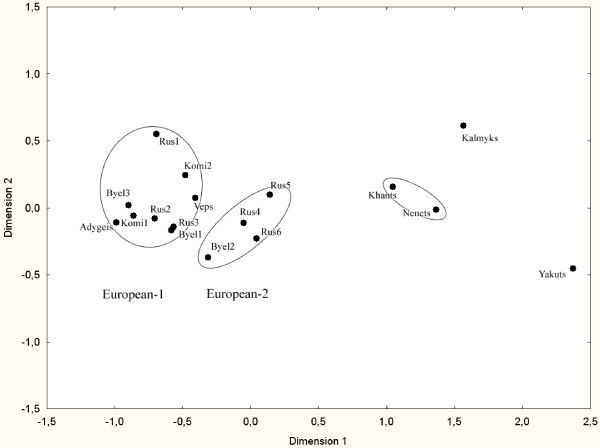
Taking into consideration perfect linkage in the TaqI B-BclI and MboI-TaqI D pairs, the BclI and MboI RFLPs were redundant for inter-population comparison, and we focused on the distribution of three-locus haplotypes TaqI B-TaqI D-TaqI A. Their frequencies are shown in Table 3. Only three haplotypes were common in the populations studied: B2-D1-A2, B2-D2-A2, and B1-D2-A1 (haplotypes termed according to [7]). The other haplotypes were extremely rare. Haplotype frequencies were used for calculation of FST-based genetic distances. The resulting distance matrix was visualized using multidimensional scaling (MDS) (Figure 2). Two large population groups can be distinguished: Asian (Khants, Nenets, Kalmyks, and Yakuts) and European (the other populations). According to UPGA cluster analysis, the European and Asian groups might be further subdivided into subclusters European-1 and European-2, Khants-Nenets group, Kalmyks, and Yakuts.
Table 3.
Frequencies of TaqI B-TaqI D-TaqI A haplotypes in the study populations
| Population | Haplotype | |||||||
| GTC | GCC | ACT | GCT | GTT | ACC | ATC | ATT | |
| B2-D1-A2 | B2-D2-A2 | B1-D2-A1 | B2-D2-A1 | B2-D1-A1 | B1-D2-A2 | B1-D1-A2 | B1-D1-A1 | |
| Adygeis 1 (Shapsugs) | 0.597 | 0.250 | 0.117 | 0.036 | ||||
| Byelorussians 1 | 0.543 | 0.314 | 0.129 | 0.007 | 0.007 | |||
| Byelorussians 2 | 0.507 | 0.373 | 0.113 | 0.007 | ||||
| Byelorussians 3 | 0.582 | 0.247 | 0.147 | 0.018 | 0.006 | |||
| Russians 1 | 0.529 | 0.209 | 0.252 | 0.002 | 0.008 | |||
| Russians 2 | 0.560 | 0.286 | 0.145 | 0.009 | ||||
| Russians 3 | 0.536 | 0.311 | 0.131 | 0.005 | 0.018 | |||
| Russians 4 | 0.442 | 0.368 | 0.158 | 0.032 | ||||
| Russians 5 | 0.401 | 0.366 | 0.211 | 0.021 | ||||
| Russians 6 | 0.442 | 0.401 | 0.153 | 0.003 | ||||
| Veps | 0.500 | 0.304 | 0.180 | 0.015 | ||||
| Komi 1 (Zyrian) | 0.585 | 0.263 | 0.138 | 0.009 | 0.004 | |||
| Komi 2 (Zyrian) | 0.505 | 0.271 | 0.206 | 0.005 | 0.014 | |||
| Khants 1 | 0.250 | 0.492 | 0.234 | |||||
| Nenets | 0.222 | 0.476 | 0.262 | 0.040 | ||||
| Yakuts 1 | 0.047 | 0.684 | 0.197 | 0.050 | 0.005 | 0.016 | ||
| Kalmyks | 0.139 | 0.500 | 0.341 | 0.005 | 0.014 | |||
Figure 2.
A two-dimensional scaling plot of FST-based genetic distances between the study populations, Russia and Belarus. Distances were inferred from haplotype frequencies. Population groups defined by UPGA cluster analysis are indicated by ellipses.
In the Asian cluster genetic distances were significant between Yakuts and Kalmyks (P < 0.00001), Khants and Yakuts (P < 0.00001), Khants and Kalmyks (P < 0.04661), Nenets and Yakuts (P < 0.00099), but not between Nenets and Kalmyks (P < 0.07438). The distance between Khants and Nenets was small and non-significant (P < 0.52033).
All populations of the European cluster had significant genetic distances from the populations of the Asian cluster. The European-1 and European-2 clusters were quite homogeneous; most distances were non-significant within the former, and all were non-significant within the latter. Only Russians 1 occupied distinct position within the European-1 cluster. In contrast, most pairwise genetic distances between the two subclusters were significant. Exceptions included Veps, which had no significant distances from the populations of the European-2 cluster, and Byelorussians 2, which, had a small number of significant distances from the populations of the European-1 cluster.
The matrix of haplotype-based genetic distances was also compared with matrices of great circle geographical distances (data not shown) to assess the possible effect of isolation by distance. The matrices including all 17 populations were highly correlated according to the Mantel test (r = 0.749, P-value = 0.0001). However, correlation of geographical and genetic distances was not observed among the populations of the European cluster, i.e., after exclusion of Khants, Nenets, Kalmyks (which have migrated from East Asia in historical times), and Yakuts (r = 0.121, P-value = 0.2999). The use of more realistic distance measurements around geographical barriers, such as the Azov Sea, the Caucasus, and some parts of the Ural Mountains had little effect on results of the test (the correlation coefficients for 17 and 13 populations were 0.754 and 0.117, respectively; P-values were 0.0001 and 0.3109, respectively). Thus, isolation by distance is not a likely cause of the genetic variation observed in the East European Plain.
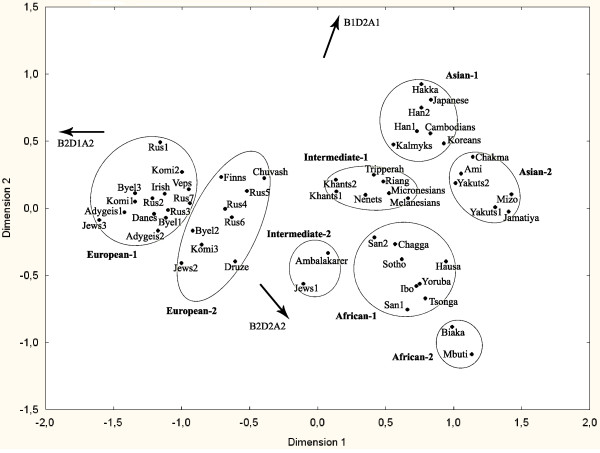
We also compared our results with those of previous authors to examine population relationships in greater detail. To do this, we analyzed haplotype frequencies in the populations studied and in 38 populations from various continents (data from [7,9], and the ALFRED database) using MDS of FST-based genetic distances (Figure 3 and Additional file 1). A good fit between the two-dimensional plot and the source data was obtained, demonstrated by the low stress value (0.062). Cluster analysis by the UPGA algorithm was performed using the same distance matrix and enabled us to define four large population clusters with two subclusters each, i.e., eight population groups in total. All clustering methods that we used (complete linkage, unweighted and weighted pair-group average, unweighted and weighted pair-group centroid, and Ward's method) revealed identical clusters at the level of eight groups. However at 'higher' and 'lower' clustering levels, some methods gave different results. To identify haplotypes responsible for the observed intercluster differences, haplotype frequencies for various clusters were compared using the Kolmogorov-Smirnov test (Table 4).
Figure 3.
A two-dimensional scaling plot of FST-based genetic distances between the study and the reference populations. Gradients of haplotype frequencies for the three most common haplotypes are indicated by arrows. Population groups identified by cluster analysis are highlighted by ellipses.
Table 4.
Comparison of some population groups according to the Kolmogorov-Smirnov test of haplotype frequencies
| B2-D1-A2 | B2-D2-A2 | B1-D2-A1 | |
| European-1/European-2 | 0.0010 | < 0.0001 | 0.4774 |
| European-2/Intermediate-1 | 0.0002 | 0.0002 | 0.0075 |
| Asian-1/Asian-2 | 0.0385 | 0.0012 | 0.0082 |
| Intermediate-1/Asian-1 | 0.0082 | 0.9441 | 0.0006 |
| Intermediate-1/Asian-2 | 0.0012 | 0.0082 | 0.3328 |
P-values are shown; those highlighted in bold indicate that the frequency distributions of the two samples are not significantly different (P > 0.05).
The following populations constituted the European-1 subcluster: Indo-European-speaking Irish, Danes, Byelorussians 1 and 3, Russians 1, 2, 3, and 7; North-Caucasian-speaking Adygeis 1 and 2; Jews 3 (Ashkenazi); Uralic-speaking Veps, Komi 1 and 2. The populations of this cluster had the following haplotype frequencies: 0.50-0.65 for the "European" haplotype B2-D1-A2, 0.21-0.31 for the "worldwide" haplotype B2-D2-A2, 0.08-0.25 for the "East Asian" haplotype B1-D2-A1. Some populations of the East European Plain (Indo-European-speaking Byelorussians 2, and Russians 4, 5, and 6; Uralic-speaking Finns and Komi 3; Altaic-speaking Chuvash) fell into the European-2 subcluster, which is characterized by the following haplotype frequencies: B2-D1-A2, 0.37-0.52; B2-D2-A2, 0.27-0.45; and B1-D2-A1, 0.04-0.25. Two European subclusters can be differentiated on the basis of their B2-D2-A2 and B2-D1-A2 frequencies (P-values according to the Kolmogorov-Smirnov test < 0.0001 and 0.001, respectively). This is also evident from Figure 3: on the MDS plot, both European subclusters occupy almost identical positions in the B1-D2-A1 frequency gradient (Figure 3, upward arrow) but have different positions in the B2-D1-A2 and B2-D2-A2 frequency gradients (Figure 3, arrows).
A low but highly significant level of population differentiation was observed between the European-1 and -2 subclusters (FCT 0.018, P < 0.00001, Table 5); 59% of genetic distances between populations of the two subclusters were significant, but only 14% and 22% of the intracluster distances were significant. Other close pairs of subclusters such as Intermediate-1 and -2 demonstrated higher levels of differentiation (FCT 0.027-0.045, Table 5). About 60% of intercluster genetic distances were significant in all instances.
Table 5.
Indexes of population structure
| Standardized variance | No. of samples | FST |
| Among populations: | ||
| Global analysis | 55 | 0.114**** |
| European | 23 | 0.013**** |
| European-1 | 14 | 0.003* |
| European-2 | 9 | 0.006* |
| Intermediate | 9 | 0.009* |
| African | 10 | 0.019*** |
| Asian | 13 | 0.026**** |
| FCT | ||
| Among groups: | ||
| European-Intermediate-Asian-African | 0.143**** | |
| European-Intermediate | 0.099**** | |
| European-African | 0.179**** | |
| European-Asian | 0.199**** | |
| Intermediate-African | 0.042**** | |
| Intermediate-Asian | 0.028**** | |
| African-Asian | 0.059**** | |
| European-1-European-2 | 0.018**** | |
| European-2-Intermediate-1-Intermediate-2 | 0.058**** | |
| European-2-Intermediate-1 | 0.065** | |
| European-2-Intermediate-2 | 0.040* | |
| European-2-Asian-1 | 0.143*** | |
| Intermediate-1-Intermediate-2 | 0.027* | |
| Intermediate-1-African-1 | 0.040** | |
| Intermediate-1-Asian-1-Asian-2 | 0.036**** | |
| Intermediate-1-Asian-1 | 0.025** | |
| Intermediate-1-Asian-2 | 0.041** | |
| African-1-African-2 | 0.031* | |
| Asian-1-Asian-2 | 0.045** | |
* P < 0.05
** P < 0.001
*** P < 0.0001
**** P < 0.00001
Ugric-speaking Khants 1 and 2 and Samoyedic-speaking Nenets were distant from the other Uralic populations (Finns, Veps, Komi 1, 2, and 3) and grouped into the Intermediate-1 subcluster with Sino-Tibetan-speaking Riang and Tripperah from India, with Melanesians and Micronesians (the results for Melanesians and Micronesians should be interpreted with caution because of a low sample size; see Additional file 1). In Khants and Nenets, the frequencies of the "European" haplotype B2-D1-A2 (0.22-0.25) and the "East Asian" haplotype B1-D2-A1 (0.23-0.26) were intermediate between those of the European-1 and Asian-1 subclusters (Figure 3). It is interesting that the frequencies of the "worldwide" haplotype B2-D2-A2 in Khants (0.48-0.49) and Nenets (0.48) fell within the range typical of the Asian-1 subcluster, 0.44-0.53 (P-value = 0.9441 according to the Kolmogorov-Smirnov test).
The Asian-1 subcluster was formed by populations of East Asian descent (Altaic-speaking Kalmyks; Koreans and Japanese; Sino-Tibetan-speaking Hakka, Han 1 and 2; and Austro-Asiatic-speaking Cambodians). They had the following frequency pattern: B2-D1-A2, 0.04-0.14; B2-D2-A2, 0.44-0.53; and B1-D2-A1, 0.34-0.46.
The global FST value, 0.11413 (Table 5), falls within the range typical of autosomal markers (0.09-0.14, [16]). This value was estimated using haplotype frequencies without taking into account the extent of molecular differences between haplotypes. Calculation of FST based on numbers of pairwise differences between haplotypes gave a nearly identical value, 0.11383. Thus, the differentiation of populations may be explained by drift only, without any significant influence of mutation.
Discussion
The East European (Russian) Plain is a region in which peoples of the Indo-European and Uralic language families have come into contact over an extended period. Uralic-speaking peoples have the longest validated archaeological record in this region [17]. The most recent large-scale migration to this region involved the movement of Slavs (the Indo-European language family) to the east and northeast of their presumed homeland in Central Europe about 500 AD [18,19]. Slavs were not the first Indo-European-speaking people who arrived in the Russian Plain: in the first millennium BC, Baltic-speaking tribes occupied a large part of the East European Plain [17]. They were later displaced by Slavic tribes. According to the widely accepted hybridization theory of the origin of Eastern Slavs [20], Slavic populations arriving in the East European Plain were mixed with indigenous Uralic- and, probably, Baltic-speaking people.
In our study, all populations of the East European Plain (excluding the Kalmyks, which are of East Asian origin) fell into a single large cluster termed European. Many populations within this cluster are indistinguishable with our genetic marker, i.e., genetic distances between them were not significant, which is in agreement with the low FST value for the European cluster (0.013). However, some populations were characterized by a large percentage of significant genetic distances from the other populations of the cluster. Most such populations fell into the so-called European-2 subcluster defined by cluster analysis; the 'core' subcluster was termed European-1, and 59% of genetic distances between populations of the two subclusters were significant. European-1 and European-2 subclusters (Figure 3) are differentiated according to the B2-D2-A2 frequency, but not according to the B1-D2-A1 frequency, which might reflect the degree of Asian admixture. Natural selection probably was not responsible for separation of the two European subclusters as there is no difference in allele frequencies at the TaqI A locus (Table 4), which is considered the most likely candidate for selection in the whole DRD2 region [2-6].
The European-2 subcluster includes two Middle Eastern populations (Jews 2 from Yemen and Druze from Israel), two Uralic-speaking populations (Finns and Komi 3), also four Slavic-speaking populations (Byelorussians 2 and Russians 4, 5, and 6), and the Altaic-speaking Chuvash. All these linguistically and geographically distant populations are differentiated to some extent from the core of the European cluster, the European-1 subcluster, because of a relatively high B2-D2-A2 frequency.
The B2-D1-A2 and B1-D2-A1 haplotypes apparently have centers of dispersal in Europe/West Asia and East Asia, respectively [7]. The B2-D2-A2 haplotype may also have a center of dispersal, the most probable location of which is in Africa. B2-D2-A2 was among the first haplotypes that evolved from the ancestral haplotype B2-D2-A1 in Africa [7] and still is the most abundant haplotype in all African populations (Additional file 1). Therefore, the first settlers of Eurasia that migrated to Arabia and Levant may mostly have carried the B2-D2-A2 haplotype and a small proportion of other haplotypes that were either subsequently eliminated or amplified by genetic drift and/or natural selection in various parts of the world.
B2-D2-A2 is the predominant haplotype in contemporary populations of Middle Eastern origin (Jews 1 from Ethiopia, Jews 2 from Yemen, and Druze from Israel). Populations of Levant began to disperse into Europe about 50,000 YBP [21,22]. People of that diaspora might have carried B2-D2-A2 as a prevalent haplotype. However, another haplotype, B2-D1-A2, is the predominant haplotype in contemporary populations of Europe, for example, in linguistically and geographically distant populations such as the Irish, Danes, Russians, and Adygeis. Amplification of this haplotype might have taken place during the initial migration to Europe or might be associated with confinement in refugia of the last glacial maximum and reexpansion. According to one of several hypotheses based on archaeological evidence (reviewed by Zvelebil [23]), Uralic-speaking groups descended from Europeans who had been confined in the East European refugium in Ukraine and Southern Russia [24-29]. It is possible that the people of the East European refugium retained a high frequency of the B2-D2-A2 haplotype, in contrast with the other European populations that have spread from other refugia.
Thus, on a global scale, the B2-D2-A2 frequency possibly reflects genetic features of the first Eurasian settlers, e.g. in Jews 2 from Yemen and Druze from Israel, and, on a local scale, genetic features of Uralic-speaking populations, e.g., in Finns and Komi 3, Khants and Nenets. Therefore, the other members of the European-2 subcluster, Byelorussians 2, Russians 4, 5, and 6, and Chuvash, probably have a certain level of Uralic admixture. Moreover, according to other genetic and historical data, these populations have Uralic substratum.
Studies on mtDNA [30-34] and Y-chromosome haplogroups [33,35,36], autosomal VNTR diversity [37,38], and autosomal haplotypes [39] consistently show a degree of Uralic admixture in populations of northern Russians. This admixture is manifested by a 'Uralic-specific' marker, the U4 mtDNA haplogroup [40-45], or by Asian-specific markers which Uralic populations acquired during earlier admixture with East Asians: Asian mtDNA haplogroups [40,42,45], N3 and N2 Y-chromosome haplogroups [46,47]. For example, the Mezen' population investigated by Balanovsky et al. [36] has high N3 frequency, resembling some Uralic populations.
Russians 5 (Oshevensk) were most closely associated with Finns and Chuvash according to the MDS results (Figure 3). In the study of Verbenko et al. [38] on polymorphic tandem repeats at the D1S80 locus, the same Oshevensk sample (as well as another northern Russian sample) clustered together with Uralic-speaking Mari, Komi, and Udmurts, whereas other Russian populations clustered with Indo-Europeans and Adygeis. Analysis of other repeat loci, 3' ApoB, DMPK, DRPLA, and SCA1, also demonstrated remoteness of some northern Russian populations (including Russians 5, Oshevensk) from the core of the European cluster [38]. Similar results were obtained using haplotypes at the TP53 locus [39]: the Oshevensk population tended to form a cluster with Uralic-speaking Mordvins and with Altaic-speaking Kalmyks and Buryats, but not with Russians from Smolensk (Russians 2) or Byelorussians from Pinsk (Byelorussians 1).
According to archaeological data, the Arkhangelsk region (including Mezen' and Oshevensk) was populated by Uralic tribes in the Middle Ages ([48]; see Figure 4). Russian colonization of this region began relatively recently (after the 12th century AD) [48]. Thus, a very high level of Uralic admixture in Mezen' and Oshevensk is not surprising.
Figure 4.
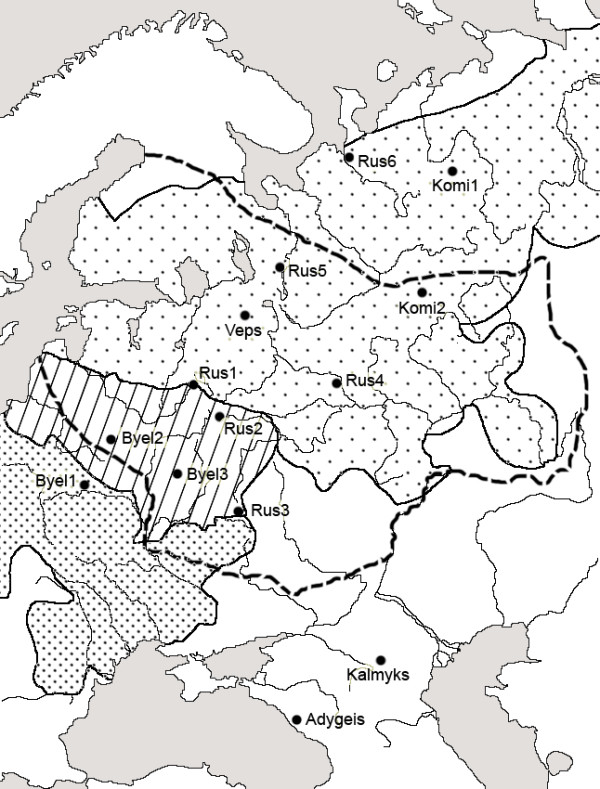
A map of archaeological cultures on the East European Plain showing the locations of the study populations. The dashed line indicates the region that was presumably occupied by Uralic-speaking peoples in the Early Neolithic era, the 5th millennium BC (according to [17]). Solid lines indicate three regions occupied by different ethnic groups in the 1st-6th centuries AD: Slavic- and Germanic-speaking tribes, densely dotted area; Baltic-speaking tribes, hatched area; Uralic-speaking tribes, sparsely dotted area [48,61].
The Uralic genetic substratum is appreciable not only in the Northeast of the East European Plain but also in its northwestern part, for example, in the Pskov [33] and Novgorod regions [38], in Latvians and Lithuanians [46,49,50]. Baltic-speaking peoples, now represented by the Latvians and Lithuanians, came into contact with Uralic groups before the Slavs did (Figure 4; [18]). That Byelorussians 2 (Mjadel') fell into the European-2 subcluster may also reflect a general tendency in the northwestern region. Mjadel' is located in the northwestern part of Belarus near the contemporary Lithuanian border. The Russians 4 (Puchezh) population is distant from the northeastern and northwestern groups, but also belongs to the European-2 subcluster (Figure 3). Uralic admixture in this population may be explained by the presence of Uralic-speaking tribes in the region of Puchezh in historical times ([51]; see Figure 4).
Russians 1 (Andreapol') and Uralic-speaking Veps are close to Uralic-speaking Komi 2 (Obyachevo) on the MDS plot (Figure 3). All these populations are located within the region occupied by Uralic peoples in the Middle Ages (Figure 4), but belong to the European-1 cluster and do not have high B2-D2-A2 frequencies typical of the European-2 cluster. However, they are shifted from the core of the European cluster because of a relatively high proportion of the "East Asian" haplotype B1-D2-A1. In fact, the Veps population has significant genetic distance only from Druze, but not from the other populations of the European-2 cluster, and Komi 2 only from Jews 2, Druze, Russians 6, and Komi 3. The Andreapol' sample had the highest B1-D2-A1 frequency of all European populations (Additional file 1), and eight of 13 genetic distances between this sample and the other populations of the European-1 subcluster are significant.
Komi populations demonstrate remarkable heterogeneity according to various marker systems. For example, Komi-Permyaks and Komi-Zyrians have rather different mtDNA haplogroup frequencies but both have a relatively high U4 frequency [40]. In our study, one of the Komi-Zyrian populations (Komi 1, Izhma) belonged to the core of the European-1 cluster. It is interesting that the craniological results of Moiseyev [52] also place Komi-Zyrians at the core of the European cluster and distant from Uralic and Asian groups. According to three-site haplotype frequencies at the TP53 locus and VNTR frequencies at the D1S80 and 3' ApoB loci, the Komi 1 and 2 populations are distant from Uralic-speaking Finns, Mordvins and Khants, East Asian groups, and Slavic groups [53]. Moreover, the Komi 2 (Obyachevo) population is distant from Komi 1 and closer to Slavic groups than Komi 1 [53]. Thus, the position of Komi in genetic gradients remains uncertain because of substantial divergence of population samples and contradictory results, which may reflect a complex history of this group or natural selection.
B2-D2-A2 haplotype frequencies in Khants (0.48-0.49) and Nenets (0.48) fell within the range typical of the Asian-1 subcluster, 0.44-0.53. Therefore, the high frequency of the B2-D2-A2 haplotype in Uralic-speaking Khants and Nenets cannot be explained only by admixture of Europeans with typical East Asians; a combination of admixture and other processes such as gene drift or selection is a more likely explanation. The Khants and Nenets may have received the B2-D2-A2 haplotype both from the East European refugium and from East Asia, e.g., from Siberian populations of East Asian origin. One of such Siberian populations, the Yakuts, has a very high frequency of haplotype B2-D2-A2, 0.6-0.7. However, DRD2 haplotype frequencies in other Siberian populations of East Asian origin are unknown, and it is not possible to draw definitive conclusions about B2-D2-A2 frequencies in Siberia based on one ethnic group alone. Moreover, because Yakuts 1 exhibited low linkage disequilibrium between RFLPs, haplotype frequencies for this sample should be interpreted with caution. Yakuts 1 and 2 apparently do not belong to the cluster of typical East Asians, East Asian-1 (Figure 3), although they are clearly of East Asian origin. As suggested by archaeological and linguistic evidence, the Yakuts probably migrated north from their original area of settlement near Lake Baykal because of the Mongol expansion from the 13th to 15th century AD [54]. Y-chromosome results reveal a very strong bottleneck in the Yakut population, which probably preceded their recent expansion [46,54]. This bottleneck effect may be responsible for the aberrant haplotype frequencies for Yakuts observed in our study.
Conclusion
Populations in the northwestern (Byelorussians 2 from Mjadel'), northern (Russians 5 from Mezen' and 6 from Oshevensk; Komi 3), and eastern parts (Russians 4 from Puchezh and Chuvash) of the East European Plain have relatively high frequencies of haplotype B2-D2-A2, which may reflect admixture with Uralic-speaking populations. Uralic genetic substratum in these regions, which were inhabited by Uralic-speaking tribes as late as the Early Middle Ages, was also shown by studies in which other genetic markers were used (mtDNA, Y-chromosome, and autosomal). Thus, the analysis of DRD2 haplotypes supports results on Slavic-Uralic admixture obtained using other markers, mainly neutral and sex-specific markers.
Methods
Populations
The linguistic affiliations and geographical locations of the studied populations are shown in Table 1 and Figure 5. These populations have been described previously (see footnotes to Table 1), except for Andreapol' (Russians 1), Mezen' (Russians 6), Veps, and Nenets. The Andreapol' sample from the Tver region, the most western of the Russian populations involved in our study, consisted of individuals born in Andreapol' (a small town), Bologovo village, and other nearby villages. The Mezen' sample from the Arkhangelsk region, the most northern of the Russian populations sampled, consisted of individuals born in the small towns, Mezen' and Kamenka, in Dorogorskoe village, and in other nearby villages. The Nenets are Samoyedic-speaking tribes, and their economy is based on reindeer herding. In 2002, the population of Nenets was about 41,000. The Nenets sample, which belongs to the Tundra group, was collected in Yar-Sale and Panaevka villages in the Yamalo-Nenets autonomous district, Tyumen' region. The Veps are Finnic-speaking and reside in small settlements scattered among Russians of the Vologda region, the Leningrad region, and Karelia. In 2002, the population of Veps was about 8,000. Samples were collected from Veps in several villages (Pyazhozero, Pyazhelka, Koloshma, and others) in the Babaevo district of the Vologda region.
Figure 5.
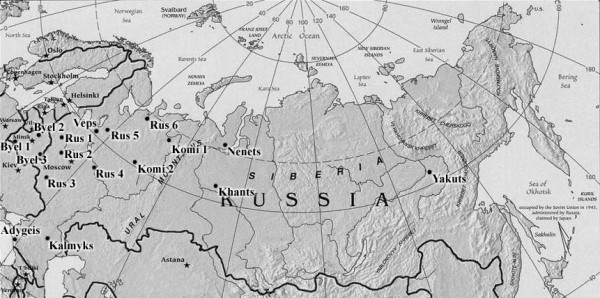
Geographical location of the study populations on the map of Russia and the neighboring countries.
DNA isolation and typing
Blood samples (8 ml) were obtained by venipuncture and collected into EDTA-coated containers. Informed consent was obtained from each individual. The research protocols and forms of informed consent have been approved by the Ethic Commission of the Medico-Genetic Scientific Centre of the Russian Academy of Medical Sciences (an approval was signed by the Head of the Ethic Commission, PhD, professor L.F. Kurilo). All individuals belonged to the native ethnic group of the region, i.e., their lineage in the region extended for at least two previous generations, and were unrelated to each other. DNA was isolated from leucocytes by proteinase K treatment and extraction with phenol-chloroform [55]. Each DNA sample was subjected to three PCR analyses: amplification of a 459 bp fragment for TaqI B and BclI RFLP analysis, amplification of a 300 bp fragment for MboI and TaqI D RFLP analysis, and amplification of a 237 bp fragment for TaqI A RFLP analysis (primer sequences and original PCR protocols were obtained from the website of K. Kidd: http://info.med.yale.edu/genetics/kkidd/Protocol_TOC_new2002.html). The locations of these polymorphisms are shown on the gene map (Figure 1). All endonuclease restriction reactions were carried out overnight. Samples containing unrestricted fragments were tested at least twice. Restriction products were separated by electrophoresis using a 2.5% agarose gel.
Statistical analyses
The general strategy of statistical analysis was similar to that used in the work of Poloni et al. [56]. Allele frequencies, correspondence to Hardy-Weinberg equilibrium, and significance of linkage disequilibrium (P-values) were evaluated using Arlequin version 2.0 software http://cmpg.unibe.ch/software/arlequin. Linkage disequilibrium values (D') between polymorphic loci were calculated as suggested by Lewontin [57]. Frequencies of haplotypes were estimated from RFLP genotype data using the expectation-maximization algorithm of Excoffier and Slatkin [58] implemented in Arlequin 2.0. Genetic affinities among populations were evaluated using coancestry coefficients, or linearized pairwise FST values [59] calculated on the basis of allele or haplotype frequencies. Significance of genetic distances was tested using permutations [60].
Correlation of geographical and genetic distances was assessed using the Mantel test and XLSTAT version 2008.6.04 software (Addinsoft). Great circle geographical distances were calculated using the haversine formula; latitudes and longitudes were determined using Google Earth software. Multidimensional scaling (MDS) and cluster analysis with the unweighted pair-group average (UPGA) method were performed using STATISTICA version 6.0 http://www.statsoft.com. Statistical comparison of haplotype frequencies in population groups was performed using the Kolmogorov-Smirnov test implemented in XLSTAT. Differentiation between population groups defined by MDS and cluster analysis was estimated using the analysis-of-molecular-variance approach, AMOVA [60], implemented in Arlequin 2.0. Conventional FST distances between haplotypes were used, i.e., all haplotypes were considered equidistant (a conservative scenario of pure drift). Significance of the genetic-structure indexes obtained with the AMOVA method was tested using a permutational procedure (1 × 106 permutations).
Authors' contributions
OVF carried out the polymorphism typing, performed the statistical analysis and drafted the manuscript. OIL participated in the polymorphism typing. AVK participated in the study design and helped with the statistical analysis and manuscript drafting. LAT, VAS, and AIM carried out sample collection and participated in the study design. SAL conceived of the study, participated in its coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Frequencies of TaqI B-TaqI D-TaqI A haplotypes in the study populations and in reference populations. This is an extended variant of Table 3 including haplotype frequencies for all populations represented in Figure 3.
Acknowledgments
Acknowledgements
The study was supported by the Russian Ministry of Science and Education program "Federal Support of Leading Scientific Schools" (grant No 3911.2008-04), by the Russian Academy of Sciences programs "Molecular and Cell Biology" and "Basic Science for the Advantage of Medicine", and by the Russian Foundation for Basic Research grant No 07-04-00027-a.
Contributor Information
Olga V Flegontova, Email: oflegontova@mail.ru.
Andrey V Khrunin, Email: khrunin@img.ras.ru.
Olga I Lylova, Email: olylova@mail.ru.
Larisa A Tarskaia, Email: tarskaia@yahoo.com.
Victor A Spitsyn, Email: ecolab@med-gen.ru.
Alexey I Mikulich, Email: mai@rambler.ru.
Svetlana A Limborska, Email: limbor@img.ras.ru.
References
- Kandel ER, Schwartz JH, Jessell TM. Principles of Neural Science. 4. New York: McGraw-Hill; 2000. [Google Scholar]
- McD Young R, Lawford BR, Nutting A, Noble EP. Advances in molecular genetics and the prevention and treatment of substance misuse: Implications of association studies of the A1 allele of the D2 dopamine receptor gene. Addict Behav. 2004;29:1275–1294. doi: 10.1016/j.addbeh.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Clark TG, Johnstone EC, Murphy MFG, Walton RT. The genetic basis for smoking behavior: A systematic review and meta-analysis. Nicotine Tob Res. 2004;6:583–597. doi: 10.1080/14622200410001734030. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Matheson IJ, Flint J. Association of the DRD2 gene Taq1A polymorphism and alcoholism: a meta-analysis of case-control studies and evidence of publication bias. Mol Psychiatry. 2007;12:454–461. doi: 10.1038/sj.mp.4001938. [DOI] [PubMed] [Google Scholar]
- Allen NC, Bagade S, McQueen MB, Ioannidis JPA, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the szGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: a novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat. 2004;23:540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Castiglione CM, Pakstis AJ, Speed WS, Bonne-Tamir B, Lu R-B, Goldman D, Lee C, Nam YS, Grandy DK, Jenkins T, Kidd JR. A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet. 1998;103:211–227. doi: 10.1007/s004390050809. [DOI] [PubMed] [Google Scholar]
- Chakrabarti CS, Roy M, Sengupta NK, Lalthantluanga R, Majumder PP. Genetic relationships among some tribal groups inhabiting the north-eastern, eastern and sub-Himalayan regions of India. Ann Hum Genet. 2002;66:361–368. doi: 10.1046/j.1469-1809.2002.00132.x. [DOI] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, Roychoudhury S, Majumder PP. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanathan H, Deepa E, Cordaux R, Stoneking M, Usha Rani MV, Majumder PP. Genetic structure and affinities among tribal populations of southern India: a study of 24 autosomal DNA markers. Ann Hum Genet. 2004;68:128–138. doi: 10.1046/j.1529-8817.2003.00083.x. [DOI] [PubMed] [Google Scholar]
- Bhaskar LV, Thangaraj K, Mulligan CJ, Rao AP, Pardhasaradhi G, Kumar KP, Shah AM, Sabeera B, Reddy AG, Singh L, Rao VR. Allelic variation and haplotype structure of the dopamine receptor gene DRD2 in nine Indian populations. Genet Test. 2008;12:153–160. doi: 10.1089/gte.2007.0073. [DOI] [PubMed] [Google Scholar]
- Castiglione CM, Deinard AS, Speed WC, Sirugo G, Rosenbaum HC, Zhang Y, Grandy DK, Grigorenko EL, Bonne-Tamir B, Pakstis AJ, Kidd JR, Kidd KK. Evolution of haplotypes at the DRD2 locus. Am J Hum Genet. 1995;57:1445–1456. [PMC free article] [PubMed] [Google Scholar]
- Kidd KK, Pakstis AJ, Castiglione CM, Kidd JR, Speed WC, Goldman D, Knowler WC, Lu R-B, Bonne-Tamir B. DRD2 haplotypes containing the TaqI A1 allele: implications for alcoholism research. Alcohol Clin Exp Res. 1996;20:697–705. doi: 10.1111/j.1530-0277.1996.tb01674.x. [DOI] [PubMed] [Google Scholar]
- Iyengar S, Seaman M, Deinard AS, Rosenbaum HC, Sirugo G, Castiglione CM, Kidd JR, Kidd KK. Analyses of cross species polymerase chain reaction products to infer the ancestral state of human polymorphisms. DNA Seq. 1998;8:317–327. doi: 10.3109/10425179809034076. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Relethford JH. Genetics and modern human origins. Evol Anthropol. 1995;4:53–63. doi: 10.1002/evan.1360040207. [DOI] [Google Scholar]
- Makkay J. An archeologist speculates on the origin of the Finno-Ugrians. Mankind Quaterly. 2003;43:235–271. [Google Scholar]
- Mallory JP. In search of the Indo-Europeans Language, Archaeology and Myth. London: Thames and Hudson; 1989. [Google Scholar]
- Sedov VV. Slavyane v rannem srednevekovye (The Slavs in the Early Middle Ages) Moscow: Archaeological Fund; 1995. (in Russian) [Google Scholar]
- Alekseeva TI. Etnogenez vostochnyh slavyan soglasno antropologicheskim dannym (Ethnogenesis of Eastern Slavs according to anthropological data) Moscow: Moscow State University; 1973. (in Russian) [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellars P. A new radiocarbon revolution and the dispersal of modern humans in Eurasia. Nature. 2006;439:931–935. doi: 10.1038/nature04521. [DOI] [PubMed] [Google Scholar]
- Zvelebil M. Revisiting Indreko's culture historical model: "Origin and area of settlement of the Finno-Ugrian peoples". TRAMES: A Journal of the Humanities and Social Sciences. 2001;5:26–47. [Google Scholar]
- Nuñez MG. Old and new ideas about the origins of the Finns and Saami. In: Julku K, Wiik K, editor. The Roots of Peoples and Languages of Northern Eurasia I (Turku 305-161997) Turku: Societas Historiae Fenno-Ugricae; 1998. pp. 151–160. [Google Scholar]
- Dolukhanov P. 'Prehistoric revolutions' and languages in Europe. In: Künnap A, editor. The roots of peoples and languages of Northern Eurasia II and III. Tartu: Tartu University Press; 2000. pp. 71–84. [Google Scholar]
- Poikalainen V. Palaeolithic art from the Danube to Lake Baikal. Folklore. 2001;18/19:7–60. [Google Scholar]
- Tambets K, Tolk H-V, Kivisild T, Metspalu E, Parik J, Reidla M, Voevoda M, Damba L, Bermisheva M, Khusnutdinova E, Golubenko M, Stepanov V, Puzyrev V, Usanga E, Rudan P, Beckmann L, Villems R. Complex signals for population expansions in Europe and beyond. In: Bellwood P, Renfrew C, editor. Examining the farming/language dispersal hypothesis. Cambridge: McDonald Institute; 2002. [Google Scholar]
- Wiik K. Some ancient and modern linguistic processes in northern Europe. In: Renfrew C, McMahon A, Trask L, editor. Time depth in historical linguistics (Papers in the prehistory of languages) Cambridge: McDonald Institute for Archaeological Research; 2000. pp. 463–479. [Google Scholar]
- Wiik K. Who are the Finns? In: Suominen M, Arppe A, Airola A, Heinämäki O, Miestamo M, Määttä U, Niemi J, Pitkänen KK, Sinnemäki K, editor. A Man of Measure: Festschrift in Honour of Fred Karlsson on his 60th Birthday. Turku: The Linguistic Association of Finland; 2006. pp. 97–108. [Google Scholar]
- Orekhov V, Poltaraus A, Zhivotovsky LA, Spitsyn V, Ivanov P, Yankovsky N. Mitochondrial DNA sequence diversity in Russians. FEBS Lett. 1999;445:197–201. doi: 10.1016/S0014-5793(99)00115-5. [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA, Derenko MV. Mitochondrial DNA variability in Russians and Ukrainians: Implication to the origin of the Eastern Slavs. Ann Hum Genet. 2001;65:63–78. doi: 10.1046/j.1469-1809.2001.6510063.x. [DOI] [PubMed] [Google Scholar]
- Belyaeva O, Bermisheva M, Khrunin A, Slominsky P, Bebyakova N, Khusnutdinova E, Mikulich A, Limborska S. Mitochondrial DNA variation in Russian and Belorussian populations. Hum Biol. 2003;75:647–660. doi: 10.1353/hub.2003.0069. [DOI] [PubMed] [Google Scholar]
- Malyarchuk B, Derenko M, Grzybowski T, Lunkina A, Czarny J, Rychkov S, Morozova I, Denisova G, Miścicka-Śliwka D. Differentiation of mitochondrial DNA and Y chromosomes in Russian populations. Hum Biol. 2004;76:877–900. doi: 10.1353/hub.2005.0021. [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA, Perkova MA, Derenko MV. On the origin of mongoloid component in the mitochondrial gene pool of Slavs. Russ J Genet. 2008;44:344–349. [PubMed] [Google Scholar]
- Khar'kov VN, Stepanov VA, Borinskaya SA, Kozhekbaeva ZhM, Gusar VA, Grechanina EYa, Puzyrev VP, Khusnutdinova EK, Yankovsky NK. Gene pool structure of eastern Ukrainians as inferred from the Y-chromosome haplogroups. Russ J Genet. 2004;40:415–421. [PubMed] [Google Scholar]
- Balanovsky O, Rootsi S, Pshenichnov A, Kivisild T, Churnosov M, Evseeva I, Pocheshkhova E, Boldyreva M, Yankovsky N, Balanovska E, Villems R. Two sources of the Russian patrilineal heritage in their Eurasian context. Am J Hum Genet. 2008;82:236–250. doi: 10.1016/j.ajhg.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbenko DA, Knjazev AN, Mikulich AI, Khusnutdinova EK, Bebyakova NA, Limborska SA. Variability of the 3'APOB minisatellite locus in eastern Slavonic populations. Hum Hered. 2005;60:10–18. doi: 10.1159/000087338. [DOI] [PubMed] [Google Scholar]
- Verbenko DA, Slominsky PA, Spitsyn VA, Bebyakova NA, Khusnutdinova EK, Mikulich AI, Tarskaia LA, Sorensen MV, Ivanov VP, Bets LV, Limborska SA. Polymorphisms at locus D1S80 and other hypervariable regions in the analysis of Eastern European ethnic group relationships. Ann Hum Biol. 2006;33:570–584. doi: 10.1080/03014460601012077. [DOI] [PubMed] [Google Scholar]
- Khrunin AV, Tarskaia LA, Spitsyn VA, Lylova OI, Bebyakova NA, Mikulich AI, Limborska SA. p53 polymorphisms in Russia and Belarus: correlation of the 2-1-1 haplotype frequency with longitude. Mol Gen Genomics. 2005;272:666–672. doi: 10.1007/s00438-004-1091-8. [DOI] [PubMed] [Google Scholar]
- Bermisheva M, Tambets K, Villems R, Khusnutdinova E. Diversity of mitochondrial DNA haplogroups in ethnic populations of the Volga-Ural region. Mol Biol (Moscow) 2002;36:990–1001. [PubMed] [Google Scholar]
- Derbeneva OA, Starikovskaya EB, Wallace DC, Sukernik RI. Traces of early Eurasians in the Mansi of Northwest Siberia revealed by mitochondrial DNA analysis. Am J Hum Genet. 2002;70:1009–1014. doi: 10.1086/339524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbeneva OA, Starikovskaya EB, Volodko NV, Wallace DC, Sukernik RI. Mitochondrial DNA Variation in the Kets and Nganasans and its implications for the initial peopling of Northern Eurasia. Russ J Genet. 2002;38:1554–1560. doi: 10.1023/A:1021111530654. [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA. Differentiation of the mitochondrial subhaplogroup U4 in the populations of Eastern Europe, Ural and West Siberia: Implication for the genetic history of the Uralic populations. Russ J Genet. 2004;40:1549–1556. doi: 10.1023/B:RUGE.0000048671.32870.cb. [DOI] [PubMed] [Google Scholar]
- Tambets K. PhD thesis. Tartu University; 2004. Towards the understanding of post-glacial spread of human mitochondrial DNA haplogroups in Europe and beyond: a phylogeographic approach. [Google Scholar]
- Starikovskaya EB, Sukernik RI, Derbeneva OA, Volodko NV, Ruiz-Pesini E, Torroni A, Bromn MD, Lott MT, Hosseini SH, Huoponen K, Wallace DC. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann Hum Genet. 2005;69:67–87. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rootsi S, Zhivotovsky LA, Baldović M, Kayser M, Kutuev IA, Khusainova R, Bermisheva MA, Gubina M, Fedorova SA, Ilumäe A-M, Khusnutdinova EK, Voevoda MI, Osipova LP, Stoneking M, Lin AA, Ferak V, Parik J, Kivisild T, Underhill PA, Villems R. A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet. 2007;15:204–211. doi: 10.1038/sj.ejhg.5201748. [DOI] [PubMed] [Google Scholar]
- Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, Dorzhu C, Luzina F, Lee HK, Vanecek T, Villems R, Zakharov I. Phylogeographic analysis of mitochondrial DNA in Northern Asian populations. Am J Hum Genet. 2007;81:1025–1041. doi: 10.1086/522933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedov VV. Finno-ugry i balty v epohu srednevekovya (Finno-Ugrians and Balts in the Middle Ages) Moscow: Nauka; 1987. (in Russian) [Google Scholar]
- Kasperavićiūtė D, Kućinskas V, Stoneking M. Y chromosome and mitochondrial DNA variation in Lithuanians. Ann Hum Genet. 2004;68:438–452. doi: 10.1046/j.1529-8817.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- Pliss L, Tambets K, Loogväli E-L, Pronina N, Lazdins M, Krumina A, Baumanis V, Villems R. Mitochondrial DNA portrait of Latvians: towards the understanding of the genetic structure of Baltic-speaking populations. Ann Hum Genet. 2006;70:439–458. doi: 10.1111/j.1469-1809.2005.00238.x. [DOI] [PubMed] [Google Scholar]
- Leont'ev AE. Arheologiya Meri. K predystorii Severo-Vostochnoj Rusi (The Archaeology of the Merya. The Early History of North-Eastern Russia) In: Afanas'ev GE, Daim F, Kidd D, editor. Russian Monographs in Migration-period and Medieval Archaeology. Vol. 4. Moscow: Institute of Archaeology, Russian Academy of Sciences; 1996. (in Russian) [Google Scholar]
- Moiseyev V. Origins of Uralic-speaking populations: craniological evidence. HOMO. 2002;52:240–253. doi: 10.1078/0018-442X-00032. [DOI] [PubMed] [Google Scholar]
- Khrunin AV, Verbenko DA, Nikitina K, Limborska SA. Regional differences in the genetic variability of Finno-Ugric speaking Komi populations. Am J Hum Biol. 2007;19:741–750. doi: 10.1002/ajhb.20620. [DOI] [PubMed] [Google Scholar]
- Pakendorf B, Novgorodov IN, Osakovskij VL, Danilova AP, Protod'jakonov AP, Stoneking M. Investigating the effects of prehistoric migrations in Siberia: genetic variation and the origins of Yakuts. Hum Genet. 2006;120:334–353. doi: 10.1007/s00439-006-0213-2. [DOI] [PubMed] [Google Scholar]
- Milligan BG. Total DNA isolation. In: Hoelzel AR, editor. Molecular genetic analysis of populations. London: Oxford University Press; 1998. pp. 29–60. [Google Scholar]
- Poloni ES, Semino O, Passarino G, Santachiara-Benerecetti AS, Dupanloup I, Langaney A, Excoffier L. Human genetic affinities for Y-chromosome P49a,f/TaqI haplotypes show strong correspondence with linguistics. Am J Hum Genet. 1997;61:1015–1035. doi: 10.1086/301602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabarov VF, Zhukovsky VYe, Krayuhin AN, et al. Natsionalny atlas Rossii (The National Atlas of Russia) Vol. 1. Moscow: Roskartografiya; 2004. (in Russian) [Google Scholar]
- Verbenko DA, Pocheshkhova EA, Balanovskaya EV, Marshanija EZ, Kvitzinija PK, Limborska SA. Polymorphisms of D1S80 and 3'ApoB minisatellite loci in Northern Caucasus populations. J Forensic Sci. 2004;49:178–180. doi: 10.1520/JFS2003253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequencies of TaqI B-TaqI D-TaqI A haplotypes in the study populations and in reference populations. This is an extended variant of Table 3 including haplotype frequencies for all populations represented in Figure 3.