Abstract
Cobalamin-dependent methionine synthase (MetH) is a modular protein that catalyzes the transfer of a methyl group from methyltetrahydrofolate to homocysteine to produce methionine and tetrahydrofolate. The cobalamin cofactor, which serves as both acceptor and donor of the methyl group, is oxidized once every ≈2,000 catalytic cycles and must be reactivated by the uptake of an electron from reduced flavodoxin and a methyl group from S-adenosyl-L-methionine (AdoMet). Previous structures of a C-terminal fragment of MetH (MetHCT) revealed a reactivation conformation that juxtaposes the cobalamin- and AdoMet-binding domains. Here we describe 2 structures of a disulfide stabilized MetHCT (s-sMetHCT) that offer further insight into the reactivation of MetH. The structure of s-sMetHCT with cob(II)alamin and S-adenosyl-L-homocysteine represents the enzyme in the reactivation step preceding electron transfer from flavodoxin. The structure supports earlier suggestions that the enzyme acts to lower the reduction potential of the Co(II)/Co(I) couple by elongating the bond between the cobalt and its upper axial water ligand, effectively making the cobalt 4-coordinate, and illuminates the role of Tyr-1139 in the stabilization of this 4-coordinate state. The structure of s-sMetHCT with aquocobalamin may represent a transient state at the end of reactivation as the newly remethylated 5-coordinate methylcobalamin returns to the 6-coordinate state, triggering the rearrangement to a catalytic conformation.
Keywords: corrinoid methyltransferase, multimodular protein, protein conformation, cob(II)alamin coordination, enzyme catalysis
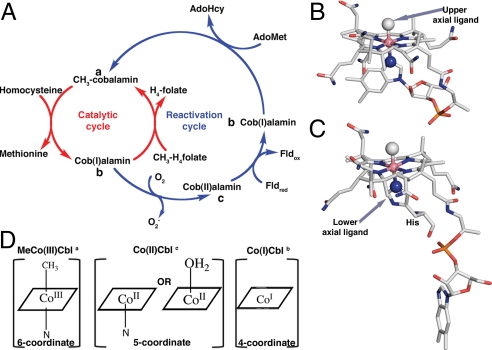
Cobalamin-dependent methionine synthase (MetH) is a modular enzyme (1) that catalyzes methyl transfer from methyltetrahydrofolate (CH3-H4folate) to homocysteine (Hcy), forming tetrahydrofolate (H4folate) and methionine. The enzyme from Escherichia coli is one of the best-studied members of a large class of cobalamin- and corrinoid-dependent methyltransferases (2). In this class of enzymes, the cobalamin cofactor plays a crucial role in catalysis, acting both as a methyl donor and acceptor. During the MetH reaction cycle the methylcobalamin form of the cofactor [MeCo(III)Cbl] is demethylated by Hcy. The electrons of the carbon–cobalt bond remain on the cofactor producing cob(I)alamin [Co(I)Cbl]. Co(I)Cbl is subsequently remethylated by CH3-H4folate regenerating the MeCo(III)Cbl cofactor (Fig. 1A). However under aerobic conditions, the cobalamin cofactor is oxidized to an inactive cob(II)alamin [Co(II)Cbl] form about once in every 2,000 turnovers (3). To avoid accumulation of this inactive species, the enzyme undergoes a reductive reactivation, in which Co(II)Cbl is reduced to Co(I)Cbl with an electron supplied by flavodoxin (Fld), and Co(I)Cbl is then remethylated using S-adenosylmethionine (AdoMet) (4, 5). Reductive reactivation is a challenging reaction because the reduction potential of Co(II)Cbl in solution is out of the range of physiological reducing agents (6–8).
Fig. 1.
(A) Reaction cycle of MetH. The primary catalytic cycle is shown in red whereas the secondary reactivation cycle is shown in blue. (B) MeCo(III)Cbl in the free state with a N atom of the dimethylbenzimidazole group coordinating Co in the lower axial position. A methyl ligand occupies the upper axial position. (C) MetH-bound MeCo(III)Cbl with H759 as the lower axial ligand (13). (D) Schematic drawings of the preferred coordination numbers and geometries for the Cbl oxidation states that are relevant to MetH function. The superscripts a, b, and c in part A indicate the locations of these forms of the cofactor in the catalytic and reactivation cycles. The corrin ring of the Cbl is depicted as a square for simplicity.
MetH has 4 functional units that bind Hcy, CH3-H4folate, Cbl, and AdoMet, respectively (1). During catalysis and reactivation, the appropriate substrate-binding module must be positioned close to the cobalamin to allow methyl transfer. A major challenge is to understand what guides the enzyme through its complex conformational changes without permitting futile cycling. Although the structure of the full-length enzyme has not yet been determined, there are structures of the individual cobalamin-binding (9) and AdoMet-binding modules (10) from E. coli. In addition, 2 multimodular fragments have been characterized: an N-terminal polypeptide (Hcy and CH3-H4folate modules) from Thermotoga maritima (11, 12) and a C-terminal polypeptide (cobalamin and AdoMet modules) from E. coli (12, 13). During catalysis the enzyme cycles between conformations in which the cobalamin-binding module is juxtaposed alternatively with the Hcy-binding module and the CH3-H4folate-binding module, whereas for the reductive reactivation the enzyme assumes a conformation in which the cobalamin-binding module is juxtaposed with the AdoMet-binding module. Access to this reactivation conformation is rigorously controlled to avoid futile cycling (14). A major factor in controlling access to the catalytic and reactivation cycles of the enzyme is the coordination of the Cbl, which in turn is responsive to the positive charge and the oxidation state of the Co.
The free Co(II)Cbl cofactor in solution is 5-coordinate, with a nitrogen ligand in the lower axial coordination site. In this state, a N atom of the dimethylbenzimidazole nucleotide substituent of the corrin macrocycle is directly coordinated to the Co atom resulting in an intramolecular linkage (Fig. 1B). At pH values <2.9, where the dimethylbenzimidazole is protonated and dissociates, it is replaced by a water ligand with the resulting shift of the reduction potential from –610 mV to −500 mV (6).
When MeCo(III)Cbl is bound to MetH, the dimethylbenzimidazole ligand that is coordinated to the cobalt in the free cofactor is displaced by a histidine from the cobalamin-binding domain (H759 in E. coli MetH) (9) (Fig. 1C). As shown in Fig. 1D, MeCo(III)Cbl, Co(II)Cbl, and Co(I)Cbl differ in their preferred coordination states. In particular, the affinity of the cobalt for a nitrogen ligand in the lower axial position is directly proportional to the net positive charge on the cobalt (15–17). Entrance into the reactivation conformation is associated with dissociation of the histidine ligand from the Co(II)Cbl (18, 19).
We previously described the generation of a C-terminal fragment of MetH (MetHCT) that contains the Cbl- and AdoMet-binding modules (18). In the MeCo(III)Cbl form this fragment undergoes temperature-dependent spectral changes, indicating that it interconverts between forms in which H759 is coordinated to the cobalt (His-on) or dissociated (His-off). To reduce conformational flexibility a disulfide cross-link was introduced by mutating Ile-690 and Gly-743 to cysteines (S-SMetHCT). The X-ray structure of the MeCo(III)Cbl form of S-SMetHCT revealed that the MeCo(III)Cbl cofactor had been photolyzed by the X-ray beam during data collection, and that the cobalt appeared 4-coordinate with no electron density evident for the methyl group (12). Four-coordinate Co(II)Cbl has never been observed in aqueous solution. Follow-up studies by Brunold and coworkers (20) of the Co(II)Cbl form of S-SMetHCT using a variety of spectroscopic approaches revealed that the Co(II)Cbl form of the isolated S-SMetHCT is 5-coordinate, with an axially coordinated water molecule. Following addition of AdoMet to S-SMetHCT, the Co–OH2 bond is dramatically lengthened such that the Co(II)Cbl is effectively 4-coordinate.
The work presented here explores the mechanisms by which MetHCT stabilizes the 4-coordinate form of Co(II)Cbl. The structure of a binary complex of S-SMetHCT in the Co(II)Cbl form with adenosylhomocysteine (AdoHcy) has been determined, revealing that Y1139 facilitates the lengthening of the Co–OH2 distance such that the cobalamin is effectively 4-coordinate. This is corroborated by the mutation of Y1139 to phenylalanine (Y1139F), which lowers the reduction potential from −490 mV to −540 mV. We have also examined the surprising finding that S-SMetHCT remains capable of converting between His-on and His-off states, even though its conformational flexibility is greatly reduced by the disulfide cross-link. We present an X-ray structure of the aquocobalamin [AquoCo(III)Cbl] form of S-SMetHCT, which crystallizes in the same conformation as the His-off MeCo(III)Cbl S-SMetHCT and which spectral studies show to be His-on. This His-on form may resemble a transient intermediate in the reductive reactivation of the enzyme.
Results
Activation Complex with Co(II)Cbl and AdoHcy Bound.
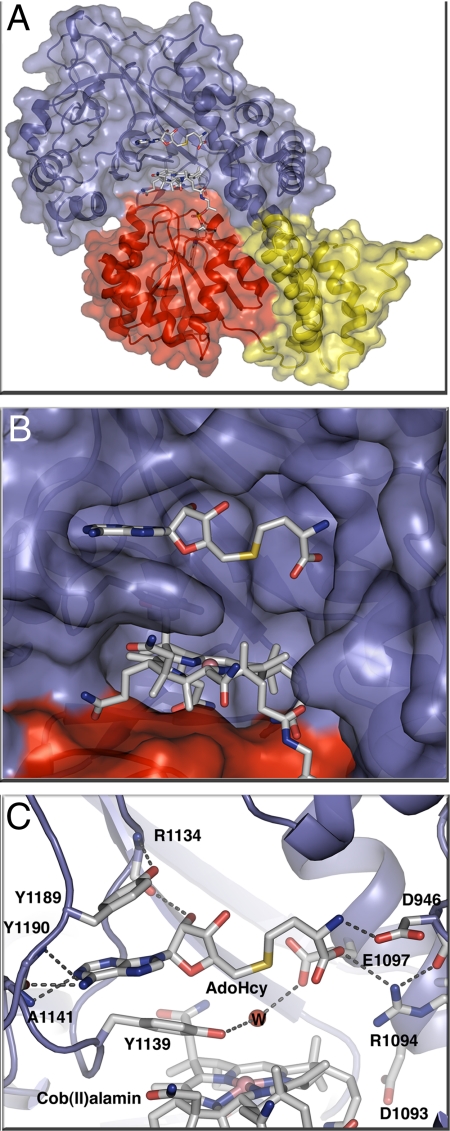
Statistics for the X-ray structure of S-SMetHCT in the Co(II)Cbl form with adenosylhomocysteine (AdoHcy) bound are shown in supporting information (SI) Table S1. This is the first structure to reveal the positioning of an AdoMet analogue with respect to the cobalamin cofactor in the reactivation complex (Fig. 2). AdoHcy was built into clear electron density at 2.7 Å resolution following molecular replacement and initial refinement without a bound ligand (Fig. 2). The orientation of the AdoHcy and its interactions with the activation domain are essentially the same as those of the AdoMet in the structure of its excised binding domain (10).
Fig. 2.
(A) S-SMetHCT:Co(II)Cbl:AdoHcy complex ribbon diagram and space-filling model with the AdoMet-binding module in blue, the Cbl-binding domain in red, and the Cap domain (the N-terminal domain of the Cbl-binding module) in yellow. (B) Surface rendering of the Cbl binding pocket below the AdoHcy binding pocket with subunits colored as in A. AdoHcy and Co(II)Cbl are shown as sticks with white C, red O, blue N, yellow S, and pink Co. (C) AdoHcy binding site. The AdoMet-binding domain is shown as a blue ribbon, with AdoHcy, Co(II)Cbl and amino acid side chains in the binding site in stick form and a water molecule as a red sphere. Hydrogen bonds are shown as dashed lines; colors as in B.
The environment of the Co(II)Cbl cofactor represents the state of the enzyme as it is ready to accept an electron from Fld (Fig. 3), and in this structure the cobalt appears to be 4-coordinate. H759, the lower axial ligand in other forms of MetH, is dissociated and interacts with the AdoMet domain, with no evidence for a water or any other ligand in the upper or lower axial position. However, density near the upper axial position was assigned to a water molecule stabilized by a hydrogen bond with the side chain of E1097. Compared to the previously determined structure of the MeCo(III)Cbl form (12), the side chain of E1097 is repositioned by AdoHcy binding, allowing formation of a hydrogen bond to the water molecule. Moreover, the distance between the cobalt and the Y1139 OH moiety has decreased from 4.5 Å to 4.0 Å. A model methyl group was added to the AdoHcy (Fig. S1), but the resulting Co–Me distance of 5.3 Å and the position of Y1139 between Cbl and AdoHcy suggest that further conformational changes would be necessary to accomplish methyl transfer.
Fig. 3.
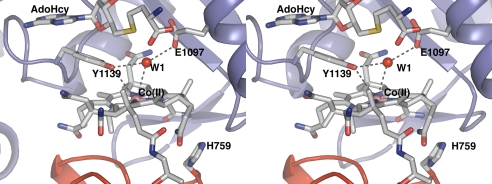
Stabilization of 4-coordinate Co(II)Cbl. The protein environment promotes transition of the metal from a 5-coordinate to a 4-coordinate state through hydrogen bonds to E1097 and Y1139 that move the water away from the Co. This cross-eyed stereoview of the active site of the S-SMetHCT:Co(II)Cbl:AdoHcy complex shows the important interactions that enable the Co–OH2 bond elongation. The water molecule formally bound to Cbl in the absence of AdoHcy is designated as W1. The distances indicated with gray dotted lines are: Y1139-OH–Co(II), 3.9 Å; W1–Co(II), 3.6 Å; Y1139-OH–W1, 3.3 Å; E1097-O–W1, 3.1 Å.
Effect of a Y1139F Mutation on Cbl Reduction Potential.
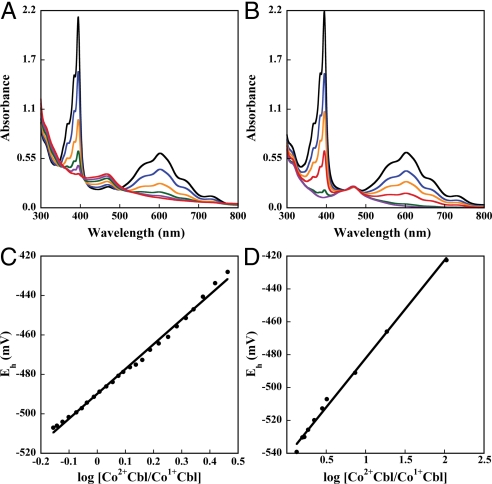
To determine the role of Y1139 in the generation of the Co(I)Cbl intermediate, a Y1139F/I690C/G743C triple mutant was constructed (S-SMetHCT/Y1139F). The Y1139F mutation was incorporated into the double mutant background (I690C/G743C). We measured the reduction midpoint potentials of the Co(II) in both the double (S-SMetHCT) and triple mutant (S-SMetHCT/Y1139F) constructs (Fig. 4 A and B). The Co(II)/Co(I) Cbl midpoint potential was −490 mV for S-SMetHCT, a value identical to that reported for the full-length wild-type enzyme. In contrast, the potential was lowered to −540 mV for the S-SMetHCT/Y1139F mutant (Fig. 4 C and D). This result can be rationalized in the context of the S-SMetHCT structure with AdoHcy bound (Fig. 3). The hydrogen bond between the Y1139 OH and the axial water observed in the structure cannot form in S-SMetHCT/Y1139F, which could thus impede the formation of 4-coordinate Co(II)Cbl and subsequent reduction of the Co(II) to Co(I)Cbl. The 50-mV decrease in reduction potential of S-SMetHCT/Y1139F is consistent with the theoretical prediction that a hydrogen bond between water and the Y1139 OH should increase the reduction potential (20).
Fig. 4.
Spectrophotometric determination of the Co(II)Cbl/Co(I)Cbl midpoint potential of S-SMetHCT (A) and Y1139F S-SMetHCT (B). The enzyme was first photoreduced with 5-deazaflavin-3-sulfonate in the presence of the indicator dye methyl viologen to generate a Co(I)Cbl-bound species (as evidenced by the decrease in absorbance at 468 nm), and spectra were then recorded to monitor the increase in absorbance at 468 nm associated with the spontaneous reoxidation to the Co(II)Cbl-bound form. From a Nernst plot the midpoint potentials were estimated to be −490 mV and −540 mV for S-SMetHCT and Y1139F S-SMetHCT (C and D).
Activation Complex with Aquocobalamin [AquoCo(III)Cbl] Bound.
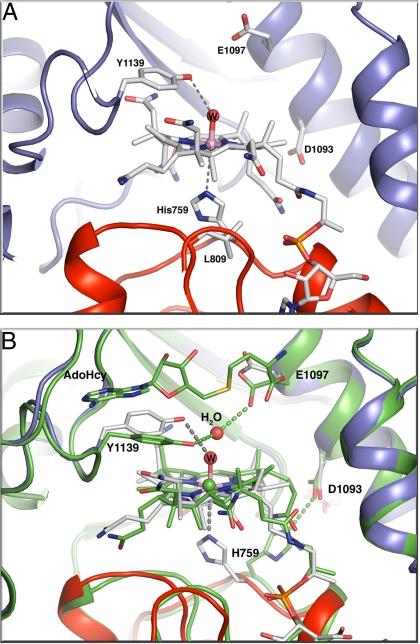
Previous attempts to crystallize the MetHCT:AquoCo(III)Cbl fragment failed, but the disulfide-stabilized form of the fragment with AquoCo(III)Cbl permitted crystallization (statistics in Table S1). These crystals do not contain the substrate/product (AdoMet/AdoHcy). The relative domain arrangement in the AquoCo(III)Cbl S-SMetHCT structure (Fig. S2) is similar to those observed in previous structures of MetHCT fragments (Fig. 2) (12, 13). Relative to other MetHCT structures, the AquoCo(III)Cbl S-SMetHCT structure has 3 distinct features (Fig. 5 A and B): (i) H759 returns to a position below the cobalt (His-on) with adjustments in the surrounding loops; (ii) Y1139 moves away from the cobalt; and (iii) the cobalamin domain exhibits a small rigid body motion relative to the reactivation domain and the corrin ring.
Fig. 5.
(A) Environment of the AquoCo(III)Cbl active site in S-SMetHCT:AquoCo(III)Cbl. The distances indicated with gray dotted lines are: Y1139-OH–W, 3.3 Å; H759-Nε2–Co(III), 3.5 Å. The Co–H2O bond distance is 2.1 Å. (B) Superposition of the active sites of the S-SMetHCT:Co(II)Cbl:AdoHcy complex (green protein and green C atoms) and the S-SMetHCT:AquoCo(III)Cbl structure (color scheme as in Fig. 1). Superpositions are based on residues from the ß-strands of the AdoMet domain. The position of the AdoMet-binding domain and the corrin ring is basically unchanged between the 2 structures. The cobalamin-binding domain, however, moves as a rigid body with respect to the AdoMet-binding domain and the corrin ring.
In solution and in the crystals used for structure determination, AquoCo(III)Cbl S-SMetHCT is red, indicating that histidine is coordinated to the cobalt. This is consistent with the His-on position of electron density for the H759 side chain. However, crystallographic refinement consistently resulted in a long distance (≈3.4 Å) from the cobalt to Nε2 of H759. The most likely explanation for the long bond is photoreduction of the Co(III) in the X-ray beam, as seen in other Co(III)Cbl structures (12, 21). Density for the Y1139 side chain indicates partial disorder, but nonetheless clear evidence of movement away from the cobalamin toward the empty AdoHcy/AdoMet binding pocket. The AquoCo(III)Cbl axial water ligand was omitted from the original model, but following refinement, prominent density above the Co atom was assigned to a water ligand. After further refinement, the 2.1-Å distance is consistent with water coordination to the Co atom.
Comparison of the AquoCo(III)Cbl S-SMetHCT structure with the structure of the MeCo(III)Cbl form of S-SMetHCT (PDB ID 3BUL) (12) reveals a small but significant shift of the cobalamin domain relative to the AdoMet domain (Fig. 5B, Fig. S2). When the AdoMet domains of the 2 structures are aligned, the average motion of the cobalamin domain is ≈1 Å, with a maximum displacement of 2.1 Å at the outer edge. Surprisingly, the corrin ring system does not move with its binding domain but maintains its position relative to the AdoMet domain.
Discussion
Activation for Electron Transfer.
In the absence of the protein scaffold, the reduction potential for a 5-coordinate Co(II)Cbl cofactor is thermodynamically unfavorable relative to those of physiological reductants. Thus, the occasional oxidation to the Co(II) state should result in very low concentrations of Co(I)Cbl at equilibrium, and extremely slow rates of reduction and subsequent methylation by AdoMet. In conjunction with recently reported spectroscopic data (20), the structure of the Co(II)Cbl S-SMetHCT provides a clear picture of how MetH is able to move the potential of the Co(II)Cbl/Co(I)Cbl couple into the physiological range. The enzyme elongates the Co–H2O bond to a formally 4-coordinate species, thereby raising the reduction potential of Co(II)Cbl (20, 22), and facilitating reduction by the physiological partner Fld.
Fig. 3 presents the crystal structure of S-SMetHCT in the conformation required for Co–H2O bond elongation. This structure reveals that elongation of the Co–H2O bond is enabled by the contributions of 2 invariant residues, Y1139 and E1097 (sequence alignment shown in Fig. S3). Following AdoMet/AdoHcy binding, these amino acids are repositioned to interact with the water molecule that was coordinated to Co in the 5-coodinate Co(II)Cbl species as seen in Fig. 3. Comparison of this structure to previously reported structures of MetHCT (12, 13) in absence of the AdoMet cofactor indicates that the Y1139/E1097 water-binding pocket is formed only when the AdoMet/AdoHcy cofactor is present. In the previous structures, E1097 was oriented differently, and only in the present structure it is poised to interact with the water molecule (Fig. 5). Further evidence for the repositioning of E1097 following AdoMet binding is found in the structure of the isolated MetH reactivation domain with AdoMet bound, in which the E1097 is pointed toward the water-binding pocket (10). The reduction potential of Y1139F S-SMetHCT is lowered and reduction of Co(II)Cbl by Fld becomes less favorable. The effects of the Y1139F mutation can be rationalized in the context of the structure and the proposed reactivation cycle (Fig. 6). The structure reveals that the AdoMet ribose moiety is perpendicular to the Y1139 aromatic group with the ribose ring oxygen pointing directly toward the center of the π-aromatic cloud. This interaction allows Y1139 to move closer to the Co atom (Co/Y-OH distance 4.0 Å), and positions it for a hydrogen bonding interaction to the water formerly bound to Co. The importance of Y1139 in lowering the reduction potential via this mechanism is corroborated by spectroscopic data (20).
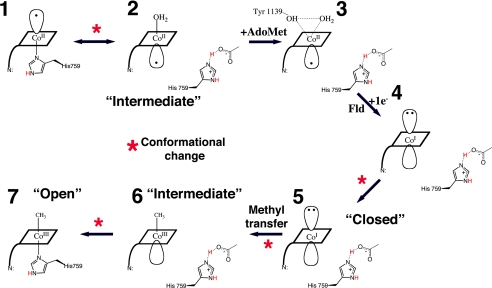
Fig. 6.
Schematic diagram of the changes in cobalt coordination oxidation state and environment during the reactivation cycle. The corrin ring of the Cbl is depicted as a rhomboid and the dimethylbenzimidazole as a line at a corner of the square. (1) Co(II)Cbl is 5-coordinate, with H759 occupying the lower axial position. (2) When Fld binds to the full-length MetH, or in S-SMetHCT, a different 5-coordinate Co(II)Cbl species is seen in which the lower axial His-ligand is replaced with an upper axial water ligand (19, 20). This step is associated with a conformational change from a catalytic conformation in which the cobalamin-binding module is juxtaposed with a Hcy-binding or CH3-H4folate-binding module to the reactivation conformation where the cobalamin-binding module is juxtaposed to the AdoMet-binding module. (3) AdoMet/AdoHcy binding leads to water-Co(II) bond elongation, represented by the AdoHcy-bound Co(II)Cbl S-SMetHCT structure presented here. Fld association and electron transfer lead to the formation of an “intermediate” 4-coordinate Co(I)Cbl species (4) and conformational changes that bring the Me group closer to the Co (5) and allow the subsequent methyl transfer in this “closed” conformation. Methyl transfer and formation of a 5-coordinate MeCo(III)Cbl species allows the domains to return to the “intermediate” state (6). Finally, domain motion is required for the His-759 to again coordinate the Co atom to form the 6-coordinate MeCo(III)Cbl species (7). This “open” conformation is represented by the AquoCo(III)Cbl structure presented here and is the last step before the Cob domain switches from the reactivation cycle back to the catalytic cycle where it can interact with substrate-binding domains and support catalysis.
Structural superpositions of the available C-terminal S-SMetH fragments demonstrate that the position of Y1139 varies significantly between structures (Fig. S4). In the absence of the AdoMet/AdoHcy ligand, the distance between the hydroxyl of Y1139 and the cobalt increases and the tyrosyl ring approaches the binding site for the adenosine of AdoHcy (Fig. 5B). The binding of AdoMet allows Y1139 to move closer to the water ligand and the Co atom, thus enabling the elongation of the H2O–Co bond. This strained state would not be possible in the absence of AdoMet or its analogue AdoHcy. This elegant safety mechanism discourages futile electron transfer from Fld, in the absence of AdoMet.
Rearrangement Following Methyl Transfer.
When MetH enters the reactivation conformation, H759 dissociates from the cobalamin and forms a hydrogen bond with D1093 in the AdoMet domain that stabilizes this conformation (11, 12). Following reduction and methyl transfer, a 5-coordinate MeCo(III)Cbl species is formed (Fig. 6). We propose that this species is in a conformation similar to the Co(II)Cbl structure presented here [Intermediate conformation (2 or 6) in Fig. 6]. After methyl transfer, the affinity of H759 for the cobalt of MeCo(III)Cbl increases (17) and allows H759 to break its interaction with the AdoMet domain and bind to Co. However, a domain motion is required to position H759 to bind the cobalt. A comparison of the Co(II)Cbl and AquoCo(III)Cbl structures provides insight into this domain motion. The AquoCo(III)Cbl structure represents an open conformation (Figs. 5, 6, Fig. S2) in which the cobalamin domain has moved away from the AdoMet domain to allow for H759 coordination. The formation of AquoCo(III)Cbl S-SMetHCT permits us to trap the enzyme in this open conformation that mimics the last step of the reactivation cycle (Fig. 6).
Conclusions
Our findings emphasize the importance of the coordination state of the cobalamin cofactor in governing entrance into and exit from the reactivation conformation. We propose a tug of war between coordination of H759 by the cobalt of Cbl and its participation in intramodular hydrogen bonding interactions. We have previously proposed that when cob(I)alamin is formed during the catalytic cycle, such intramodular interactions are formed between H759 and the CH3-H4folate module, locking the enzyme into this conformation until methyl transfer has occurred (12). The increased positive charge on cobalt following either methyl transfer or oxidation would allow breakage of these intramodular bonds and recoordination of H759 to the cobalt, allowing conformational transitions. In the case of reactivation, the binding of Fld (19) and AdoMet then shift the equilibrium to favor histidine dissociation and stabilization of the reactivation conformation by intramodular hydrogen bonding to D1093 in the AdoMet module (12), allowing reduction of Co(II)Cbl and subsequent methyl transfer from AdoMet. The increased positive charge on MeCo(III)Cbl then allows breakage of the intramodular bonds and recoordination of H759, which in turn permits a conformational transition to the catalytic cycle.
Methods
Chemicals/Cofactors.
AdoMet, glycerol, hydrochloric acid (HCl), 2,2,6,6-tetramethylpiperidine-1-oxyl, MeCbl, and potassium ferricyanide were purchased from Sigma-Aldrich and used without further purification. The 5-deazaflavin-3-sulfonate was a gift from the late Vincent Massey (University of Michigan). E. coli Fld was purified according to published procedures (23, 24).
Construction of an Expression Plasmid for S-SMetHCT and Y1139F S-SMetHCT.
The I690C/G743C double mutation was introduced into the pVB8 vector containing an amino-terminal His6-tagged E. coli wild-type C-terminal MetH, using the QuikChange site-directed mutagenesis kit (Stratagene). The primers were obtained from Invitrogen). The sequence was confirmed by complete sequencing at the Biomedical Research Core Facility of the University of Michigan. The resulting plasmid was named pSD-3.
The Y1139F mutation was introduced into pSD-3 by ligation of a fragment of DNA from the pCS-15 vector containing the mutation. Briefly, pCS-15 was digested with HindIII and BbvcI at 37 °C. The 330-bp fragment was purified from an agarose gel and ligated into the similarly digested pSD-3 vector, yielding pSD-4. The presence of this mutation was verified by sequencing at the Biomedical Research Core Facility of the University of Michigan.
Expression and Purification of S-SMetHCT and Y1139F S-SMetHCT.
The enzymes were expressed in cells of E. coli Hms174(DE3) (Novagen) and purified by nickel affinity chromatography using a 5-mL Hi-trap and a MonoQ 16/10 column (Amersham Biosciences).
Reduction and Photolysis of Cbl Bound to S-SMetHCT.
S-SMetHCT was converted to the MeCo(III)Cbl form through reductive methylation by AdoMet in an electrochemical cell. The Co(II)Cbl-bound MetH forms were generated via photolysis of the corresponding MeCo(III)Cbl-bound species, as described previously (25). Protein concentrations were determined spectrophotometrically by using the extinction coefficients of the enzyme-bound MeCo(III)Cbl cofactor (ε452[S-SMetHCT] = 10.2 mM−1 cm−1) (26). The AquoCo(III)Cbl forms of the mutants were prepared by treating the enzyme with 50 μM of potassium ferricyanide in 10 mM phosphate buffer, pH 7.2 in a cuvette immersed in ice-water and exposing to a tungsten-halogen lamp (650W) at a distance of 5 cm. The formation of the aquo species was monitored in a spectrophotometer (352 nm) for completion of the reaction. The excess reagents were removed by the use of a gel filtration column. Protein concentrations were determined spectrophotometrically by using the extinction coefficients of the enzyme-bound AquoCo(III)Cbl cofactor (ε352[S-SMetHCT] = 21.2 mM−1 cm−1 and ε352[Y1139FS-SMetHCT] = 20 mM−1 cm−1).
Spectrophotometric Determination of the Co(II)/Co(I)Cbl Midpoint Potential.
The midpoint potential was measured as described previously (3). A 1-mL solution of 30 μM enzyme, 100 μM methyl viologen, and 5 μM 5-deazaflavin-3-sulfonate in buffer (100 mM potassium phosphate, 100 mM KCl, and 25 mM EDTA, pH 7.2) was equilibrated with Ar(g) in an anaerobic glass cuvette. The enzyme was reduced by irradiation with a 600 W tungsten/halogen lamp, and the slow oxidation at 37 °C was monitored by absorption spectroscopy. The concentration of reduced methyl viologen was calculated on the basis of the absorbance at 600 nm (ε600 = 13,600 M−1 cm−1) (27), taking into account that the enzyme also weakly absorbs at this wavelength. These values were used in the Nernst equation, along with the midpoint potential for methyl viologen (−446 mV vs. SHE), to calculate the system potential at each time point. Then, the absorbance at 468 nm was corrected for contributions from methyl viologen and used to calculate the concentrations of Co(II)Cbl [ε468 = 10800 M−1 cm−1 (S-SMetHCT), ε468 = 10,000 M−1 cm−1 (Y1139F S-SMetHCT)] and Co(I)Cbl enzyme [ε468 = 2,000 M−1 cm−1]. The midpoint potential was then determined graphically from a Nernst plot of the cell potential vs. log [Co(II)Cbl/Co(I)Cbl].
Crystallization and Cryoprotection.
The 65 kDa S-SMetHCT:Co(II)Cbl, and S-SMetHCT:AquoCo(III)Cbl fragments in 50 mM Tris buffer at pH 7.2 were concentrated to about 15 mg/mL. Crystals of S-SMetHCT:Co(II)Cbl:AdoHcy were grown by the hanging drop vapor diffusion method by mixing 2 μL of protein solution with 2 μL of reservoir solution (0.2 M potassium nitrate, 18% (wt/vol) PEG3350). Crystals of S-SMetHCT:AquoCo(III)Cbl were grown at 22 °C by the microbatch vapor diffusion method under a 1:1 mixture of paraffin to silicon oil by mixing 2 μL of protein with 2 μL of 0.2 M potassium nitrate, 20% (wt/vol) PEG3350, 50 mM Hepes pH 7.5. Crystals were cryoprotected by transfer to a solution of 18% (wt/vol) PEG3350, 0.2 M potassium nitrate, 15% propylene glycol in 25 mM Tris buffer pH 7.0 for the S-SMetHCT:Co(II)Cbl:AdoHcy crystals and in 25 mM Hepes pH 7.5 for the S-SMetHCT:AquoCo(III)Cbl crystals, for a few minutes before flash freezing in liquid N2.
Data Collection and Structure Determination.
Diffraction data were collected at GM/CA-CAT IDB (APS) on a Mar300 detector and processed with XDS (28). Phases for both data sets were obtained by molecular replacement using the individual protein domains of the S-SMetHCT:MeCo(III)Cbl structure (PDB ID 3BUL). EPMR (29) was used to determine initial phases for the S-SMetHCT:Co(II)Cbl:AdoHcy structure, and the AutoMR routine of the PHENIX package (30) was used to determine the AquoCo(III)Cbl phases. Initial density allowed for cofactors/ligand to be added. Both structures were refined with CNS (31) including rigid body refinement of the individual domains followed by simulated annealing in torsional space, coordination minimization, and restrained individual B-factor adjustment with maximum-likelihood targets. CNS recognized and restrained the disulfide between C690 and C743. To eliminate bias in the S-SMetHCT:AquoCo(III)Cbl structure, both H759 and Y1139 were replaced with alanine in the model used for molecular replacement and waters were omitted. Positive electron density below the cobalamin ring indicated the presence of the histidine side chain, but after H759 was modeled and refined, the distance from the cobalt to Nε2 of H759 was 3.4 Å. The histidine is in the expected “on” location, but too far from the cobalt to be considered coordinated (Fig. 5A, Fig. S5). To determine whether this could be an artifact of relatively low resolution, the refinement was repeated with the Co–Nε2 bond restrained to 2.0 Å. The resulting difference maps show negative density at the new position of the histidine ring verifying the long Nε2–Co distance and confirming that the original histidine was modeled properly. Model modification was performed with Coot (32), and the geometric quality of the model and its agreement with the structure factors were assessed with MolProbity (33). Figs. 1 B and C, 2–5, S1, S2, S4, and S5 were generated with PyMOL (34).
Supplementary Material
Acknowledgments.
This research was supported in part by National Institutes of Health Grants GM29408 (to R.G.M.) and GM16429 (formerly to M.L.L., currently to J.L.S.). Diffraction data were collected at GM/CA CAT at the Advanced Photon Source. Our preliminary studies for this paper were conducted in collaboration with the late Professor Martha L. Ludwig, to whom this contribution is dedicated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure coordinates reported in this study have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 3IVA for the Co(II)Cbl I690C/G743C MetH(649–1227) and 3IV9 for the AquoCo(III)Cbl I690C/G743C MetH(649–1227)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0906132106/DCSupplemental.
References
- 1.Goulding CW, Postigo D, Matthews RG. Cobalamin-dependent methionine synthase is a modular protein with distinct regions for binding homocysteine, methyltetrahydrofolate, cobalamin, and adenosylmethionine. Biochemistry. 1997;36:8082–8091. doi: 10.1021/bi9705164. [DOI] [PubMed] [Google Scholar]
- 2.Matthews RG, Koutmos M, Datta S. Cobalamin-dependent and cobamide-dependent methyltransferases. Curr Opin Struct Biol. 2008;18:658–666. doi: 10.1016/j.sbi.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drummond JT, Huang S, Blumenthal RM, Matthews RG. Assignment of enzymatic function to specific protein regions of cobalamin-dependent methionine synthase from Escherichia coli. Biochemistry. 1993;32:9290–9295. doi: 10.1021/bi00087a005. [DOI] [PubMed] [Google Scholar]
- 4.Fujii K, Galivan JH, Huennekens FM. Activation of methionine synthase: Further characterization of flavoprotein system. Arch Biochem Biophys. 1977;178:662–670. doi: 10.1016/0003-9861(77)90238-7. [DOI] [PubMed] [Google Scholar]
- 5.Taylor RT, Weissbach H. N5-methyltetrahydrofolate-homocysteine transmethylase: Partial purification and properties. J Biol Chem. 1967;242:1502–1508. [PubMed] [Google Scholar]
- 6.Lexa D, Saveant JM. Electrochemistry of vitamin B12. I. Role of the base-on/base-off reaction in the oxidoreduction mechanism of the B12r-B12s system. J Am Chem Soc. 1976;98:2652–2658. doi: 10.1021/ja00425a039. [DOI] [PubMed] [Google Scholar]
- 7.Vetter H, Jr, Knappe J. Flavodoxin and ferredoxin of Escherichia coli. Hoppe Seylers Z Physiol Chem. 1971;352:433–446. doi: 10.1515/bchm2.1971.352.1.433. [DOI] [PubMed] [Google Scholar]
- 8.Wolthers KR, Scrutton NS. Cobalamin uptake and reactivation occurs through specific protein interactions in the methionine synthase-methionine synthase reductase complex. FEBS J. 2009;276:1942–1951. doi: 10.1111/j.1742-4658.2009.06919.x. [DOI] [PubMed] [Google Scholar]
- 9.Drennan CL, Huang S, Drummond JT, Matthews RG, Lidwig ML. How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 10.Dixon MM, Huang S, Matthews RG, Ludwig M. The structure of the C-terminal domain of methionine synthase: Presenting S-adenosylmethionine for reductive methylation of B12. Structure. 1996;4:1263–1275. doi: 10.1016/s0969-2126(96)00135-9. [DOI] [PubMed] [Google Scholar]
- 11.Evans JC, et al. Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc Natl Acad Sci USA. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta S, Koutmos M, Pattridge KA, Ludwig ML, Matthews RG. A disulfide-stabilized conformer of methionine synthase reveals an unexpected role for the histidine ligand of the cobalamin cofactor. Proc Natl Acad Sci USA. 2008;105:4115–4120. doi: 10.1073/pnas.0800329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandarian V, et al. Domain alternation switches B(12)-dependent methionine synthase to the activation conformation. Nat Struct Biol. 2002;9:53–56. doi: 10.1038/nsb738. [DOI] [PubMed] [Google Scholar]
- 14.Jarrett JT, Huang S, Matthews RG. Methionine synthase exists in two distinct conformations that differ in reactivity toward methyltetrahydrofolate, adenosylmethionine, and flavodoxin. Biochemistry. 1998;37:5372–5382. doi: 10.1021/bi9730893. [DOI] [PubMed] [Google Scholar]
- 15.Brown KL, Hakimi JM, Nuss DM, Montejano YD, Jacobsen DW. Acid-base properties of a-ribazole and the thermodynamics of dimethylbenzimidazole association in alkylcobalamins. Inorg Chem. 1984;23:1463–1471. [Google Scholar]
- 16.Brown KL, Pecksiler S. Heteronuclear NMR-studies of Cobalamins. 9. Temperature-dependent NMR of organocobalt corrins enriched in C-13 in the organic ligand and the thermodynamics of the base-on base-off reaction. Inorg Chem. 1988;27:3548–3555. [Google Scholar]
- 17.Fleischhacker AS, Matthews RG. Ligand trans influence governs conformation in cobalamin-dependent methionine synthase. Biochemistry. 2007;46:12382–12392. doi: 10.1021/bi701367c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandarian V, Ludwig ML, Matthews RG. Factors modulating conformational equilibria in large modular proteins: A case study with cobalamin-dependent methionine synthase. Proc Natl Acad Sci USA. 2003;100:8156–8163. doi: 10.1073/pnas.1133218100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoover DM, et al. Interaction of Escherichia coli cobalamin-dependent methionine synthase and its physiological partner flavodoxin: Binding of flavodoxin leads to axial ligand dissociation from the cobalamin cofactor. Biochemistry. 1997;36:127–138. doi: 10.1021/bi961693s. [DOI] [PubMed] [Google Scholar]
- 20.Liptak MD, Datta S, Matthews RG, Brunold TC. Spectroscopic study of the cobalamin-dependent methionine synthase in the activation conformation: Effects of the Y1139 residue and S-adenosylmethionine on the B12 cofactor. J Am Chem Soc. 2008;130:16374–16381. doi: 10.1021/ja8038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Champloy F, Gruber K, Jogl G, Kratky C. XAS spectroscopy reveals X-ray-induced photoreduction of free and protein-bound B12 cofactors. J Synchrotron Rad. 2000;7:267–273. doi: 10.1107/S0909049500006336. [DOI] [PubMed] [Google Scholar]
- 22.Stich TA, Yamanishi M, Banerjee R, Brunold TC. Spectroscopic evidence for the formation of a four-coordinate Co2+ cobalamin species upon binding to the human ATP:cobalamin adenosyltransferase. J Am Chem Soc. 2005;127:7660–7661. doi: 10.1021/ja050546r. [DOI] [PubMed] [Google Scholar]
- 23.Hall DA, Jordan-Starck TC, Loo RO, Ludwig ML, Matthews RG. Interaction of flavodoxin with cobalamin-dependent methionine synthase. Biochemistry. 2000;39:10711–10719. doi: 10.1021/bi001096c. [DOI] [PubMed] [Google Scholar]
- 24.Hall DA, et al. Mapping the interactions between flavodoxin and its physiological partners flavodoxin reductase and cobalamin-dependent methionine synthase. Proc Natl Acad Sci USA. 2001;98:9521–9526. doi: 10.1073/pnas.171168898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarrett JT, Goulding CW, Fluhr K, Huang S, Matthews RG. Purification and assay of cobalamin-dependent methionine synthase from Escherichia coli. Methods Enzymol. 1997;281:196–213. doi: 10.1016/s0076-6879(97)81026-9. [DOI] [PubMed] [Google Scholar]
- 26.Jarrett JT, et al. Mutations in the B12-binding region of methionine synthase: How the protein controls methylcobalamin reactivity. Biochemistry. 1996;35:2464–2475. doi: 10.1021/bi952389m. [DOI] [PubMed] [Google Scholar]
- 27.Mayhew SG. The redox potential of dithionite and SO-2 from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur J Biochem. 1978;85:535–547. doi: 10.1111/j.1432-1033.1978.tb12269.x. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 29.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D Biol Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 31.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano WL. Palo Alto, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.