Abstract
The PUR protein family is a distinct and highly conserved class that is characterized by its sequence-specific RNA- and DNA-binding. Its best-studied family member, Pur-α, acts as a transcriptional regulator, as host factor for viral replication, and as cofactor for mRNP localization in dendrites. Pur-α-deficient mice show severe neurologic defects and die after birth. Nucleic-acid binding by Pur-α is mediated by its central core region, for which no structural information is available. We determined the x-ray structure of residues 40 to 185 from Drosophila melanogaster Pur-α, which constitutes a major part of the core region. We found that this region contains two almost identical structural motifs, termed “PUR repeats,” which interact with each other to form a PUR domain. DNA- and RNA-binding studies confirmed that PUR domains are indeed functional nucleic-acid binding domains. Database analysis show that PUR domains share a fold with the Whirly class of nucleic-acid binding proteins. Structural analysis combined with mutational studies suggest that a PUR domain binds nucleic acids through two independent surface regions involving concave β-sheets. Structure-based sequence alignment revealed that the core region harbors a third PUR repeat at its C terminus. Subsequent characterization by small-angle x-ray scattering (SAXS) and size-exclusion chromatography indicated that PUR repeat III mediates dimerization of Pur-α. Surface envelopes calculated from SAXS data show that the Pur-α dimer consisting of repeats I to III is arranged in a Z-like shape. This unexpected domain organization of the entire core domain of Pur-α has direct implications for ssDNA/ssRNA and dsDNA binding.
Keywords: crystal structure, DNA binding, RNA binding, fragile X-associated tremor, ataxia syndrome
Pur-α, Pur-β, and Pur-γ are the three members of the purine-rich element binding protein (PUR) family, mainly found in higher eukaryotes (1, 2). Studies on human and mouse Pur-α revealed a regulatory function in the transcription of neuronal genes with TATA-less promoters (1, 2). More recent evidence indicates that Pur-α is an integral factor of actively transported neuronal mRNPs (3, 4). Furthermore, Drosophila Pur-α binds to CGG repeats in the 5′UTR of FMR1 mRNA, and thereby contributes to the occurrence of fragile X-associated tremor/ataxia syndrome (5). Given these multiple roles for Pur-α, it is not surprising that Pur-α-deficient mice die within the first weeks after birth, with severe neurologic pathologies (6). Pur-α also serves as cellular host factor for the infection of RNA viruses like JC virus and HIV (7–10), most likely by augmenting viral replication (11, 12). The function of Pur-β and -γ is less well understood.
PUR proteins consist of a glycine-rich flexible N terminus, a central core region, and a C-terminal, potentially phosphorylated protein-interaction region of variable length (Fig. 1A) (1). The core region shows the highest sequence conservation [supporting information (SI) Fig. S1], defines the family of PUR proteins, and mediates sequence-specific binding to ssDNA, dsDNA, and ssRNA, with a preference for (GGN)-repeats. PUR proteins lack sequence homology to proteins with known structure. Thus, it is unclear which three-dimensional-fold PUR proteins adopt in their core region and how they interact with nucleic acids.
Fig. 1.
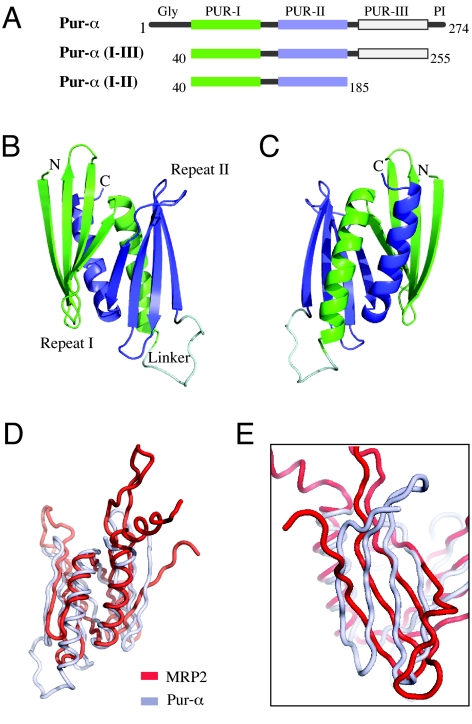
The structure of Drosophila Pur-α identifies the PUR domain as an MRP1/MRP2/P24-like nucleic-acid binding protein. (A) Schematic drawing of the distinct protein regions of Pur-α (Top) and the two protein fragments used in this study (Middle and Bottom). PUR repeats have been identified in this study by structure determination. (B) Ribbon backbone model of the globular domain formed by two PUR repeats, termed PUR domain. Similar to the schematic drawing in (A), PUR repeat I is shown in green, PUR repeat II in blue, and the linker connecting both repeats in gray. (C) View from (B) rotated by 180° around the vertical axis. (D) Superposition of the structure of Pur-α (gray) with the structure of MRP2 (red). Orientation is identical to (C). (E) Rotated and magnified superposition from (D) showing the orientation of the β-sheets from Pur-α and MRP2 with respect to each other.
We have determined the crystal structure of a major part of the core region from Drosophila melanogaster Pur-α at 2.1 Å resolution. The structure reveals that this region is constituted by two highly homologous repeats, which interact with each other to form a PUR domain. Each repeat consists of an antiparallel β-sheet and one α-helix. Size-exclusion chromatography and small-angle x-ray scattering (SAXS) studies confirmed that both repeats also interact to form a PUR domain in solution. Systematic database searches revealed that the PUR domain is homologous to the “Whirly” class of nucleic-acid binding folds. EMSA confirmed that the PUR domain is also a functional DNA- and RNA-binding domain. Structural analysis and DNA- and RNA-interaction studies suggest two nucleic-acid binding surfaces per PUR domain. Sequence alignment and structural prediction indicate that the core region harbors an additional third repeat. A longer fragment containing all three repeats dimerizes Pur-α. Surface envelope calculations from SAXS measurements with a fragment containing repeats I to III show an unusual Z-like conformation of the Pur-α dimer.
Results
Overall Structure of Pur-α Repeats I and II.
Various fragments of Pur-α from different species were expressed and used for crystallization trials. A fragment consisting of residues 40 to 185 from Drosophila melanogaster Pur-α isoform 1 [Pur-α (I–II)] (see Fig. 1A) resulted in initial crystals. After optimization of crystallization conditions, Selenomethionine-derivatized protein crystals were produced and phases determined by multiple wavelength anomalous dispersion (Tables S1 and S2). The structural model was built from a native dataset at 2.1 Å resolution (see Tables S1 and S2). The final model with Rwork = 22.3% and Rfree = 24.0% revealed that the protein structure consists of two repeats. We refer to them as PUR repeats I and II. Each of the PUR repeats consist of a four-stranded antiparallel β-sheet, followed by a C-terminal α-helix (Fig. 1 B and C). Both repeats are connected through a short linker region and form a globular domain we refer to as “PUR domain” (see Fig. 1 B and C). The intercalation of the α-helix from one repeat into the other repeat results in a hydrophobic buried surface interface of 1,830 Å2 per monomer (approximately 33% of each monomer's surface). Such a large buried surface significantly exceeds the areas masked in average by protein-protein interactions (13, 14) and strongly suggests a stable interaction of PUR repeats I and II in Pur-α.
PUR Domains Are Members of the Whirly-Class of Nucleic-Acid Binding Domains.
A systematic structural comparison using DaliLite (15) revealed significant similarities to a set of annotated proteins. Among them, the highest scores were obtained for the mitochondrial RNA-binding protein-2 (MRP-2) (PDB-ID: 2GIA; Z-score: 8.2; rmsd: 4.7 Å for 114 α-carbon pairs), the mitochondrial RNA-binding protein-1 (MRP-1) (PDB-ID: 2GJE; Z-score: 7.0; rmsd: 4.7 Å for 108 α-carbon pairs), and the plant transcriptional regulator PBF-2 (P24) (PDB-ID: 1L3A; Z-score: 6.9; rmsd: 4.7 Å for 113 α-carbon pairs). Even higher scores were obtained for two hypothetical proteins with no known function from cyanobacteria (PDB-ID: 2IT9; Z-score: 11.4; rmsd: 3.5 Å for 116 α-carbon pairs/PDB-ID: 2NVN; Z-score: 10.9; rmsd: 3.7 Å for 118 α-carbon pairs). For none of the mentioned proteins did sequence identities with Pur-α exceed 16%.
MRP-2 and MRP-1 are RNA-binding proteins (16), whereas P24 binds to dsDNA and ssDNA (17). All three proteins contain a so-called Whirly domain (Fig. S2) that matches the overall ββββα-ββββα topology of PUR domains and nicely superposes with them (Fig. 1 D and E). The main differences to PUR domains are longer loop regions, a rotated orientation of the β-sheets (see Fig. 1E), and a longer kinked α-helix at the C terminus for MRP2. In summary, the structural comparison uncovers intriguing similarities to the Whirly domains of MRP1, MRP2, and P24.
Examination of the crystal contacts of Pur-α shows a pronounced difference to the quarternary structure of the known Whirly proteins (see Fig. S2). Whereas Whirly proteins tetramerize via interactions of their α-helices, such an arrangement is not observed in the crystal lattice of Pur-α.
PUR Domains Show Positively Charged Surface Regions Around Their β-Sheets with High Sequence Conservation.
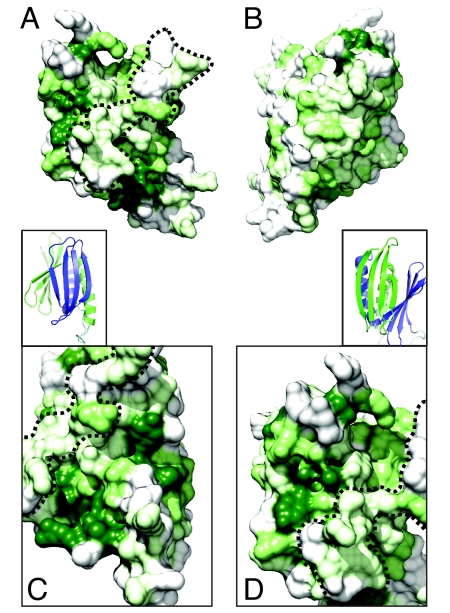
A surface plot of sequence conservation reveals great differences at the opposing sides of the PUR domain (Fig. 2 A and B). Whereas the side showing both α-helices is almost devoid of highly conserved residues (see Fig. 2B), the opposing side reveals considerable sequence conservation at and around the two β-sheets (Fig. 2 A, C, and D). Both conserved β-sheets are clearly separated by an unconserved rim (dashed line in Fig. 2 A, C, and D). These findings suggest two independent surface areas of functional importance.
Fig. 2.
Surface conservation of the PUR domain. (A) Surface representation of the sequence conservation of the solvent-accessible surface of the PUR domain. Surface plot is based on the interspecies alignment shown in Fig. S1. Dark green coloration indicates complete conservation, whereas light green shows partially conserved and gray unconserved residues. Orientation is identical to Fig. 1B. Dashed line indicates the unconserved rim that separates the two highly conserved surface regions around the β-sheets. (B) Representation as in (A) rotated by 180° around the vertical axis (identical to orientation in Fig. 1C). (Insets) Cartoon representation of a close-up of the β-sheet from PUR repeats I and PUR repeat II (Fig. 1B). Color-coding is identical to (Fig. 1 B and C). (C) Close-up of surface conservation from Pur repeat II with the view identical to inset above (C). (D) Close-up of surface charge of Pur repeat I with the view identical to inset above (D).
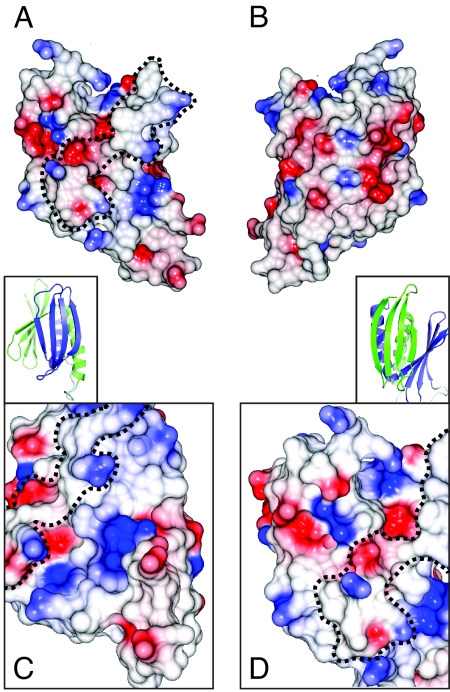
Analysis of the surface charges of Pur-α (I–II) (Fig. 3 A and B) shows moderate negative charges at the α-helical side (see Fig. 3B). In contrast, the surface side with both β-sheets shows a number of positively charged residues around the β-sheet of PUR repeat II (K121, R132, K138, R142, R147, R155, R158) (see Fig. 3 A and C). Positive surface charges at the β-sheet of PUR repeat I appear slightly less pronounced (R55, K61, R65, K70, R80, R131) (see Fig. 3 A and D). These positive charges at both β-sheets hint at potential nucleic-acid binding surfaces.
Fig. 3.
Surface charges of the PUR domain. (A) Representation of the electrostatic potentials of the solvent-accessible surface of the PUR domain. Red and blue coloration indicate negative and positive electrostatic potentials, respectively. Orientation is identical to Figs. 1B and 2A. Dashed line indicates the unconserved rim that separates the two highly conserved surface regions around the β-sheets. (B) Representation as in (A) rotated by 180° around the vertical axis (identical to orientation in Figs. 1C and 2B). (Insets) Cartoon representation of a close-up of the β-sheet from PUR repeats I (A) and PUR repeat II (Fig. 1B). (C) Close-up of surface charges of Pur repeat II with the view identical to inset above (C). (D) Close-up of surface charges of Pur repeat I with the view identical to inset above (D).
PUR Domains Are Functional Nucleic-Acid Binding Domains.
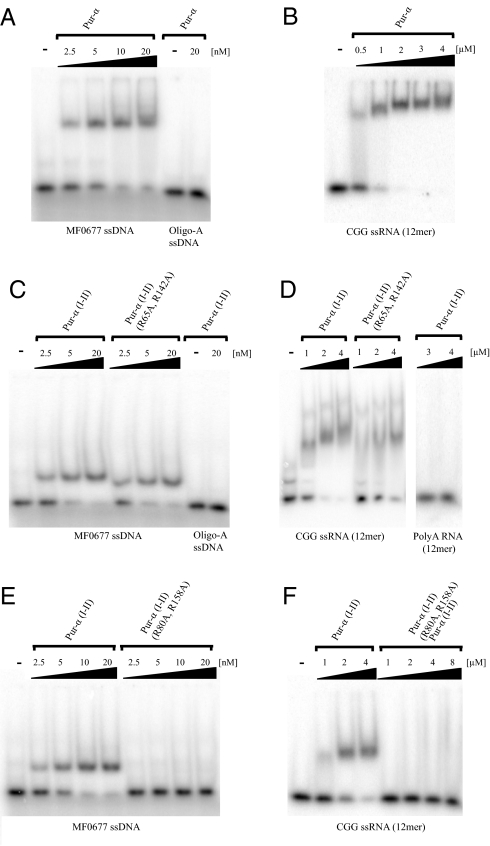
As shown for human Pur-α (18), EMSA with full-length Pur-α showed strong and specific binding to the previously reported MF0677 ssDNA (18) (Fig. 4A), indicating an equilibrium dissociation constant (Kd) of about 5 nM. Binding to a CGG RNA oligomer (5) was considerably weaker (Fig. 4B) and indicates a Kd around 1 μM. To test what the contribution of the PUR domain observed in the crystal structure is to this binding, we repeated EMSAs with Pur-α (I–II). Mobility shifts with ssDNA and ssRNA (Fig. 4 C and D) were comparable to shifts observed with full-length Pur-α. This finding indicates that the crystallized PUR domain represents a functional nucleic-acid binding domain that is sufficient for the main binding affinity of the protein. No unspecific binding to oligoA ssDNA or oligoA ssRNA was observed (see Fig. 4 C and D).
Fig. 4.
DNA EMSAs with different Pur-α fragments. (A) Full-length Pur-α binds with high affinity to MF0677 24mer ssDNA but not to control A(24)-mer ssDNA. (B) Full-length Pur-α also binds to a (CGG)(12)-mer ssRNA. Pur-α (I-II) binds to the ssDNA (C) and to the ssRNA (D) with affinities comparable to full-length Pur-α. The double-mutation R65A and R142A in Pur-α (I-II) does not affect its binding to ssDNA (C) but showed a moderate reduction in ssRNA binding (D). The double-mutant R80A and R158A abolished binding of Pur-α (I-II) to ssDNA (E) as well as to ssRNA (F).
Surface Areas Around the β-Sheets Are Involved in Nucleic-Acid Binding.
It has been previously reported that in mouse Pur-α the mutation of arginine 71 into glutamic acid results in reduced ssDNA binding and dsDNA unwinding (19). The corresponding residue arginine 55 in Drosophila Pur-α is located in the second β-strand of PUR repeat I (Fig. S3 A and B), suggesting that the β-sheets are indeed sites of nucleic-acid binding. We generated mutations to probe whether both ends of the conserved β-sheets are involved in nucleic-acid binding (see Fig. S3 A and B). The first Pur-α (I–II) construct has mutated a conserved arginine in the loop connecting β-strand 2 with 3 in both PUR repeats [Pur-α (I–II/R65A, R142A)]. This double mutant did not show any defect in ssDNA binding (see Fig. 4C), whereas ssRNA binding was moderately impaired (see Fig. 4D). The second set of mutations affect arginines at the fourth β-strand of PUR repeats I and II [Pur-α (I–II/R80A, R158A)]. This double mutation abolishes ssDNA binding (Fig. 4E) as well as ssRNA binding (Fig. 4F). Correct folding of both double mutants was verified by circular dichroism spectroscopy (Fig. S3C). In summary, these results indicate that the β-sheets and nearby residues are the interaction surfaces for ssDNA and ssRNA binding.
Pur-α (I–II) Is Monomeric in Solution.
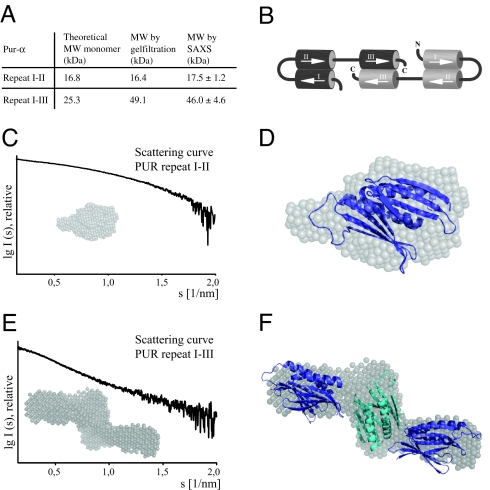
We examined if the crystallographic, intramolecular interaction of PUR repeats I and II is consistent with the oligomeric state of Pur-α (I–II) in solution. Using size-exclusion chromatography (Fig. 5A) and SAXS experiments (see Fig. S4A and Fig. 5A), we confirmed the monomeric state of Pur-α (I–II). In addition, the p(r)-distribution of Pur-α (I–II) is rather symmetric with a single peak and shows the typical shape of a globular molecule (Fig. S4B). From the SAXS data, a surface envelope was calculated (Fig. 5C) into which the crystal structure could be nicely fitted (Fig. 5D). In addition, the theoretical scattering curve for the PUR domain was calculated from the crystal structure and was found to match well with the measured data (Fig. S4 C and D). The buried surface interface of 1,830 Å2 per PUR repeat in the PUR domain is exceedingly large for such a small protein. From all these findings, we rationalize that the PUR domain consisting of PUR repeats I and II from the same Pur-α molecule is also found in solution and is likely to exist also in context of the full-length protein. As already suggested by our analysis of the crystal contacts (see Fig. S2), we conclude that these PUR domains do not adopt the tetrameric quarternary structure (see Fig. S2) observed in the known Whirly proteins (16, 17).
Fig. 5.
Analysis by SAXS. (A) Table summarizing molecular-weight calculations by size-exclusion chromatography and by SAXS. (B) Schematic drawing depicting the role of individual PUR repeats in the formation of PUR domains and Pur-α dimerization. (C) Scattering curve of Pur-α (I–II) and representative surface envelope calculated from SAXS measurements. (D) A fit of the crystal structure into the surface envelope of Pur-α (I–II) confirms the presence of intramolecular PUR domains in solution. (E) Scattering curve of Pur-α (I–III) and representative surface envelope calculated from SAXS measurements. These envelopes adopt a Z-like shape. (F) A fit of three PUR domains into the Z-like envelope is consistent with the presence of two intramolecular PUR domains and one intermolecular PUR domain (B). Structural models of Pur-α (I-II) were positioned manually in the envelopes to demonstrate the compatibility of envelopes with the size of PUR domains.
Pur-α Has Three PUR Repeats in Its Central Region.
An alignment of the central nucleic-acid binding region of Pur-α homologs from different species reveals a moderate level of sequence identity but a strong conservation of key residues that define PUR repeats (see Fig. S1).
These residues were also found in the Drosophila core region C-terminal to PUR repeat II (see Fig. S1), indicating the existence of a third PUR repeat (see Fig. 1A). This third PUR repeat shares 25% (53%) and 32% (56%) sequence identity (similarity) with repeats I and II, respectively (Table S3). It shares an almost identical secondary-structure prediction and shows strong conservation in residues that are buried in the interface between PUR repeats I and II. Therefore, we hypothesize that the third PUR repeat might also adopt a three-dimensional fold similar to repeats I and II.
Pur-α (I–III) Forms a Dimer in Solution.
The PUR-domain structure (see Fig. 1 B and C) suggests that an isolated, monomeric PUR repeat is unstable. For stabilization of PUR repeat III an interaction with another PUR repeat forming a PUR domain might be necessary. The most likely interaction is an intermolecular PUR domain by two repeats III, resulting in Pur-α dimers. By using size-exclusion chromatography and SAXS measurements, we found that Pur-α (I–III) does indeed form dimers (see Fig. S4A and Fig. 5A). This finding suggests that PUR repeat III induces dimerization of Pur-α, most likely through its interaction with a PUR repeat III from a different peptide chain (Fig. 5B). Consistently, the pair-distribution function of Pur-α (I-III) shows the typical skewed shape of an elongated molecule (see Fig. S4B). The elongated arrangement of PUR domains contributed by two polypeptide chains appears to be well suited to recognize repetitive sequences as reported for Pur-α. Because Pur-α dimers are stable at experimental concentrations (20) as low as 30 nM (Table S4), we anticipate that they might be present also in vivo.
SAXS Indicates that Dimeric Pur-α (I–III) Adopts a Z-Like Shape.
As expected from the p(r)-distribution (see Fig. S4B), structural calculations with SAXS data from Pur-α (I–III) yielded surface envelopes with an elongated shape (Fig. 5E). However, Pur-α (I–III) envelopes have an unexpected Z-like shape into which three PUR domains can be placed (Fig. 5F). For a good fit into the envelope, the PUR-domain in the middle has to be oriented perpendicular to the two flanking PUR domains.
Discussion
We performed structural analysis on the core region from Drosophila Pur-α and determined that it consists of three PUR repeats. The interaction of two repeats results in a PUR domain with strong structural similarity to the MRP1/MRP2 and P24 class of Whirly-domain proteins. On the other hand, no significant sequence similarity is detectable between Pur-α and these proteins. We also noted a similarity in topology to the transcriptional co-activator PC4 (21). However, PC4 was not detected in DaliLite searches and, in contrast to Pur-α (Ι–ΙΙ), forms intermolecular dimers.
Whereas MRP1/MRP2 binds ssRNA, P24 has been shown to bind ssDNA and dsDNA as well as to unwind dsDNA. Intriguingly, Pur-α combines the functions of both protein classes. For example, Drosophila Pur-α binds to CGG repeats of the 5′UTR of FMR1 mRNA (5), but also interacts with dsDNA and ssDNA (19, 22). ATP-independent short-range unwinding of dsDNA has also been reported for Pur-α (22). EMSA with DNA and RNA indicate that the PUR domain consisting of PUR repeats I and II is the main nucleic-acid binding domain of this protein (see Fig. 4 A–D). In summary, the similarities are not limited to the structural level, but also extend to their functional properties. There are, however, also significant differences between these proteins. PUR repeats I and II do not tetramerize like their Whirly-domain counterparts; instead, they are monomeric.
Analysis of the crystallographic contact of the two hypothetical proteins with similarity to Pur-α revealed no tetrameric organization (see Fig. S2). An assessment of their crystallographic contacts (EBI-PISA server; www.ebi.ac.uk) failed to suggest surfaces that are likely to mediate oligomerization in solution. Like Pur-α, these potential members of the Whirly family seem to fail to form tetramers.
Analysis of the surface conservation and charges suggests that both conserved regions around the two β-sheets of Pur-α (I–II) are functionally important. The corresponding regions of MRP1/MRP2 and P24 have been mapped to bind ssRNA and DNA, respectively (16, 17). A mutation in the fourth β-strand in both PUR repeats abolished DNA and RNA binding (see Fig. 4 E and F), confirming that the conserved regions around the concave β-sheets are indeed nucleic-acid binding surfaces. The observed selective reduction of RNA binding by Pur-α (I–II/R65A, R142A) (see Fig. 4D) indicates that the tested RNA and DNA target are not bound in an identical fashion.
One unexpected feature of the surface analysis of Pur-α is the significant difference in patterns of surface conservation and charges between the areas around the β-sheets of PUR repeats I and II (see Figs. 2 A, C, and D and 3 A, C, and D). It is possible that both surfaces have evolved to bind distinct nucleic-acid targets. Alternatively, one of the potential binding regions might lack nucleic-acid binding properties and instead plays a different functional role in Pur-α. Consistent with the latter hypothesis is the finding that in the MRP1/MRP2 complex, only one of the two β-sheets binds RNA (16).
One other difference found between Pur-α and MRP1/MRP2 is that Pur-α binds to specific RNA sequences (5), whereas the MRP1/MRP2 complex does not (16, 23–25). In the crystal structure of the gRNA co-complex with MRP1/MRP2, the bases of the RNA point away from the MRP1/MRP2 surface. This orientation prevents sequence-specific interaction with the protein complex. Thus, in the case of sequence-specific binding by Pur-α, the RNA or DNA is likely to be oriented differently.
Size-exclusion chromatography and SAXS analyses (see Fig. S4A and Fig. 5A) indicated that the third PUR repeat mediates dimerization of two Pur-α molecules. Surface envelopes calculated from SAXS data show that dimeric Pur-α adopts a Z-like conformation in solution (see Fig. 5E). We were able to accommodate three PUR domains within the envelope (see Fig. 5F), further indicating that PUR repeat III might dimerize to form a PUR domain (see Fig. 5B). However, we should note that our data provide no direct evidence for the existence of a PUR domain formed by two PUR repeats III.
Pur-α binds to (NGG)n and r(CGG)n repeats (1, 5). The elongated organization of the Pur-α dimer seems well suited to accommodate the binding to such repetitive single-stranded sequences. Such a domain arrangement also allows for the simultaneous binding of more than one independent RNA molecule per full-length Pur-α dimer. Assuming that the Z-like arrangement is stable also in the nucleotide-bound state, a binding of dsDNA by more than one PUR domain is likely to require extreme bending of the DNA, and therefore appears unlikely. However, we cannot rule out the possibility that upon dsDNA binding Pur-α undergoes conformational changes. Alternatively, the previously reported unwinding of dsDNA upon binding (19, 22) could generate the conformational flexibility required for full DNA binding.
The structural study on MRP1/MRP2 revealed that hairpin RNA is unwound in the bound state. Because, to our knowledge for Pur-α, only unwinding of dsDNA has been reported (19, 22), it might be interesting to investigate whether Pur-α has similar capabilities in unwinding dsRNA. Regardless of the nucleic-acid target, the question arises how Pur-α achieves and maintains unwinding of double-stranded nucleic acids. It has been suggested that its structural homolog P24 unwinds dsDNA first by exploiting melted regions of dsDNA and then maintains unwinding by intercalating the entire tetramer into an otherwise double-stranded DNA (17). An ATP-independent exploitation of melted dsDNA regions appears to be a likely first step also for Pur-α-dependent unwinding. In addition, the C terminus of Pur-α may be required for the unwinding of dsDNA (19, 22). However, an intercalation of Pur-α analogous to P24 is unlikely because it does not form tetramers. Instead, one could imagine that Pur-α separates complementing DNA strands by binding with both β-sheets of a PUR domain to the individual DNA strands. The unconserved rim between these β-sheets (see Figs. 2A and 3A) could serve as a spacer between both DNA strands. Our SAXS analyses revealed that Pur-α (I-III) has a Dmax value of about 135 Å. This distance corresponds to the length of about 25 DNA bases and should roughly match the maximum sequence stretch Pur-α would be able to intercalate via such a mechanism. Indeed, it has been reported that only short oligomeric stretches of dsDNA are unwound by Pur-α (19, 22). Albeit intriguing, such hypotheses require new sets of experiments to be tested in future. The Pur-α structure reported here and its structural and functional similarity to the Whirly class of nucleic-acid binding proteins will provide a rational basis for such studies and will help to understand the mechanistic roles this protein plays in transcription, mRNA localization, fragile X-associated tremor/ataxia syndrome, and viral replication.
Materials and Methods
Protein Expression and Purification.
GST-tagged protein fragments were expressed in Escherichia coli and purified using standard conditions (26). After protease cleavage of the GST tag, GST was subtracted using a GST column and nucleic acids removed by a Q column. Pur-α was further purified by Heparin column. The final purification step was performed by size-exclusion chromatography. Selenomethionine-substituted protein was expressed as described (27).
Crystallization and Structure Determination.
Crystals were grown at 21 °C by hanging-drop vapor-diffusion using a 1:1 mixture of protein (3–5 mg/ml) and crystallization solutions containing 100 mM Hepes or Mes pH 5.9 to 7.8, 200 mM MgCl2, and 20 to 26% PEG 3350. Crystals appeared within 2 to 4 days. Cryo protection was achieved by adding 20% glycerol. Multiple wavelength anomalous dispersion experiments were recorded at beamline ID14–4 [European Synchrotron Radiation Facility (ESRF) Grenoble, France] and native datasets at beamline ID14–1 (ESRF, Grenoble, France). Data were integrated and scaled with XDS (28). Phases were obtained with Crank (Crunch2/BP3/Solomon) (29, 30), and extended to 2.1 Å resolution. Parts of the final model were automatically build with Buccaneer (31) and manually completed using COOT (32). Refinement of the native data was performed with Refmac (33, 34), using noncrystallographic symmetry. The final model was analyzed using SFCHECK (35).
Structure Visualization and Analysis.
Images of the crystal structures were prepared with PyMol (DELano), electrostatic surface potentials represented with CCP4 mg, and buried surface areas calculated with Areaimol (29). Surface plot of sequence conservation was prepared with Chimera (36).
DNA- and RNA-Binding Assays.
DNAs and RNAs were obtained by total chemical synthesis. EMSAs were essentially performed as previously described (37). Protein was transferred to DEPC-treated Binding Buffer [50 mM Tris-HCl (pH 7.4 at 4 °C), 150 mM KCl, 1 mM EDTA, 1 mM MgCl2, and 1 mM DTT] and monitored for proper folding by size-exclusion chromatography. Serial protein dilutions and a constant amount of radiolabeled DNA/RNA (2.5 nM) were incubated in Binding Buffer on ice for 20 min. DNA-binding experiments contained 25 μg/ml PolydI/PolydC competitor, and RNA-binding experiments contained 25 μg/ml yeast tRNA as well as 200 U/ml RiboLock RNase inhibitor. Reaction mixtures (15 μl) were loaded onto 6% TBE polyacrylamide gels and analyzed after electrophoresis (35 min, 110 V) by phosphoimaging. Sequences of oligonucleotides were as follows: MF0677 ssDNA, 5′-GGA GGT GGT GGA GGG AGA GAA AAG-3′; Oligo-A ssDNA, A(24); CGG ssRNA, 5′-CGG CGG CGG CGG-3′; Oligo-A ssRNA, A(12).
Size-Exclusion Chromatography.
Pur-α (0.5 ml) was loaded onto Superose 12 10/300 GL column in buffer with 20 mM Hepes (pH 8.0) and 500 mM NaCl (flow rate: 0.9 ml/min). Column calibration was performed with a size-exclusion calibration kit (BioRad). Absence of nucleic acid contamination was confirmed by measuring the A260/A280 ratio.
Small-Angle X-Ray Scattering.
Synchrotron SAXS data were collected at the X33 beamline (European Molecular Biology Laboratory/Deutsches Elektronen Synchrotron) and at the ID14–3 beamline (ESRF, Grenoble, France). Scattering curves were measured in 20 mM Hepes (pH 8.0), 500 mM NaCl, and 3 mM DTT with exposure times of 2 min (X-33) and 10 times 30 s (ID14–3), respectively. Data analysis was performed using ATSAS (38). For molecular-mass determination, scattering intensities were extrapolated to zero angle (I0), using BSA and lysozyme as references. The radius of gyration Rg was calculated using the Guinier approximation with the constraint s*Rg < 1.3. GNOM was used to calculate p(r) and Dmax. The correct Dmax was iteratively determined by evaluating the resulting Rg value and the shape of the p(r)-distribution. Bead models were calculated with GASBORp, and for Pur-α (I–II) overlayed with the crystal structure using SUPCOMB. CRYSOL was used to determine the theoretical scattering curve based on the Pur-α crystal structure.
Supplementary Material
Acknowledgments.
We thank Verena Aumiller, Thomas Carell, Klaus Förstemann, Daniela Hüls, Andrea Hildebrand, Sigrun Jaklin, Marisa Müller, Johannes Soeding, Sabine Ströbl, and Gregor Witte for their help; Stephen K. Burley for his support during the early stage of this project; Dirk Kostrewa for his continuous support during structure determination; and the crystallization facility of the Max Planck Institute for Biochemistry (Martinsried, Germany). We acknowledge the European Molecular Biology Laboratory/Deutsches Elektronen Synchrotron and European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and thank Dmitry Svergun at DESY-X33 for support and Adam Round and Petra Pernot for assistance in using beamline ID14–3. This work was supported by the Helmholtz Association (VG-NH 142 to D.N.) and the Deutsche Forschungsgemeinschaft (FOR855 and SFB646 to D.N.). Stéphane Roche is a fellow of the Human Frontiers Science Program Organization.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3K44).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907990106/DCSupplemental.
References
- 1.Gallia GL, Johnson EM, Khalili K. Puralpha: A multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White MK, Johnson EM, Khalili K. Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle. 2009;8:1–7. doi: 10.4161/cc.8.3.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Ohashi S, et al. Identification of mRNA/protein (mRNP) complexes containing Pur-alpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 5.Jin P, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalili K, et al. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen NN, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 9.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krachmarov CP, Chepenik LG, Barr-Vagell S, Khalili K, Johnson EM. Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Pur alpha. Proc Natl Acad Sci USA. 1996;93:14112–14117. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chepenik LG, Tretiakova AP, Krachmarov CP, Johnson EM, Khalili K. The single-stranded DNA binding protein, Pur-alpha, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene. 1998;210:37–44. doi: 10.1016/s0378-1119(98)00033-x. [DOI] [PubMed] [Google Scholar]
- 12.Daniel DC, et al. Coordinate effects of human immunodeficiency virus type 1 protein Tat and cellular protein Puralpha on DNA replication initiated at the JC virus origin. J Gen Virol. 2001;82:1543–1553. doi: 10.1099/0022-1317-82-7-1543. [DOI] [PubMed] [Google Scholar]
- 13.Conte LL, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 14.Dasgupta S, Iyer GH, Bryant SH, Lawrence CE, Bell JA. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins. 1997;28:494–514. doi: 10.1002/(sici)1097-0134(199708)28:4<494::aid-prot4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 15.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 17.Desveaux D, Allard J, Brisson N, Sygusch J. A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat Struct Biol. 2002;9:512–517. doi: 10.1038/nsb814. [DOI] [PubMed] [Google Scholar]
- 18.Bergemann AD, Ma ZW, Johnson EM. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol Cell Biol. 1992;12:5673–5682. doi: 10.1128/mcb.12.12.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wortman MJ, Johnson EM, Bergemann AD. Mechanism of DNA binding and localized strand separation by Pur alpha and comparison with Pur family member, Pur beta. Biochim Biophys Acta. 2005;1743:64–78. doi: 10.1016/j.bbamcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Muller M, et al. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA. 2009 doi: 10.1261/rna.1753309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandsen J, et al. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat Struct Biol. 1997;4:900–903. doi: 10.1038/nsb1197-900. [DOI] [PubMed] [Google Scholar]
- 22.Darbinian N, Gallia GL, Khalili K. Helix-destabilizing properties of the human single-stranded DNA- and RNA-binding protein Puralpha. J Cell Biochem. 2001;80:589–595. [PubMed] [Google Scholar]
- 23.Lambert L, Muller UF, Souza AE, Goringer HU. The involvement of gRNA-binding protein gBP21 in RNA editing-an in vitro and in vivo analysis. Nucleic Acids Res. 1999;27:1429–1436. doi: 10.1093/nar/27.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller UF, Lambert L, Goringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koller J, et al. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J Biol Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- 26.Heuck A, et al. Monomeric myosin V uses two binding regions for the assembly of stable translocation complexes. Proc Natl Acad Sci USA. 2007;104:19778–19783. doi: 10.1073/pnas.0706780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doublie S. Preparation of selenomethionyl proteins for phase determination. In: Charles W, Carter J, Sweet RM, editors. Methods in Enzymology. Vol 276. San Diego: Academic Press; 1997. pp. 523–537. [PubMed] [Google Scholar]
- 28.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 29.Collaborative Computational Project N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Ness SR, de Graaff RA, Abrahams JP, Pannu NS. CRANK: New methods for automated macromolecular crystal structure solution. Structure. 2004;12:1753–1761. doi: 10.1016/j.str.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr D. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 35.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 36.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Third Edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 38.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS2.1, a program package for small-angle scattering data analysis. J Appl Cryst. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.