Abstract
Technological developments make mass spectrometry (MS)-based proteomics a central pillar of biochemical research. MS has been very successful in cell culture systems, where sample amounts are not limiting. To extend its capabilities to extremely small, physiologically distinct cell types isolated from tissue, we developed a high sensitivity chromatographic system that measures nanogram protein mixtures for 8 h with very high resolution. This technology is based on splitting gradient effluents into a capture capillary and provides an inherent technical replicate. In a single analysis, this allowed us to characterize kidney glomeruli isolated by laser capture microdissection to a depth of more than 2,400 proteins. From pooled pancreatic islets of Langerhans, another type of “miniorgan,” we obtained an in-depth proteome of 6,873 proteins, many of them involved in diabetes. We quantitatively compared the proteome of single islets, containing 2,000–4,000 cells, treated with high or low glucose levels, and covered most of the characteristic functions of beta cells. Our ultrasensitive analysis recapitulated known hyperglycemic changes but we also find components up-regulated such as the mitochondrial stress regulator Park7. Direct proteomic analysis of functionally distinct cellular structures opens up perspectives in physiology and pathology.
Keywords: hyperglycemia, liquid chromatography-mass spectrometry, quantitative proteomics, replay technology
Mass spectrometry (MS) is inherently an extremely sensitive analysis technique: single proteins have been analyzed at the attomole level many years ago (1, 2). Similarly, it is now possible to identify thousands of proteins in single experiments and identification and quantitation of a comprehensive proteome has been reported (3). However, the complexity of proteomic mixtures typically necessitates micrograms or milligrams of starting material—protein amounts that can readily be extracted from cell or tissue homogenates (4, 5). The analysis of small tissue substructures with specialized functions is medically important but has remained much more difficult. Ideally these structures are separated from the surrounding tissue to enrich proteins specific to these areas. Although such selection is possible by laser-capture microdissection (LCM), successful use of this technology has so far overwhelmingly been in the areas of RNA and DNA assays. In the few cases where LCM was combined with MS analysis, many thousands cells were collected (taking presumably several hours or days of selection), to obtain the protein amounts typical for MS (6). Only a few investigators have attempted to measure smaller cell numbers. Luider and coworkers reported 1,000 identified proteins from 3,000 cells and a similar number from LCM-selected breast cancer tissues, but they did not sequence peptides by mass spectrometry, relying instead on mass measurements only (7, 8).
Main obstacles in analyzing small protein amounts are the considerable sample losses during preparation and the limited “dynamic range” of liquid chromatography-tandem mass spectrometry (LC-MS/MS) systems used in proteomics. Sample preparation efficiency has improved by the advent of polished sample tubes (e.g., Eppendorf Protein LoBind) and MS-friendly detergents (9, 10), thereby allowing shorter extraction and digestion protocols and reduced unspecific surface binding.
The second important obstacle, the dynamic range, refers to the number of different ion species that can be observed simultaneously. With highly complex samples, as are typical for proteomic experiments, low intensity ions are likely to be masked by more abundant species. To also detect these low abundance proteins, extensive fractionation is commonly applied. But this is not possible with very low starting amounts as sample losses would be prohibitive.
Recently, we reported the development of a chromatographic system that allows direct reanalysis of the injected sample without losing signal intensity (11). In this LC-MS interface, termed RePlay, post-column effluent is split with one part directed to on-line LC-MS/MS analysis and the other one stored in a capture capillary for another round of analysis after the first one is completed. We envisioned that this setup should also be valuable for analyses where starting amount is limiting, and therefore developed the system for high sensitivity “single shot” analysis. We used optimized sample preparation methods and the RePlay system in combination with the sensitive LTQ-Orbitrap mass spectrometer. Due to the split, flow rates could be reduced 5-fold compared to normal operation in our laboratory while maintaining normal backpressures and a continuous stable flow and spray. Importantly, the re-analysis of the sample does not lose signal intensity because the peptide concentration is not affected by the split. Thus, we inherently obtain a technical replicate, increasing quantitation accuracy, even from the smallest proteomic samples. In addition the doubled gradient time increases peptide sequencing events per sample.
Here we demonstrate that our workflow enables the analysis of small “miniorgans:” we examined mouse glomeruli selected by LCM and single islets of Langerhans, handpicked after purification. In both cases the starting material was less than 400 ng of proteins, —but still resulted in more than 2,000 protein identifications. Besides high confidence identifications, we demonstrate quantitation of protein expression levels in single islets of Langerhans. These islets were stimulated with high or low glucose before LC-MS/MS and we identified many responders, like anti-oxidants and proteins involved in glucose catabolism or vesicle secretion processes.
Results and Discussion
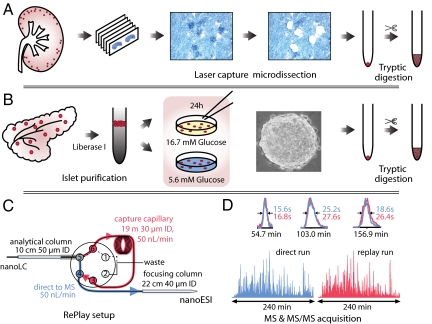
There are two challenges in the measurement of very small in vivo cell populations by liquid chromatography-tandem mass spectrometry (LC-MS/MS): efficient recovery of the proteins and high analytical sensitivity. To meet the first challenge, we implemented a workflow with a minimal number of handling steps starting from LCM (Fig. 1A) or miniorgan purification (Fig. 1B) and resulting in a trypsin digested proteome (Materials and Methods). LC-MS/MS sensitivity was improved in three ways (Fig. 1C): (i) by splitting the flow after chromatographic peptide separation, thereby providing a technical replicate from a single sample loading without loss of signal [RePlay (11)]; (ii) by reducing the effective flow rate to 50 nL/min and employing smaller diameter columns; and (iii) by separating the peptide mixture with long gradients (4 h) to increase peptide sequencing events by the high-resolution LTQ-Orbitrap mass spectrometer.
Fig. 1.
Schematic workflow for high-sensitive LC-MS analysis with direct replicate measurement. (A) Workflow for laser capture microdissection for mouse kidney glomeruli isolation. (B) Purification of islets of Langerhans from mouse pancreas by Liberase I treatment and gradient centrifugation. Islets were stimulated with high or low glucose, individually collected, and proteins solubilized and digested by trypsin. (C) High sensitivity nanoLC-MS setup with the RePlay to obtain two measurements per injection. (D) Total ion chromatogram in direct and replicate analysis are very similar. Insets show identical resolution and intensity for three typical peptide peaks.
First, we tested this high-sensitivity workflow on mouse glomeruli, about 50 of which we isolated by LCM from histological slices (Materials and Methods). Glomeruli are the filtering units in the kidney and consist of only a few hundred cells in mice. Automated analysis proved robust and the chromatographic setup retained excellent average peak width (< 22s) in direct and replicate (“replay”) runs (Fig. 1D). Overall signal was the same in both runs and individual peptide intensities, which ranged over more than four orders of magnitude, correlated highly between them (R = 0.92) (Fig. S1A). Data processing with MaxQuant (12) resulted in the identification of 2406 distinct proteins from a single injection. Virtually all known glomeruli proteins are contained in this dataset (Table S1). Repeat measurements resulted in 2,402 and 2,377 proteins identified and the three datasets shared 86% of protein identifications (Fig. S1B). For comparison, our recent proteomic analysis of 20 fractions of mouse liver tissues using 1,000-fold more material resulted in a similar number of identifications (13).
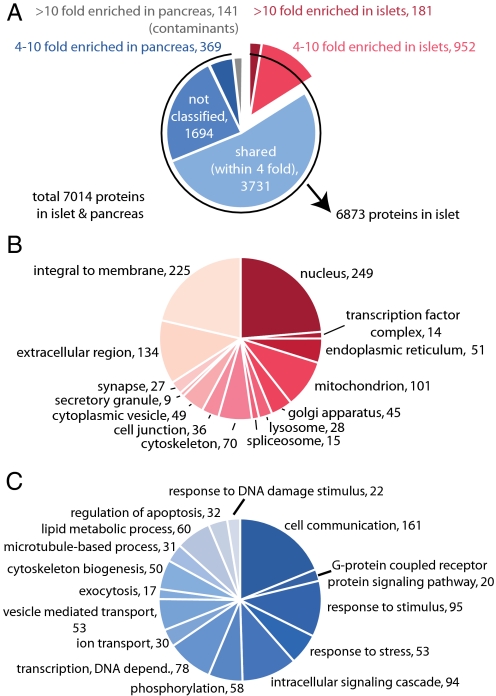
Islets of Langerhans are another type of miniorgan essential for human health. In mice, 2,000 islets are dispersed throughout the pancreas of which the insulin producing β cell is the most predominant (14). To obtain an in-depth islet proteome we pooled purified islets and analyzed them after peptide fractionation (Materials and Methods). Triplicate analysis employing RePlay and material from six mice resulted in the identification of 7,014 proteins with at least two peptides and a False Discovery Rate (FDR) of 1% (Table S2). The largest previous islet proteome is less than half the size and almost completely contained in our proteome (94%) (15). This list consists of (i) islet specific proteins, (ii) proteins shared between islets and the rest of the pancreas, and (iii) proteins from the rest of the pancreas that contaminate the islet preparation. To distinguish these classes we measured an in-depth proteome of complete pancreas homogenate, of which the islets constitute less than 2%, and quantified proteins against the islet proteome by label-free quantification in MaxQuant. Elastase, trypsin, and other digestive enzymes produced by the exocrine pancreas were more than 30-fold more abundant in the total pancreas proteome compared to purified islets. After filtering out proteins with a similar behavior, we retain a high quality islet proteome of 6,873 proteins. Our data indicate that islets, which are highly specialized tissue structures, use at least a third of the genes in the genome. Moreover, several proteins were identified as the pancreas specific isoform of the gene, and our data provides isoform specific tissue expression information for many more.
The known endocrine hormones produced by this miniorgan, such as insulin 1 and 2, glucagon, and secretogranins are among the most highly expressed proteins in the proteome judged by added peptide intensity (3) (Table S2). To contrast islet specific expression to expression in whole pancreas we categorized islet proteins by their enrichment factors (Fig. 2A). More than half of the islet proteome is within a 4-fold abundance range compared to whole pancreas, and these proteins are predominantly associated with house-hold functions. The pancreatic islet is an intricately assembled structure connecting the endocrine cells to the circulation and neural control. Concordantly, proteins connected to cell communication and cell adhesion make up much of the specific islet proteome. In the 1,133 proteins enriched more than 4-fold in islets, membrane and extracellular proteins comprise a large part (Fig. 2B and Table S3). This class of proteins contains various G protein coupled receptors, cAMP regulators, receptor tyrosine kinases and ion channels–potential and established targets for diabetes drugs, as well as autoantigens in type I diabetes (Fig. 2 B and C and Fig. S1C). Interestingly, 86 of 396 identified kinases are highly enriched in the islets. Our dataset also contains many unannotated proteins, especially among those that were close to the limit of detection and therefore not quantified and categorized (Fig. 2A).
Fig. 2.
The proteome of pooled islets of Langerhans. (A) Classification of purified islet proteome versus pancreatic proteome, based on protein ratios between the two samples. After subtracting pancreas contaminants (141), a high confidence list of 6,873 islet proteins was obtained. Note that for proteins with less than three peptides and close to the limit of detection, accurate ratios and hence classification were not assigned (“not classified”). (B) Proteins enriched more than 4-fold (total 1,133) were categorized by GO cellular compartmentalization and (C) GO biological process. Numbers indicate proteins per category in the enriched dataset.
High fat and high glucose conditions are reported to negatively affect β cell function, a process that is central to the development of type 2 diabetes (16). Dynamics of β-cells are frequently studied in cell culture; however, it is physiologically more relevant to investigate these cells in their native cellular context (17). We wondered if our high sensitivity workflow was capable of analyzing single pancreatic islets. To this end, we determined single, isolated islet proteomes after 24 h at 16.7 and 5.6 mM glucose, representing hyperglycemic and basal conditions, respectively. One day of elevated glucose concentration significantly affects metabolic functions at the mRNA level (18), but does not yet evoke irreversible changes (19).
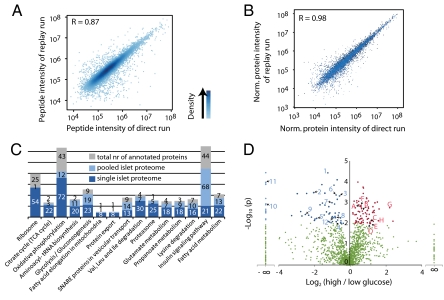
First we assessed the proteome coverage in single islet experiments. Similar to the case of glomeruli we achieved very good correlation on the peptide level between direct and replicate runs (Fig. 3A). Protein quantitation integrates several peptide measurements and therefore the quantitative reproducibility is even higher on the protein level (R = 0.98) (Fig. 3B). We sequenced 15,193 different peptides originating from 2,013 unique proteins (2.5% FDR). Per biological replicate we identified on average 84% of the total number of proteins. Ninety-seven percent of the single islet proteins (1,949) were also present in the large islet proteome determined above (Fig. S1D). The single islet proteome contained all of the islet specific hormones as well as known β cell proteins like glucose transporter 2, glucokinase, and prohormone convertase 1 and 2. Furthermore, we covered the majority of proteins involved in classical β cell functions (Fig. 3C and Fig. S1 E–G); for example, between 40 and 85% of the annotated proteins (irrespective of tissue) in glycolysis, TCA cycle, and oxidative phosphorylation, ribosomes, and secretory granules (Fig. 4).
Fig. 3.
Quantitative analysis of single islets by MS-based proteomics. (A) Correlation of peptide intensities between direct and replay runs of one single islet measurement. (B) Comparison of six direct and replay runs indicates very high quantitative reproducibility at the protein level. (C) Annotation of single islet and pooled-islet proteome to KEGG pathways important for β cell function. Numbers indicate identified proteins within annotated group. The single islet proteome covers most essential β cell processes like glycolysis, TCA cycle and oxidative phosphorylation. (D) Volcano plot showing P values (-log10) versus protein ratio of high/low glucose-treated islets (log2) of all 1,482 proteins fulfilling strict quantitation criteria. (Red, 77 up-regulated proteins; blue, 65 down-regulated proteins; green, not significantly changed upon 24-h glucose stimulation; ANOVA with P < 0.01). Gene names of proteins discussed in the text: A, ANT1; B, Ero1lα; C, Prdx3; D, Park7; E, Gsn; F, Vil1; G, Actn1; H, Itgb1; 1, Pcsk1n; 2, Pdyn; 3, ATP2a2; 4, ATP2a3; 5, Ins2; 6, Sytl4; 7, Vamp2; 8, Vamp8; 9, Scamp3; 10, Rab3b; 11, Capza1; 12, Capza2.
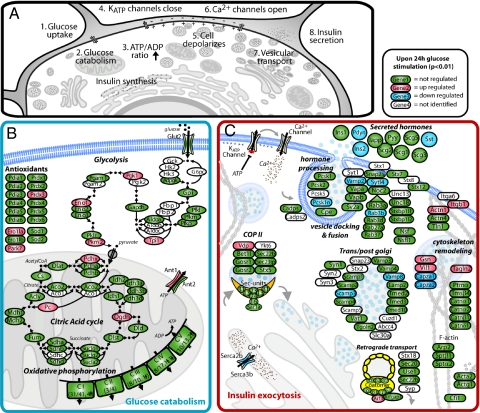
Fig. 4.
Main differentially expressed proteins upon 24-h glucose stimulation in the context of physiological processes related to glucose stimulated insulin secretion. (A) Cellular events leading to insulin secretion after normal glucose stimulation. Two of the affected processes are presented in further detail, showing proteins (gene name) with the following color-coding: green, identified, red, up-regulated, blue, down-regulated, and white, not identified. (B) Glucose uptake, glycolysis, TCA cycle, oxidative phosphorylation, and antioxidants. (C) Vesicle transport and cytoskeleton remodeling.
Next we determined changes of the proteome after 24-h glucose stimulation. Quantitative comparison was based on proteins identified with greater than or equal to two unique peptides and detected in at least four out of eight runs (1,487 quantifiable proteins). With these stringent criteria and ANOVA P < 0.01, there were 77 up- and 65 down-regulated proteins. Some of the most significantly affected proteins are highlighted in the volcano plot in Fig. 3D. Many regulated proteins have previously been associated with hyperglycemia in animal models for diabetes or in patients (Table S4). Concordant with a recently published microarray study (18) we find a general up-regulation of glycolysis, the TCA cycle and ATP translocation (Fig. 4). In contrast to elevated insulin transcription, we observed reduced insulin protein levels, presumably due to depletion of insulin from internal stores as a consequence of continuous secretion. This assumption is indirectly supported by the finding of decreased levels of two hormone-processing inhibitors, prodynorphin (Pdyn) and ProSAAS (Pcsk1n) (20).
Increased glucose catabolism induces oxidative stress, a process detrimental to β-cells. We find molecular evidence for this in the form of antioxidant proteins, supporting and expanding previous observations (e.g., ERO1l-α and Prdx3) (Figs. 3 and 4) (21). Prdx3, up-regulated 2.4-fold, protects β-cells from mitochondrial hydrogen peroxide stress and mice overexpressing Prdx3 are more glucose tolerant upon high-fat diet feeding and better protected against hyperglycemia (22). Interestingly, DJ-1 (Park7) and Mn2+-superoxide dismutase (Sod2) were up-regulated 2- and 1.7-fold (P < 0.05), respectively. DJ-1 is a mitochondrial oxidative stress and apoptotic regulator implicated in Parkinson disease (23, 24) but has not yet been associated with β cell responses in hyperglycemia. It induces mitochondrial antioxidant proteins like Sod2 (25). To explore DJ-1's connection to β cell dysfunction, we quantified its expression in mouse islets after high-fat diet and found the protein up-regulated 1.7-fold (Fig. S1H)–similar to the short term hyperglycemic stimulation, suggesting that it is involved in β cell regulation under multiple stress conditions.
In contrast to the increased protein levels in glucose catabolism and induced stress mechanisms described above, proteins associated with vesicle secretion are reduced (Fig. 4). In the case of vesicle-associated membrane protein (VAMP)-2, synaptotagmin-like protein 4 (Sytl4), and Rab3b, who interact upon Ca2+ increase, this has been observed before (26, 27). Decreased expression of Sytl4 reduces the number of docked vesicles to the plasma membrane, which is suggested to effect insulin secretion (19, 28). Decreased mRNA and protein expression of VAMP2 and various other secretory proteins have also been associated with impaired protein secretion in diabetic patients (29) and animal models for diabetes (30). Furthermore, we observed depletion of all three members of the sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA) complex, which has also been correlated with insulin secretion deficiencies in diabetic animal models (31). In contrast, mRNA levels of SERCA increase after 18 h of glucose stimulation (18), possibly pointing to a compensatory mechanism for acute protein depletion.
Vesicle access to the plasma membrane is thought to be hindered by dense cortical F-actin and depolymerization of F-actin upon glucose stimulation potentiates insulin secretion (32). Supporting this view, we observed altered levels of proteins involved in remodeling the cortical cytoskeleton network—the depolymerizing enzymes Villin 1 and Gelsolin are up and inhibitory enzymes Capza 1 and 2 are down-regulated. Gelsolin's role in this process has already been demonstrated (33). Moreover, α-actinin-1 and 4 were up-regulated. Actinin-1 associates with cell adhesion molecules like integrin-β1, which was increased 3-fold and plays an important role in β cell extracellular matrix interaction (34). β1-integrin is an essential factor for β cell proliferation and communicates with laminins in the basement membrane (35). Consistently, there is a direct relation between its expression level and the rate of insulin secretion upon glucose stimulation (36). Thus, our single islet analysis directly supports an important role of cytoskeletal remodeling in high glucose conditions.
In conclusion, by optimizing the proteomic workflow, using reduced flow rates, direct replicate measurements and extremely accurate and highly sensitive mass spectrometry, in vivo structures comprising a few thousand cells can now be quantitatively analyzed. Our study highlights a beneficial aspect of this directed analysis: the functionally relevant proteome turns out to be expressed highly in these specialized cells and is, by avoiding the dilution effect of whole tissue analysis, more readily accessible to proteomic measurements. Already, the method described here could be used to evaluate single islets developed from in vitro differentiated stem cells or disease control of a few islets taken from human biopsies. The flow rate and column diameter can be significantly lowered (37) and MS sensitivity and scan speed increased (38). Thus, a few hundred cells are analyzable or alternatively depth of coverage can be increased significantly. This allows in depth proteomic analysis of virtually all features visible by standard histological methods.
Materials and Methods
LCM Workflow.
Cryosections of mouse kidney were cut (12–16 μm thick), placed on glass sections and stained with methylene blue. Per sample, about 50 glomeruli were selected (tissue areas of ≈500,000 μm2) and dissected by a MicroBeam laser (Carl Zeiss MicroImaging).
Pooled Islets and Total Pancreas Homogenate.
Pancreatic islets were isolated from 12–16-week-old C57Bl6/J male mice. Islet preparation was done as described in (35). Before removal of the pancreas, Liberase I solution was injected into the pancreatic duct (≈2 mL/mouse). The pancreas was incubated at 37 °C for 23 min after which the digestion was stopped by adding media supplemented with 10% FBS, followed by 3 min centrifugation at 1,000 × g. Low glucose DMEM was added to the pellet, mixed vigorously, and filtered although a 400-μm diameter wire mesh. Remaining islets were washed once. The islets were spun down and resuspended in histopaque 1077 (Sigma). Low glucose DMEM was added carefully, before centrifugation for 50 min at 1,100 × g at 10 °C without acceleration or braking. The islet layer was collected and washed twice with DMEM to remove histopaque. Total pancreas tissue was obtained by dissecting directly after pancreas excision.
Single Islet Preparation.
Pancreatic islets were isolated from C57Bl6/J male mice, 8–10 weeks old as described above. Purified islets were recovered overnight in 11 mM glucose CMRL medium containing 15% heat inactivated fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, 1 mM sodium bicarbonate, and 0.001% beta-mercaptoethanol. Subsequently, the cells were incubated for 24 h with either 16.7 mM (high) or 5.6 mM (low) glucose in Krebs-Ringer-Hepes buffer (25 mM Hepes, pH 7.4, 115 mM NaCl, 24 mM NaHCO3, 5 mM KCl, 2.5 mM CaCl2, 1 mM MgCl2, and 0.1% BSA). Single islets were handpicked.
Sample Digestion.
LCM and single islet samples were dissolved in 0.1% PPS Silent Surfactant (Protein Discovery) in 50 mM ammonium bicarbonate (ABC) with 1 mM DTT, heated to 95 °C for 3 min, and sonicated for 2 min. Subsequently proteins were alkylated in 1 mM chloroacetamide in ABC for 30 min and digested overnight with porcine trypsin (Promega) in an enzyme:protein ratio of 1:50. After acidification by trifluoroacetic acid, peptide mixtures were concentrated and desalted on StageTips (39) and eluted directly before LC-MS measurement. To prevent protein losses during sample preparation LoBind tubes (Eppendorf) and Maxymum Recovery tips (Axygen) were used and sample contact was minimized.
For pooled islet and pancreas proteome the peptide samples were additionally separated into 12 fractions by peptide isoelectric focusing as described (40).
High Sensitivity LC-MS Interface.
The RePlay system was based on the one described in (11), but developed for high sensitivity purposes: the capture capillary was 19 m long × 30 μm inner diameter (ID); the focusing column, a pulled capillary of 22 cm × 40 μm ID was packed with 3 μm ReproSil-Pur C18-AQ resin (Dr. Maisch GmbH), as described in (41). For the LCM and single islet measurements we used a split ratio of 1:1 (MS:capture capillary), an effective flow rate of 50 nL to the MS in both direct and replicate (or replay) run, and 240 min gradients. For the pooled islet and pancreas proteome gradients of 2 h were used with 150 nL/min effective flow rates. The flow sensor was placed in the waste line to prevent potential dilution effects.
The gradient generated by an EasyLC (Proxeon) was coupled, via the RePlay system and an electrospray interface, to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). MS and MS/MS were acquired data-dependently of the ten most intense ions per survey spectrum, using MS/MS target values of 5,000, with a minimal intensity of 1,000 ions and a maximum fill time of 150 ms. For survey spectra one million ions were accumulated with maximum fill times of 500 ms.
Data Analysis.
MS/MS data were analyzed by in-house developed MaxQuant software (v 1.0.12.27), essentially as described (12). The data were searched using Mascot (v 2.1.04, Matrix Science Ltd) against the mouse IPI database (v 3.37) supplemented with frequently observed contaminants and concatenated with reversed copies of all sequences (2 × 51,467 entries). Enzyme specificity was set to trypsin, allowing for cleavage N-terminal to proline and between aspartic acid and proline and a maximum of two missed cleavages. Carbamidomethylcysteine was set as fixed and N- acetylation and methionine oxidation were set as variable modifications. The initial maximum allowed mass deviation was set to 7 ppm for monoisotopic precursor ions and 0.5 Da for MS/MS peaks. The required minimum peptide length was six amino acids. If the identified peptide sequence set of one protein was equal to or contained another protein's peptide set, these two proteins were grouped together and the proteins were not counted as independent hits. Proteins were considered identified when at least two peptides were identified of which at least one was uniquely assignable to the respective sequence. The false discovery rate (FDR) at the peptide level and protein level were set to 1% for the glomeruli, pooled islet, and pancreas proteome. Based on the additional identification confidence provided by the pooled islet dataset, we accepted single unique peptide identifications with 5% FDR on peptide and 2.5% on protein level. For quantitation proteins were additionally filtered, and were required to be detected in four out of eight most intense LC-MS/MS runs with at least two unique peptides.
Label-free quantitation was performed in MaxQuant. Briefly, total peptide signals were determined in the mass to charge, elution time and intensity space. For every peptide, corresponding total signals from multiple runs were compared to determine peptide ratios. Pair-wise peptide ratios were only determined when the corresponding peak is detected in both LC-MS runs. Median values of all peptide ratios of one protein then represent a robust estimate of the protein ratio. MaxQuant determines these ratios after first applying a normalization procedure to determine total peptide signal. Because proteins with low intensity signal are typically determined less accurately than high abundant proteins, we applied “Analysis of Variance.” ANOVA correlates the variation within the replicates of the treated and non-treated samples with the variation between the sample groups. This is superior to defining arbitrary significance to fold-changes depending on the protein signal. By application of a very stringent cut-off value (P < 0.01) low intense peptides and proteins are efficiently removed from the “significant” dataset.
Supplementary Material
Acknowledgments.
We thank Tami Geiger, Ana Velic, and Gabriele Friedemann for assisting with glomeruli collection and data analysis; Jürgen Cox for help with the data analysis; Irena Constantinova for providing the single islet picture in Fig. 1; members of the Proteomics and Signal Transduction group for help and discussions; and Tobias Walther, Maximiliane Hilger, Nina Hubner, and Tami Geiger for critical reading of the manuscript. This project was supported by European Commission's 7th Framework Program PROteomics SPECification in Time and Space (PROSPECTS, HEALTH-F4-2008-021648), and German Research Council Grant DFG LA1216 (to K.C. and E.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908351106/DCSupplemental.
References
- 1.Valaskovic GA, Kelleher NL, McLafferty FW. Attomole protein characterization by capillary electrophoresis-mass spectrometry. Science. 1996;273:1199–1202. doi: 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 3.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 4.Yates JR, 3rd, Gilchrist A, Howell KE, Bergeron JJ. Proteomics of organelles and large cellular structures. Nat Rev Mol Cell Biol. 2005;6:702–714. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- 5.Mann M, Kelleher NL. Precision proteomics: The case for high resolution and high mass accuracy. Proc Natl Acad Sci USA. 2008;105:18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espina V, Heiby M, Pierobon M, Liotta LA. Laser capture microdissection technology. Exp rev mol diagn. 2007;7:647–657. doi: 10.1586/14737159.7.5.647. [DOI] [PubMed] [Google Scholar]
- 7.Umar A, Luider TM, Foekens JA, Pasa-Tolic L. NanoLC-FT-ICR MS improves proteome coverage attainable for ≈3,000 laser microdissected breast carcinoma cells. Proteomics. 2007;7:323–329. doi: 10.1002/pmic.200600293. [DOI] [PubMed] [Google Scholar]
- 8.Umar A, et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics. 2009;8:1278–1294. doi: 10.1074/mcp.M800493-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norris JL, Porter NA, Caprioli RM. Methods for direct biomolecule identification by matrix-assisted laser desorption ionization (maldi) mass spectrometry. Anal Chem. 2003;75:6642–6647. doi: 10.1021/ac034802z. [DOI] [PubMed] [Google Scholar]
- 10.Yu YQ, Gilar M, Kaska J, Gebler JC. A rapid sample preparation method for mass spectrometric characterization of N-linked glycans. Rapid Commun Mass Spectrom. 2005;19:2331–2336. doi: 10.1002/rcm.2067. [DOI] [PubMed] [Google Scholar]
- 11.Waanders LF, et al. A novel chromatographic method allows on-line reanalysis of the proteome. Mol Cell Proteomics. 2008;7:1452–1459. doi: 10.1074/mcp.M800141-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 13.Shi R, et al. Analysis of the mouse liver proteome using advanced mass spectrometry. J Proteome Res. 2007;6:2963–2972. doi: 10.1021/pr0605668. [DOI] [PubMed] [Google Scholar]
- 14.Kulkarni RN. The islet β-cell. Int J Biochem Cell Biol. 2004;36:365–371. doi: 10.1016/j.biocel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Petyuk VA, et al. Characterization of the mouse pancreatic islet proteome and comparative analysis with other mouse tissues. J Proteome Res. 2008;7:3114–3126. doi: 10.1021/pr800205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahn SE. The importance of the beta-cell in the pathogenesis of type 2 diabetes mellitus. Am J Med. 2000;108:2S–8S. doi: 10.1016/s0002-9343(00)00336-3. [DOI] [PubMed] [Google Scholar]
- 17.Kinard TA, de Vries G, Sherman A, Satin LS. Modulation of the bursting properties of single mouse pancreatic—Cells by artificial conductances. Biophys J. 1999;76:1423–1435. doi: 10.1016/S0006-3495(99)77303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bensellam M, Van Lommel L, Overbergh L, Schuit FC, Jonas JC. Cluster analysis of rat pancreatic islet gene mRNA levels after culture in low-, intermediate-, and high-glucose concentrations. Diabetologia. 2009;52:463–476. doi: 10.1007/s00125-008-1245-z. [DOI] [PubMed] [Google Scholar]
- 19.Tsuboi T, Ravier MA, Parton LE, Rutter GA. Sustained exposure to high glucose concentrations modifies glucose signaling and the mechanics of secretory vesicle Fusion in primary rat pancreatic β-cells. Diabetes. 2006;55:1057–1065. doi: 10.2337/diabetes.55.04.06.db05-1577. [DOI] [PubMed] [Google Scholar]
- 20.Fricker LD, et al. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J Neurosci. 2000;20:639–648. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekkers DH, et al. Identification by a differential proteomic approach of the induced stress and redox proteins by resveratrol in the normal and diabetic rat heart. J Cell Mol Med. 2008;12:1677–1689. doi: 10.1111/j.1582-4934.2008.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, et al. Reduction of mitochondrial H2O2 by overexpressing peroxiredoxin 3 improves glucose tolerance in mice. Aging Cell. 2008;7:866–878. doi: 10.1111/j.1474-9726.2008.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hague S, et al. Early-onset Parkinson's disease caused by a compound heterozygous DJ-1 mutation. Ann Neurol. 2003;54:271–274. doi: 10.1002/ana.10663. [DOI] [PubMed] [Google Scholar]
- 24.Yokota T, et al. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312:1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 25.Zhong N, Xu J. Synergistic activation of the human MnSOD promoter by DJ-1 and PGC-1alpha: Regulation by SUMOylation and oxidation. Hum Mol Genet. 2008;17:3357–3367. doi: 10.1093/hmg/ddn230. [DOI] [PubMed] [Google Scholar]
- 26.Abderrahmani A, et al. ICER induced by hyperglycemia represses the expression of genes essential for insulin exocytosis. EMBO J. 2006;25:977–986. doi: 10.1038/sj.emboj.7601008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois M, et al. Glucotoxicity inhibits late steps of insulin exocytosis. Endocrinology. 2007;148:1605–1614. doi: 10.1210/en.2006-1022. [DOI] [PubMed] [Google Scholar]
- 28.Torii S, Zhao S, Yi Z, Takeuchi T, Izumi T. Granuphilin modulates the exocytosis of secretory granules through interaction with syntaxin 1a. Mol Cell Biol. 2002;22:5518–5526. doi: 10.1128/MCB.22.15.5518-5526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostenson CG, Gaisano H, Sheu L, Tibell A, Bartfai T. Impaired gene and protein expression of exocytotic soluble N-ethylmaleimide attachment protein receptor complex proteins in pancreatic islets of type 2 diabetic patients. Diabetes. 2006;55:435–440. doi: 10.2337/diabetes.55.02.06.db04-1575. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Khan A, Östenson C-G, Berggren P-O, Efendic S, Meister B. Down-regulated expression of exocytotic proteins in pancreatic islets of diabetic GK rats. Biochem Biophys Res Commun. 2002;291:1038–1044. doi: 10.1006/bbrc.2002.6555. [DOI] [PubMed] [Google Scholar]
- 31.Varadi A, Molnar E, Ostenson CG, Ashcroft SJ. Isoforms of endoplasmic reticulum Ca(2+)-ATPase are differentially expressed in normal and diabetic islets of Langerhans. Biochem J. 1996;319(2):521–527. doi: 10.1042/bj3190521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurmond DC, Gonelle-Gispert C, Furukawa M, Halban PA, Pessin JE. Glucose-stimulated insulin secretion is coupled to the interaction of actin with the t-SNARE (target membrane soluble N-ethylmaleimide-sensitive factor attachment protein receptor protein) complex. Mol Endocrinol. 2003;17:732–742. doi: 10.1210/me.2002-0333. [DOI] [PubMed] [Google Scholar]
- 33.Tomas A, Yermen B, Min L, Pessin JE, Halban PA. Regulation of pancreatic beta-cell insulin secretion by actin cytoskeleton remodelling: Role of gelsolin and cooperation with the MAPK signaling pathway. J Cell Sci. 2006;119:2156–2167. doi: 10.1242/jcs.02942. [DOI] [PubMed] [Google Scholar]
- 34.Honda K, et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolova G, et al. The vascular basement membrane: A niche for insulin gene expression and beta cell proliferation. Dev Cell. 2006;10:397–405. doi: 10.1016/j.devcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Bosco D, Meda P, Halban PA, Rouiller DG. Importance of cell-matrix interactions in rat islet beta-cell secretion in vitro: Role of alpha6beta1 integrin. Diabetes. 2000;49:233–243. doi: 10.2337/diabetes.49.2.233. [DOI] [PubMed] [Google Scholar]
- 37.Ficarro SB, et al. Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal Chem. 2009;81:3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz JC, Syka JEP, Remes PM, Quarnby ST. Proc 56th ASMS Conf Mass Spectrom and Allied Topics. 2008 WPAA039. For additional information, contact the corresponding author of this article. [Google Scholar]
- 39.Rappsilber J, Ishihama Y, Mann M. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Anal Chem. 2003;75:663–670. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 40.Graumann J, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Ishihama Y, Rappsilber J, Andersen JS, Mann M. Microcolumns with self-assembled particle frits for proteomics. J Chromatogr A. 2002;979:233–239. doi: 10.1016/s0021-9673(02)01402-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.