Abstract
Patients with neurofibromatosis type 1 (NF1) carry approximately a 10% lifetime risk of developing a malignant peripheral nerve sheath tumor (MPNST). Although the molecular mechanisms underlying NF1 to MPNST malignant transformation remain unclear, alterations of both the RAS/RAF/MAPK and PI3K/AKT/mTOR signaling pathways have been implicated. In a series of genetically engineered murine models, we perturbed RAS/RAF/MAPK or/and PTEN/PI3K/AKT pathway, individually or simultaneously, via conditional activation of K-ras oncogene or deletion of Nf1 or Pten tumor suppressor genes. Only K-Ras activation in combination with a single Pten allele deletion led to 100% penetrable development of NF lesions and subsequent progression to MPNST. Importantly, loss or decrease in PTEN expression was found in all murine MPNSTs and a majority of human NF1-associated MPNST lesions, suggesting that PTEN dosage and its controlled signaling pathways are critical for transformation of NFs to MPNST. Using noninvasive in vivo PET-CT imaging, we demonstrated that FDG can be used to identify the malignant transformation in both murine and human MPNSTs. Our data suggest that combined inhibition of RAS/RAF/MAPK and PTEN/PI3K/AKT pathways may be beneficial for patients with MPNST.
Keywords: in vivo PET imaging, peripheral nerve sheath tumor, tumor suppressor
Neurofibromatosis type 1 (NF1) is one of the most common inherited disorders with an estimated birth incidence of 1 in 2,500. An autosomal dominant disorder, NF1 is clinically characterized by peripheral neurofibromas (NF), café-au-lait spots, axillary freckling, optic nerve gliomas, and hamartomas of the iris. NFs are benign peripheral nerve sheath tumors that develop as cutaneous or s.c. masses, deep soft tissue lesions, plexiform NFs, or intraneural tumors (1–4).
NF1 patients carry approximately a 10% lifetime risk of developing malignant peripheral nerve sheath tumors (MPNSTs) compared to less than 0.1% in the general population (5, 6). MPNSTs are malignant soft tissue sarcomas with particularly poor survival rates. The molecular mechanisms behind the malignant transformation of NF into MPNST remain unclear. Although NF1 is the most important known risk factor for the development of MPNST and the loss of the second copy of the NF1 allele is found in MPNST cells possibly contributing to malignant transformation, loss of both NF1 alleles is not sufficient for malignant transformation of benign NFs (5).
Mutations in the NF1 tumor suppressor gene are believed to be one of the earliest events contributing to peripheral nerve tumor development in NF1 patients. Neurofibromin, the protein product of NF1, is a RAS-GTPase-activating protein (RAS-GAP) that negatively regulates RAS activity (1, 7, 8). Recent studies demonstrate a role of neurofibromin in controlling mammalian target of rapamycin (mTOR), indicating the involvement of PI3K/AKT/mTOR pathway in the etiology of NF1 (9–11). Although no mutations have been identified in the RAS- and PI3K-controlled pathways, both the RAS/MAPK and PI3K/AKT/mTOR signaling pathways may play critical roles in the development of NF1-related tumors.
The murine homolog for Nf1 has been knocked out via homologous recombination (12). Nf1−/− mice die during embryogenesis due to cardiac development failure, while Nf1+/− mice show no hallmark of the human NF1 phenotype (13, 14). Chimeric mice bearing Nf1−/− cells develop plexiform NFs, suggesting that Nf1 loss of heterozygosity (LOH) or NF1 gene dosage is essential for NF1 initiation. However, no dermal NFs were reported (15). Nf1 conditional knockout mice were since generated by multiple groups. Schwann cell- and astrocyte-specific ablation of Nf1 leads to plexiform NFs, confirming loss of NF1 expression is sufficient for formation of tumors with pathological features of NFs, whereas MPNST development may require alterations of additional genes or signaling pathways. In the search for pathways responsible for the malignant transformation, Nf1+/− mice were crossed onto p53 null background. Although neither p53 null nor heterozygous mice develop MPNSTs, mice with mutations in both genes do develop soft tissue tumors resembling MPNSTs (15, 16). In addition, genetic studies also suggest that other cell types, such as Nf1+/− mast cells or fibroblasts, may also be critical for disease development (17–19).
Since the RAS/RAF/MAPK and PI3K/AKT/mTOR are the two major signaling pathways involved in tumorigenesis of both the peripheral and central nervous systems (7), we reasoned that perturbation of these pathways, individually and in combination, may be a more efficient way of modeling human NF1 and its associated MPNST development. By creating a series of mouse models harboring conditionally deletable Pten (20) or Nf1 allele (21) and conditionally activatable mutant LSL-K-rasG12D alleles (22), we demonstrated that loss of expression of Pten, the second most frequently mutated tumor suppressor gene in all human cancers, in combination with K-RAS activation, led to the development of NFs with 100% penetrance, followed by MPNST transformation. Importantly, MPNST development correlates with loss of PTEN expression in both our murine model as well as human NF1 patients and can be visualized via noninvasive [18F]-2-fluoro-2-deoxy-D-glucose positron emission tomography (FDG-PET) imaging.
Results
Conditional Deletion of Pten and Activation of K-ras Leads to NF and MPNST Development.
To genetically test the contribution of PTEN/PI3K/AKT and RAS/RAF/MAPK pathways in NF and MPNST development, we crossed the Pten (Ptenloxp/loxp) (20) or the Nf1 conditional deletion allele (Nf1loxp/loxp) (23) and the K-ras conditional activatable allele (LSL-K-rasG12D/+) (22) with mGFAP-Cre+ mice (line 77.6). The resulting mice with Pten or Nf1 single gene conditional deletion or K-Ras activation were tumor-free with normal life spans and indistinguishable from their wild-type (WT) littermates (Fig. 1A). Therefore, K-RAS activation and NF1 or PTEN loss alone are not sufficient for NF1 development even though both K-RAS activation and NF1 loss can activate the mTOR pathway (9–11).
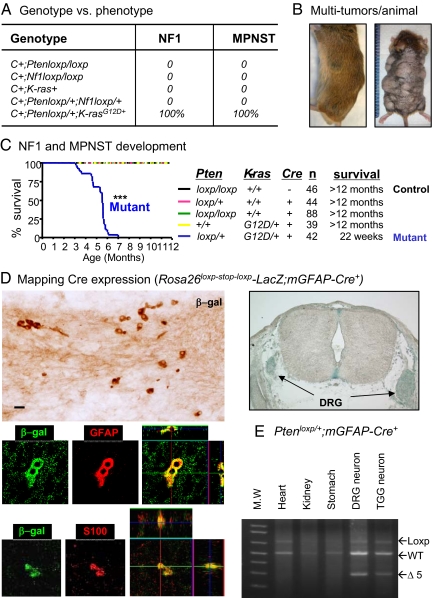
Fig. 1.
Genotypes and phenotypes of mice generated. (A) Overview of genotype and phenotype of mice generated. (B) Representative photo of a mutant mouse bearing multiple tumors. Additional tumors were revealed upon removal of body hair (right; ranging from two to 25 per animal). Ruler ticks, 1 mm. (C) Kaplan Meier curve for duration of survival of mice with five genotypes. The mortality rate of mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ mice (mutant in blue; P < 0.01) was significantly earlier than that of control (black). Genetic composition, number of mice, and mean survival are shown on the right. (D) Mapping Cre expression. Left, colocalization of β-gal+ cells with endogenous GFAP and S100 expression on E13.5 embryo; right, X-gal staining of DRG of E13.5 embryo. (E) PCR genotype for Pten exon 5 excision (Δ5) in adult DRG and TGG neurons C+, mGFAP-Cre+.
A single activated K-ras allele or Nf1 deleted allele was then introduced into the Pten heterozygous background (mGFAP-Cre+;Ptenloxp/+). Although mGFAP-Cre+;Nf1loxp/+;Ptenloxp/+ compound heterozygous animals showed no tumor development, the resulting mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ mice developed multiple visible s.c. tumors with 100% penetrance, starting from postnatal 4 months (Fig. 1 B and C; n = 42). These tumors varied in size and location with the majority located on the back and sides of the animal body (Fig. 1B). When exposed, almost all of the nodules were locally confined and solid. Detailed histopathological analyses demonstrated that each mutant mouse carried more than one lesion with pathological features indistinguishable from human NFs, including the plexiform NF characteristic of NF1 patients, and MPNSTs (Fig. S1). Murine and human NFs are comprised of a mixture of cell types, including Schwann cells, mast cells, perineural cells, and fibroblasts (18, 24), and we thus tested for these cells in the murine tumors. Special stains or antibodies known to be positive in murine and human NFs (18, 24), including the Schwann cell marker S100, mast cell markers toluidine blue, or c-Kit showed classic staining patterns for NFs (Fig. S2). In addition, benign NFs had fewer numbers of Ki-67+ cells, whereas MPNST lesions showed an increased Ki-67 labeling index (Fig. S2 and Fig. 2B). Importantly, MPNST lesions were found in 100% of mutant mice when followed for 7 months. However, when mutant mice were killed at 12 weeks of age, multiple small foci of benign NFs and plexiform NFs were found without any MPNSTs, indicating that progression from NFs to MPNST is likely a time-dependent event. This result suggests that Pten haploinsufficiency, i.e., loss of one allele of the Pten tumor suppressor gene, is critical for NF initiation caused by K-Ras activation.
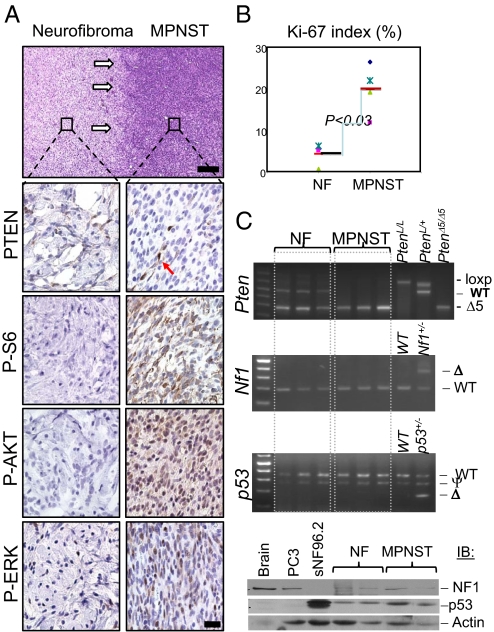
Fig. 2.
PTEN loss is critical for malignant transformation of benign NF in mice. (A) Lesions containing both benign NFs and MPNSTs were harvested from mutant mice and subjected to immunohistochemical analysis (white arrows indicate “transition zone” from benign NF to MPNST). Compared to benign NF, MPNSTs showed marked reduction in immunoreactivity for PTEN in tumor cells, with PTEN+ stained endothelial cells as an internal positive control (arrow). MPNSTs also show intensive staining with antibodies specific for surrogate markers of activated PI3K/AKT (P-AKT, P-S6) and RAS/MAPK (P-ERK) pathways. [Scale bars, 150 μm (H&E) and 25 μm (IHC).] (B) MPNST tumors have higher Ki-67 labeling index than NF tumors (P < 0.03). (C) Upper three panels: PCR analysis of NF and MPNST tumor DNA from mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ mice, indicating loss of Pten WT allele and retaining of both p53 and Nf1 genes in MPNST lesions; lower panel, Western blot analysis showing NF1 and p53 proteins in NF and MPNST samples. PC3 and sNF96.2 are human cell lines used here as controls that are null for p53 and NF1, respectively.
Mapping the NF Initiating Cells in mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ Model.
The cell-of-origin of NF is a subject of current debate. Although several recent studies agree that Nf1-null neural crest cells are not the NF-initiating cells, they differ as to what particular glial differential stage gives rise to the neoplastic lesions (25). To define the NF-initiating cells in our model, we crossed mGFAP-Cre 77.6 line with the Rosa26loxp-stop-loxp-LacZ reporter mouse (26). mGFAP-Cre+;Rosa26loxp-stop-loxp-LacZ embryos were harvested from E12.5 to P0 and cryoprotected before staining with either X-gal or anti-β-gal antibody (Fig. 1D, upper panels). No LacZ expression could be detected at E12.5, 0.5–1 days before the onset of endogenous GFAP expression (25). On the other hand, Cre expression could be detected in E13.5 intercostal nerve (left) and dorsal root ganglions (DRG; right). Detailed fluorescent immunohistochemistry analysis (27) further confirmed that all β-gal expressing cells also expressed endogenous GFAP and S100 expression (Fig. 1D, lower left panels; Fig. S3), suggesting that the oncogenic initiation event in our model begins at the stage between Schwann precursor and immature Schwann cells (25). Cre expression could be detected throughout embryogenesis and in adult peripheral nerve tissues, such as DRG and trigeminal ganglion (TGG) neurons, as indicated by PCR-aid genotype analysis (Fig. 1E) and immunohistochemistry staining. Collectively, this analysis suggest that Schwann precursor or immature Schwann cells are the cell-of-origin in mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ model.
Pten LOH Correlates with MPNST Transformation in the NF Murine Model.
Since all MPNSTs are developed within existing NF in mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ mice, we focused our attention on the “transition zone” between NF and MPNST lesions. As shown in the upper panel of Fig. 2A, a benign lesion (left) is clearly separated from MPNST (right; white arrows) within the same tumor mass. Pathologically, NFs showed relatively uniform, ovoid to spindle-shaped cells, many with tapered ends, intermixed with collagen fibers and mast cells (Figs. S1 and S2). In contrast, MPNSTs showed a marked increase in cellularity, moderate to severe nuclear pleomorphism (cellular anaplasia), obvious mitoses, including atypical mitoses (Fig. S2) and focal areas of necrosis. These histological features, particularly that of MPNSTs arising from preexisting NFs, are classic features of human NF1-associated MPNSTs (3, 28).
Importantly, loss of PTEN expression was detected in all (n = 15) MPNST tumors (Fig. 2A, second right; arrow points to a PTEN+ blood vessel). P-AKT and P-S6 levels, two surrogate markers for PTEN-controlled PI3K pathway activation, were also elevated in MPNST lesions (Fig. 2A, lower right panels). The corresponding NFs, from which the MPSNT developed, showed relatively normal PTEN expression with no significant increase in P-AKT or P-S6 levels (Fig. 2A, lower left panels). P-ERK expression could be detected in both lesions, although significantly increased in MPNSTs (Fig. 2A, bottom panels). Consistent with the malignant nature of MPNST and with previous reports in humans (29), the proliferation index, as measured by Ki-67 staining (Fig. S2), was significantly higher than that seen in NF (Fig. 2B; P < 0.03), suggesting that PTEN controls NF to MPNST malignant transformation, at least in part, via its role in negatively regulating cell proliferation.
Loss of PTEN expression in MPNSTs could be due to either genetic loss of the second Pten allele, mutations that destabilize PTEN protein, or epigenetic silencing of Pten mRNA expression. To determine the molecular mechanisms involved in loss of PTEN expression, we first tested whether the WT allele of Pten was lost during tumor progression (LOH) by PCR analysis. As shown in Fig. 2C, the WT allele of Pten is completely deleted in three independent NF1-associated MPNST lesions (Fig. 2C, top panel). On the other hand, we did not detect obvious defects, either by PCR or via Western blot analysis, in Nf1 and p53 genes, although we cannot rule out possible interstitial deletions or point mutations (Fig. 2C, lower three panels). This study clearly demonstrates that Pten LOH is critical for the malignant transformation of NF to MPNST in this mouse model.
Reduction of PTEN Expression in Human MPNSTs.
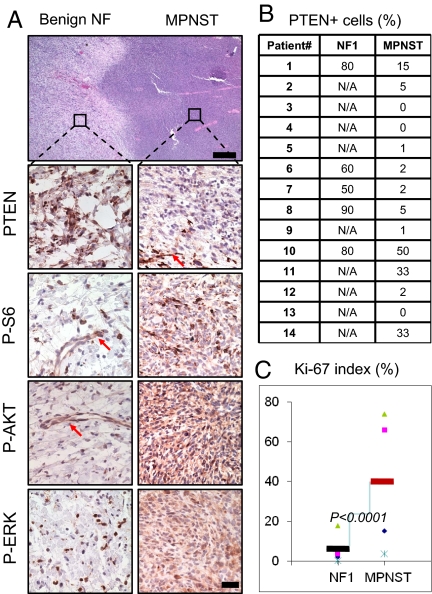
The correlation of Pten loss and MPNST development in our NF murine model prompted us to investigate whether PTEN expression is similarly reduced in the transition of human NFs to MPNST. For this, we surveyed the status of PTEN in human NF1-associated high grade MPNSTs and if possible the associated NF from which they arose (for details, please see Materials and Methods). As was seen in the murine model, marked reduction of PTEN protein levels was detected in all human MPNSTs samples examined (Fig. 3A; n = 14). Detailed quantitative analysis showed less than 20% PTEN positive tumor cells in 11 of 14 samples (Fig. 3B), with tumor vascular endothelial cells retaining PTEN staining and thus serving as an internal positive control (Fig. 3A, arrows). Similar to the murine model, increased P-S6, P-AKT, and P-ERK levels (Fig. 3A, lower right panels) were detected in the human MPNST samples. All corresponding NFs (n = 5) demonstrated higher PTEN expression levels, as well as low and sporadic P-S6, P-AKT, and P-ERK staining patterns (Fig. 3B, lower left panels). In addition, human MPNSTs had significantly increased Ki-67 proliferation index (Fig. 3C; P < 0.0001). These observations suggest that, similar to the murine model, reduced PTEN expression may be important for the malignant transformation of NFs to MPSNTs in human NF1 patients.
Fig. 3.
Decreased PTEN expression in human NF1-associated MPNSTs. (A) Histological and immunohistological analyses of transition zone of human NF1-MPNST lesions demonstrate reduced PTEN expression (second right panel; arrows indicates PTEN+ vascular endothelial cells) and activated PI3K and RAS/MAPK pathways. [Scale bars, 150 μm (H&E) and 25 μm (IHC).] (B) Summary of PTEN IHC staining results, presented as percentages of PTEN+ tumor cells, of 14 NF1 patients with MPNST lesions. N/A, only MPNST samples are available. (C) Human MPNST tumors have higher Ki-67 labeling index than NF tumors (P < 0.0001).
Differentiating NF from MPNST Using Noninvasive PET/CT Imaging.
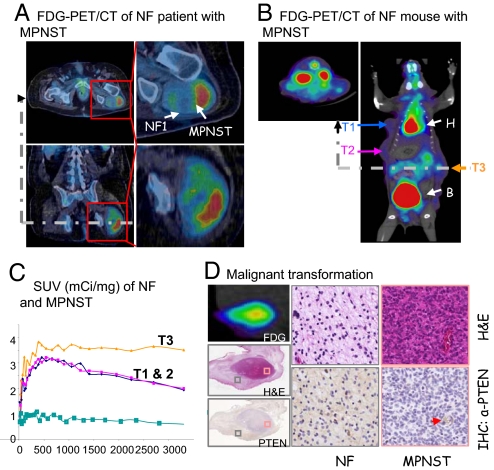
Currently the diagnosis of MPNST is made pathologically. Unfortunately, targeted needle core biopsies are often not technically possible or falsely negative due to the heterogeneous nature of these tumors. Noninvasive modalities that can reliably differentiate NF from MPNST would significantly contribute to the management of NF1 patients. Recently, FDG-PET has been shown by us and other investigators to be a promising noninvasive modality with the potential to differentiate malignant from benign tumors (30, 31). Fig. 4A shows FDG-PET/CT images of a NF1 patient with a left gluteal MPNST arising within a NF lesion. The MPNST component of the tumor demonstrated high FDG uptake (standardized uptake value SUVmax of MPNST = 6.3 g/mL), which represents 4.85-fold increase over NF region (SUVmax of the NF = 1.3 g/mL).
Fig. 4.
Noninvasive in vivo imaging of MPNSTs transformation within NF lesions. (A) FDG-PET/CT image of a NF1 patient with MPNST. (B) FDG-PET/CT image of a NF mouse with MPNST. mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ mouse with three independent tumors, T1 and T2 are NF with low FDG intake, while T3 has increased FDG intake. (C) The FDG-PET SUVs, measured from the ROI for each tumor, compared to baseline uptake (blue) measured from limb muscle without tumor. (D) Correlation of FDG uptake, H&E staining, and PTEN expression (left panels) in a NF lesion (gray boxed sample area, middle panels) with MPNST transformation (red boxed sample area, right panels). Red arrow points to PTEN+ vessel. T, tumor; H, heart; B, bladder.
Similar to the human studies, we also monitored MPNST malignant transformation in our NF murine model using noninvasive in vivo PET imaging in conjunction with CT. While MPNSTs could be sensitively detected with FDG (n = 10), similar to human MPNSTs, benign NF lesions were low or negative for FDG (n = 29). Fig. 4B showed a mouse with three independent lesions, two NFs with low FDG uptake (T1 and T2) and one MPNST with higher FDG uptake (T3; expressed as SUV in Fig. 4C). Importantly, malignant transformation can be positively confirmed on FDG-positive NFs by high cellularity, loss of PTEN expression (Fig. 4D), high proliferation index, necrosis, and massive angiogenesis (Fig. S4A). The percentage of animals with detectable FDGlow and FDGhigh lesions appears to be age-dependent, similar to our histopathological analysis (Fig. S4B). These data suggest that noninvasive FDG-PET imaging may be used longitudinally to follow the NF to MPNST malignant transformation in animal models, as well as to provide a valuable and noninvasive/nonlethal diagnostic strategy to assess potential therapeutic response.
Discussion
The transition from benign NF to MPNST represents the most lethal complication of NF1. Currently, there are no treatment strategies that prevent this transformation. Here, we describe a mouse model for NF and MPNST, which recapitulates the essential clinical features observed in human NF1 and NF-associated MPNSTs. Our study demonstrates the importance of PTEN loss in the NF to MPNST transformation. This finding provides support for potential therapeutic strategies that are both rational and selective.
Although RAS or PTEN mutations per se have not been identified in human NFs, several reports support the alterations of RAS or PTEN controlled signaling pathways in NF and MPNST development. A recent study showed that five of six human MPNST samples contain a hypermethylated PTEN promoter (32), providing a mechanistic explanation of loss of PTEN expression observed in our study. Several groups have identified constitutive RAS activation as a result of NF1 or NF2 deletion (33–36). Previous studies have also shown that RAS activity and its downstream effectors are elevated in the cortex and hippocampus of NF1+/− mice (37, 38). Furthermore, the learning deficits in Nf1+/− mice can be rescued by decreasing RAS function either genetically (crossing with the K-Ras+/− heterozygote) or pharmacologically (treated with farnesyl-transferase inhibitors of Ras) (37, 39). Although mice with oncogenic N-Ras expression in nerve and neural crest-derived cells mimic two main symptoms of human NF1 and/or NF2, namely pigmentary abnormality and dermal NFs, plexiform NF, Schwannoma, astrocytoma, and pheochromocytoma were not detected (40). Since the three forms of Ras, (H, K, N) are expressed in different cells, this could explain the absence of some of these tumor types in our model and the presence of various other tumors.
Mutations in the NF1 tumor suppressor gene is inherited as an autosomal-dominant trait, suggesting a possible gene dosage effect. In fact, haploinsufficiency is apparently enough to bring about many of the clinical manifestations seen in NF1 patients. However, the development of MPNST in NF1 individuals requires acquisition of additional genetic aberrations, whether it is inactivation of TP53, CDKN2A, or amplification of platelet-derived growth factor receptor or epidermal growth factor receptor (18, 41–43). Reminiscent of the malignancies seen in NF1 patients, compound heterozygous mice for both Nf1 and p53 develop MPNSTs with full penetrance (15, 16). In addition, as NF1 has a spectrum of specific tumors, modifying genes and epigenetic phenomena have been shown to play a role in modulating Nf1-associated tumor susceptibility (44, 45).
Consistent with multigene targets hypothesis, when we crossed the same mGFAP-Cre line to conditional knockout Pten (46) or Nf1 or constitutively activate K-ras (this study), we did not observe any tumor development. However, NFs and MPNSTs were found in compound mGFAP-Cre+;Ptenloxp/+;LSL-K-rasG12D/+ mice. Interestingly, NFs had Pten expression, while MPNSTs within the NF lesions had lost the WT allele of Pten. When human samples were tested for PTEN expression, the results matched those found in mice, demonstrating that PTEN loss or PI3K/AKT activation is the rate-limiting step in murine and human NF1 malignant transformation and the development of MPNST.
Distinguishing MPNST from benign NFs is often difficult, particularly in patients with NF1. Optimal clinical management rests on correct pretreatment classification. Patients with benign NFs can either be followed with serial imaging or undergo nerve sparing surgery. Patients with MPNST require radical resection, radiation therapy, and frequently, chemotherapy. Currently, histologic features alone determine diagnosis. However, even targeted needle core biopsies can be inaccurate and are often not technically possible. Diagnostic methods that will reliably differentiate between MPNST and benign NFs would be essential in the management of these tumors. Our parallel comparison of mouse model and human patients indicates that FDG-PET can distinguish benign NF from MPNST with high sensitivity and specificity. Our study further suggests that such a noninvasive functional imaging strategy may serve as a valuable modality to assess potential therapeutic response.
Materials and Methods
Animals.
mGFAP-Cre+;Ptenloxp/loxp line was generated previously (21) and crossed to LSL-KrasG12D/+ mice (26) on a C57, 129/BALB/c background. Since LSL-K-rasG12D/G12D mice are embryonically lethal (27), Ptenloxp/loxp;LSL-KrasG12D/+ males were backcrossed to mGFAP-Cre+;Ptenloxp/+ females to produce experimental animals. Similarly, Nf1loxp/loxp mice (47) were crossed with mGFAP-Cre+;Ptenloxp/loxp, and compound heterozygous animals were backcrossed with mGFAP-Cre+;Ptenloxp/loxp mice to produce experimental animals. Mice were observed daily for evidence of illness or tumor formation. If palpable tumors exceeded 1.5 cm in diameter or interfered with feeding and grooming, mice were killed. Animals were housed in a temperature-, humidity-, and light-controlled room (12-h light/dark cycle), and allowed free access to food and water. All experiments were conducted according to the research guidelines of the University of California, Los Angeles (UCLA) Chancellor's Animal Research Committee.
Polymerase Chain Reaction.
Pten, Nf1, or p53 deletion, K-ras activation, and Cre expression were evaluated by PCR using genomic DNA from tail clip biopsy as described previously using standard techniques (14, 47, 48). PCR was performed in 20-μL reactions using standard procedures for 40 cycles; each cycle consisted of denaturing at 94 °C for 30 s, annealing at 60 °C for 1 min 30 s, and extension at 72 °C for 1 min, followed by a single 5-min extension at 72 °C. The PCR products were resolved on 2% agarose gels.
Histology and Immunohistochemistry of Tissue Sections.
All tumors were graded according to World Health Organization (WHO) histopathological criteria (32). IHC staining was performed on age-matched control and mutant sections. Five-micrometer sections that were prepared from paraffin-embedded blocks were placed on charged glass slides. The slides were deparaffinized with xylene and rehydrated in descending grades (100–70%) of ethanol. The endogenous peroxidase activity was inactivated in 3% hydrogen peroxide (H2O2). After washing in deionized water, antigen retrieval was performed by incubating the slides in 0.01 M citric acid buffer (pH 6.0) at 95 °C for 13.5 min. Slides were then allowed to cool for 30 min in citric acid buffer. After washing in deionized water, the slides were then transferred to either PBS (pH 7.4) or TBST for 5 min. For DAB staining, slides were first blocked with 5% normal goat serum, then incubated with primary antibody overnight at 4 °C. Following three 5-min washes in either PBS or TBST, slides were incubated with biotinylated secondary antibody (1:200; Biogenex) for 30 min at room temperature. Amplification was performed with a horseradish peroxidase system (Vectastain ABC kit, PK-6100; Vector Laboratories) using a liquid DAB peroxidase substrate (HK130–5K; Biogenex). Slides were counterstained in Gill's hematoxylin, dehydrated, cleared, and coverslipped. Negative control slides were run without primary antibody. Primary antibodies used were rabbit anti-PTEN (1:100, 9552; Cell Signaling), mouse anti-PTEN (1:100, 9556; Cell Signaling), rabbit anti-pAKT (1:50, 3787; Cell Signaling), pERK, pS6, S100beta (1:100, 9101 and 2215; Cell Signaling and 1:400, Z0311; Dako).
To map Cre-expressing cells, embryos were cryoprotected in buffered 30% sucrose overnight, and 20-μm frozen sections were prepared. Bright field and fluorescence immunohistochemistry were performed as described previously (27) using biotinylated secondary antibodies (Vector Laboratories), biotin-avidin-peroxidase complex (Vector Laboratories), and diaminobenzidine (Vector Laboratories) as the bright field developing agent or Alexa Fluor-tagged secondary antibodies Alexa 488 (green), Alexa 568 (red) (Invitrogen). Primary antibodies were: Rabbit anti-β-gal (1:200 or 1:6,000; Millipore), rat anti-GFAP (1:500; Zymed Laboratories), sheep anti-S100 (1:200; QED Bioscience). Stained sections were examined and photographed using bright field and fluorescence microscopy (Zeiss) and scanning confocal laser microscopy (Leica).
microPET/CT Imaging and Analysis.
microPET/CT imaging was performed with a microPET FOCUS 220 PET scanner (Siemens Preclinical Solutions) and microCAT II CT scanner (Siemens Preclinical Solutions) with UCLA Chancellor's Animal Research Committee approval. Briefly, mice are anesthetized with isofluorane 15 min before receiving 18F-fluoro-D-glucose (200 μCi 18FDG per mouse) via tail vein. The mice are then placed in the imaging chamber and imaged over 1 h in the microPET scanner, followed by a 10-min microCAT scan for anatomical localization. PET images were analyzed with the AMIDE software. Regions of interest (ROIs) were manually drawn on the area of tumor with maximal tracer uptake at 1 mm in diameter. Activity concentrations were quantified as SUVs normalized to injected dose per weight of mouse (mCi/g).
Human NF1-MPNST Analysis.
Following UCLA IRB approval for studies on human MPNSTs, the UCLA Sarcoma and Pathology databases were used to identify patients with NF1 who underwent surgical treatment for a MPNST. Slides from the selected cases were reviewed. Whenever possible, we chose cases in which we could identify a MPNST arising from the associated NF and selected sections demonstrating a transition from NF to MPNST. Anonymously labeled sections for immunohistochemistry studies, as detailed above, were prepared by the UCLA Department of Pathology Translational Pathology Laboratory. Additionally, an H&E slide was made from each block to confirm the diagnosis.
Supplementary Material
Acknowledgments.
We thank members of our laboratories for helpful comments on the manuscript. C.G. and K.B.S. are predoctoral trainees supported by USHHS Ruth L. Kirschstein Institutional National Research Service Award no. T32 CA09056. J.N. is supported by National Cancer Institute (NCI) P50 (P50 CA086306) Career Development Award. This work is supported by the following grants and awards: Miriam and Sheldon Adelson Program (to H.W. and X.L.), National Institutes of Health Grants NS057624 (to M.V.S.) and NS50151 (to P.S.M.), Brain Tumor Funders' Collaborative (to P.S.M., I.K.M., and H.W.), University of California, Los Angeles' Jonsson Comprehensive Cancer Center Foundation (to F.C.E. and S.M.D.), American Cancer Society and National Institutes of Health Grant P50NS052606 (to L.F.P.), Brain Tumor Society Award (to X.L.), and Henry Singleton Brain Cancer Research Program and a James S. McDonnell Foundation Award (to H.W. and P.L.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910398106/DCSupplemental.
References
- 1.Ferner RE, et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. 2007;44:81–88. doi: 10.1136/jmg.2006.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Listernick R, Louis DN, Packer RJ, Gutmann DH. Optic pathway gliomas in children with neurofibromatosis 1: Consensus statement from the NF1 Optic Pathway Glioma Task Force. Ann Neurol. 1997;41:143–149. doi: 10.1002/ana.410410204. [DOI] [PubMed] [Google Scholar]
- 3.Ferner RE. Neurofibromatosis 1. Eur J Hum Genet. 2007;15:131–138. doi: 10.1038/sj.ejhg.5201676. [DOI] [PubMed] [Google Scholar]
- 4.Williams VC, et al. Neurofibromatosis type 1 revisited. Pediatrics. 2009;123:124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 5.Ferner RE, Gutmann DH. International consensus statement on malignant peripheral nerve sheath tumors in neurofibromatosis. Cancer Res. 2002;62:1573–1577. [PubMed] [Google Scholar]
- 6.Evans DG, et al. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002;39:311–314. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Parada LF. The molecular and genetic basis of neurological tumours. Nat Rev Cancer. 2002;2:616–626. doi: 10.1038/nrc866. [DOI] [PubMed] [Google Scholar]
- 8.Dasgupta B, Li W, Perry A, Gutmann DH. Glioma formation in neurofibromatosis 1 reflects preferential activation of K-RAS in astrocytes. Cancer Res. 2005;65:236–245. [PubMed] [Google Scholar]
- 9.Dasgupta B, Yi Y, Chen DY, Weber JD, Gutmann DH. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer Res. 2005;65:2755–2760. doi: 10.1158/0008-5472.CAN-04-4058. [DOI] [PubMed] [Google Scholar]
- 10.Johannessen CM, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008;18:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 11.Johannessen CM, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci USA. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutmann DH, Giovannini M. Mouse models of neurofibromatosis 1 and 2. Neoplasia. 2002;4:279–290. doi: 10.1038/sj.neo.7900249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brannan CI, et al. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 14.Jacks T, et al. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 15.Cichowski K, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 16.Vogel KS, et al. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajenaru ML, et al. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 2003;63:8573–8577. [PubMed] [Google Scholar]
- 18.Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang FC, et al. Nf1-dependent tumors require a microenvironment containing Nf1+/− and c-kit-dependent bone marrow. Cell. 2008;135:437–448. doi: 10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesche R, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaughlin ME, Jacks T. Thinking beyond the tumor cell: Nf1 haploinsufficiency in the tumor environment. Cancer Cell. 2002;1:408–410. doi: 10.1016/s1535-6108(02)00078-8. [DOI] [PubMed] [Google Scholar]
- 25.Carroll SL, Ratner N. How does the Schwann cell lineage form tumors in NF1? Glia. 2008;56:1590–1605. doi: 10.1002/glia.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 27.Faulkner JR, et al. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci. 2004;24:2143–2155. doi: 10.1523/JNEUROSCI.3547-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferner RE. Neurofibromatosis 1 and neurofibromatosis 2: A twenty first century perspective. Lancet Neurol. 2007;6:340–351. doi: 10.1016/S1474-4422(07)70075-3. [DOI] [PubMed] [Google Scholar]
- 29.Lin BT, Weiss LM, Medeiros LJ. Neurofibroma and cellular neurofibroma with atypia: A report of 14 tumors. Am J Surg Pathol. 1997;21:1443–1449. doi: 10.1097/00000478-199712000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Benz MR, et al. Quantitative FDG-PET accurately characterizes peripheral nerve sheath tumors as malignant or benign. Cancer. 2009 doi: 10.1002/cncr.24755. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferner RE, et al. [18F]2-fluoro-2-deoxy-D-glucose positron emission tomography (FDG PET) as a diagnostic tool for neurofibromatosis 1 (NF1) associated malignant peripheral nerve sheath tumours (MPNSTs): A long-term clinical study. Ann Oncol. 2008;19:390–394. doi: 10.1093/annonc/mdm450. [DOI] [PubMed] [Google Scholar]
- 32.Weiss SW, Goldblum JR. Soft Tissue Tumors. 5th Ed. Philadelphia: Elsevier; 2008. [Google Scholar]
- 33.Bollag G, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 34.Basu TN, et al. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 35.DeClue JE, et al. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 36.Murray SK, Breau RH, Guha AK, Gupta R. Spread of prostate carcinoma to the perirectal lymph node basin: Analysis of 112 rectal resections over a 10-year span for primary rectal adenocarcinoma. Am J Surg Pathol. 2004;28:1154–1162. doi: 10.1097/01.pas.0000131543.80147.3d. [DOI] [PubMed] [Google Scholar]
- 37.Li W, et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 38.Costa RM, et al. Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat Genet. 2001;27:399–405. doi: 10.1038/86898. [DOI] [PubMed] [Google Scholar]
- 39.Costa RM, et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 40.Saito H, Yoshida T, Yamazaki H, Suzuki N. Conditional N-rasG12V expression promotes manifestations of neurofibromatosis in a mouse model. Oncogene. 2007;26:4714–4719. doi: 10.1038/sj.onc.1210250. [DOI] [PubMed] [Google Scholar]
- 41.Levy P, et al. Molecular profiling of malignant peripheral nerve sheath tumors associated with neurofibromatosis type 1, based on large-scale real-time RT-PCR. Mol Cancer. 2004;3:20. doi: 10.1186/1476-4598-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castle B, Baser ME, Huson SM, Cooper DN, Upadhyaya M. Evaluation of genotype-phenotype correlations in neurofibromatosis type 1. J Med Genet. 2003;40:e109. doi: 10.1136/jmg.40.10.e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stemmer-Rachamimov AO, et al. Comparative pathology of nerve sheath tumors in mouse models and humans. Cancer Res. 2004;64:3718–3724. doi: 10.1158/0008-5472.CAN-03-4079. [DOI] [PubMed] [Google Scholar]
- 44.Easton DF, Ponder MA, Huson SM, Ponder BA. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): Evidence for modifying genes. Am J Hum Genet. 1993;53:305–313. [PMC free article] [PubMed] [Google Scholar]
- 45.Reilly KM, et al. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. Proc Natl Acad Sci USA. 2004;101:13008–13013. doi: 10.1073/pnas.0401236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregorian C, et al. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J Neurosci. 2009;29:1874–1886. doi: 10.1523/JNEUROSCI.3095-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Y, Parada LF. Neurofibromin, a tumor suppressor in the nervous system. Exp Cell Res. 2001;264:19–28. doi: 10.1006/excr.2000.5138. [DOI] [PubMed] [Google Scholar]
- 48.Freeman DJ, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.