Abstract
Soluble guanylate cyclase (sGC), a hemoprotein, is the primary nitric oxide (NO) receptor in higher eukaryotes. The binding of NO to sGC leads to the formation of a 5-coordinate ferrous-nitrosyl complex, and a several hundred-fold increase in cGMP synthesis. NO activation of sGC is influenced by GTP and the allosteric activators YC-1 and BAY 41-2272. Electron paramagnetic resonance (EPR) spectroscopy shows that the spectrum of the sGC ferrous-nitrosyl complex shifts in the presence of YC-1, BAY 41-2272, or GTP in the presence of excess NO relative to the heme. These molecules shift the EPR signal from one characterized by g1 = 2.083, g2 = 2.036, g3 = 2.012 to a signal characterized by g1 = 2.106, g2 = 2.029, g3 = 2.010. The truncated heme domain constructs β1(1-194) and β2(1-217) were compared to the full-length enzyme. The EPR spectrum of the β2(1-217)-NO complex is characterized by g1 = 2.106, g2 = 2.025, g3 = 2.010 indicating the protein is a good model for the sGC-NO complex in the presence of the activators, while the β1(1-194)-NO complex resembles the EPR spectrum of sGC in the absence of the activators. Low-temperature resonance Raman spectra of the β1(1-194)-NO and β2(1-217)-NO complexes show that the Fe-NO stretching vibration of the β2(1-217)-NO complex (535 cm−1) is significantly different than that of the β1(1-194)-NO complex (527 cm−1). This shows that sGC can adopt different 5-coordinate ferrous nitrosyl conformations, and suggests that the Fe-NO conformation characterized by this unique EPR signal and Fe-NO stretching vibration represents a highly active sGC state.
Nitric oxide (NO)1 is the physiologically relevant activator of the hemoprotein soluble guanylate cyclase (sGC). Synthesis of cGMP by sGC is critical to several signaling pathways including those that regulate vasodilation, neurotransmission, and platelet aggregation (1-4). In addition to NO, the purine nucleotides GTP and ATP are known to regulate sGC activity (5, 6). Numerous studies have attempted to elucidate the mechanism of sGC activation, however, this mechanism remains unclear. It is critical to acquire molecular details about the binding of NO to sGC, and the influence of nucleotides on the protein to understand the mechanisms of enzyme regulation.
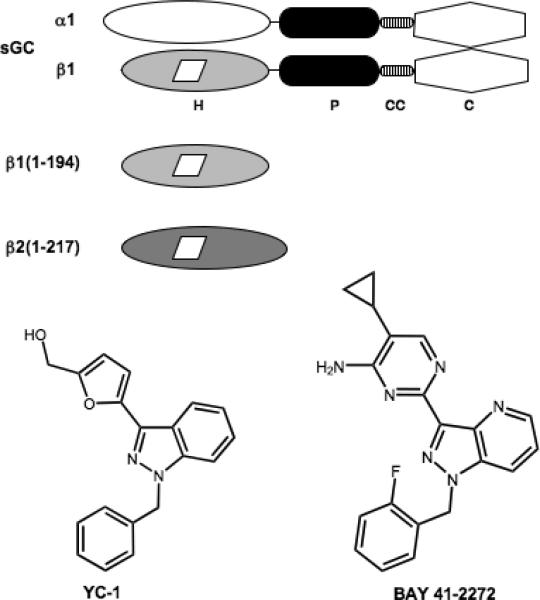
sGC is a heterodimeric protein consisting of two homologous subunits, α and β. While the most commonly studied isoform is the α1β1 protein, α2 and β2 subunits have been identified (7, 8). Architectural information about sGC has been advanced by the expression and isolation of functional domains. Truncations of the β subunits have identified the minimal heme binding domains as β1(1-194)2 and β2(1-217) (9) and the catalytic domains have been localized to the C-terminal 467-690 and 414-619 residues of the α1 and β1 subunits, respectively (10) (Figure 1). As purified, the heme binding domains are monomeric (9), and the catalytic domains are dimeric (10). While the β2 isoform has not yet been purified and characterized, transient expression of the full-length protein in insect cells suggests it functions as a homodimer (11). These functional domains of sGC have facilitated the study of NO binding and activation.
Figure 1.
Schematic models of full-length α1β1 sGC, β1(1-194), and β2(1-217). sGC consists of H-NOX (H), PAS-like (P), coiled-coil (CC), and catalytic domains (C). The heme cofactor is represented by a white parallelogram. The structures of the allosteric activators YC-1 and BAY 41-2272 are shown.
The steps involved in NO binding to the sGC heme are well established. NO binds to sGC to form an initial 6-coordinate intermediate complex, which rapidly converts to a 5-coordinate ferrous nitrosyl complex (12, 13). Generally, heme ligands that form 6-coordinate complexes with sGC activate the enzyme 2 to 4 fold (12, 14). These results have been interpreted to support the proposal that the breaking of the Fe-His bond is crucial for full activity, however, some 6-coordinate sGC complexes can be fully activated in the presence of the allosteric activators YC-1 and BAY 41-2272 (15, 16) without complete conversion to a 5-coordinate complex (17-19). Additionally, a low activity 5-coordinate FeII-NO complex can be isolated in the presence of stoichiometric amounts of NO (5, 20). When excess NO or YC-1 is added to this low activity species, a highly active ferrous nitrosyl species is formed. Interestingly, pre-incubation of sGC with GTP before NO addition also leads to maximal enzyme activity, an effect blocked by the presence of ATP (5). These results exemplify the complicated roles of NO and nucleotides in regulating sGC activity.
Electron paramagnetic resonance (EPR) spectroscopy is a sensitive method for studying the environment of a protein-bound radical and has been used to investigate the sGC FeII-NO complex (17, 21, 22). Resonance Raman spectroscopy provides information about the protein environment via the position of the N-O and Fe-NO stretching vibrations (23). In this report EPR and low-temperature resonance Raman spectroscopy were used to study the FeII-NO environment in the truncated heme binding domains of sGC and the full-length protein. Our results show that the β1(1-194)-NO complex is similar to the previously reported sGC-NO complex (21), but the β2(1-217)-NO complex has a unique EPR signal and ν(Fe-NO) stretch. This suggests that β2(1-217) has a different FeII-NO conformation. The effects of YC-1, BAY 41-2272, GTP and excess NO on the sGC-NO EPR spectrum were also examined. These molecules shift the sGC-NO EPR signal to one that resembles the β2(1-217)-NO complex. These results show that sGC can adopt different 5-coordinate FeII-NO conformations, and these conformations may be relevant for regulation of NO-stimulated activity.
Experimental Section
Material
Rat α1β1 sGC was expressed using a baculovirus/Sf9 expression system and purified as described previously (24). The rat sGC heme domains β1(1-194) and β2(1-217) were prepared as previously described (9). 15N-Labeled sodium nitrite was from Cambridge Isotope Laboratories. Diethylammonium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate (DEA/NO) was from Cayman Chemical Co. The non-cyclizable GTP analogue α,β-methylene guanosine 5′-triphosphate (GMPCPP) was from Jena Biosciences. All other reagents were from Sigma, unless otherwise noted.
Preparation of EPR samples
NO complexes of β1(1-194) and β2(1-217), as well as the full length α1β1 heterodimer, were characterized by EPR. Samples were 20 μM and 22 μM for β1(1-194) and β2(1-217), respectively, in 50 mM HEPES, pH 7.4, 50 mM NaCl, 1 mM DTT. For experiments with isotopically-labeled NO, the gas was generated from acidified sodium nitrite (Na14NO2 or Na15NO2). For all experiments with sGC (5-10 μM) 100 μM DEA/NO was used to form the nitrosyl complex. DEA/NO stocks were prepared in 10 mM NaOH. When present, YC-1 and BAY 41-2272 (both in DMSO) were at 150 μM and 20 μM, respectively, and the final concentration of DMSO was 2 % (v/v). sGC-NO samples with 1 equivalent of NO per heme were made by adding 100 μM DEA/NO to the protein and then removing excess NO by 3 cycles of dilution/concentration using a 10K Ultrafree-0.5 centrifugal filter device (Millipore) into 50 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM MgCl2, 1 mM DTT. GTP (1 mM), GMPCPP (0.9 mM) or cGMP (1 mM) was either added immediately after removal of excess NO for samples with 1 equivalent of NO, or immediately after addition of 100 μM DEA/NO for samples with excess NO. The electronic absorption spectrum of each sample was collected before freezing with liquid nitrogen. The time between nucleotide addition and freezing was ~ 1 min. The final volume of all protein samples was 150 μL.
EPR spectroscopy
Continuous wave (CW) EPR spectra were obtained with the Bruker ECS106 X-band spectrometer in the UC Davis CalEPR facility, equipped with an Oxford Instruments ESR-900 helium flow cryostat. Experimental conditions were as follows: temperature, 25 K; microwave frequency, 9.66 GHz; microwave power, 5.09 mW; modulation frequency, 100 kHz; modulation amplitude, 10.1 G; and time constant, 81.92 msec. The microwave frequency was determined from the spectrum of BDPA (data not shown). Each spectrum is the signal average of 12 or 16 scans, as indicated in the figure legends. Each set of EPR experiments was repeated 2-4 times with different protein preps and sample preparations to ensure reproducibility.
Simulations of EPR spectra
EPR spectra were simulated using EasySpin 2.5.1 (25). Each signal is simulated using the Hamiltonian
| (1) |
The three terms are the electron Zeeman interaction, HeZ =βB0·g·S, the Zeeman interaction of a single 14N or 15N nucleus, HnZ =βnB0·gn·I, and the hyperfine interaction between the unpaired electron and the 14N or 15N nucleus, HHF =S·A·I. The parameters g and A are both third rank tensors with principal values g1, g2, and g3, and A1, A2, and A3 respectively. The microwave frequency used for the simulations was 9.66 GHz.
Only the g-values were varied to fit each spectrum simulated. The principal values for both g and 14N hyperfine tensors are given in Tables 1 and 2. For isotopic comparisons, spectra from both 14NO and 15NO complexes were simulated with a single set of parameters, changing only the nuclear g-value and spin, and scaling the hyperfine values by the ratio of the appropriate g-values.
| (2) |
Table 1.
EPR parameters of sGC and sGC heme domain constructs.a
| Protein | 1 | 2 | 3 | |
|---|---|---|---|---|
| sGC | g-value | 2.083 | 2.036 | 2.012 |
| g-strain | 0.0345 | 0.0341 | 0.0047 | |
| A | 67 | 43 | 45 | |
| β1(1-194) | g-value | 2.083 | 2.036 | 2.012 |
| g-strain | 0.0345 | 0.0341 | 0.0047 | |
| A | 67 | 43 | 45 | |
| β2(1-217) | g-value | 2.106 | 2.025 | 2.010 |
| g-strain | 0.0088 | 0.0090 | 0.0035 | |
| A | 42 | 54 | 48 |
Values are for 14NO complexes.
Table 2.
Effects of allosteric activators on sGC-NO EPR parameters.a
| Compound | 1 | 2 | 3 | |
|---|---|---|---|---|
| - | g-value | 2.083 | 2.036 | 2.012 |
| g-strain | 0.0345 | 0.0341 | 0.0047 | |
| A | 67 | 43 | 45 | |
| YC-1 | g-value | 2.106 | 2.030 | 2.011 |
| g-strain | 0.0088 | 0.0090 | 0.0035 | |
| A | 42 | 54 | 48 | |
| BAY 41-2272 | g-value | 2.106 | 2.029 | 2.010 |
| g-strain | 0.0088 | 0.0090 | 0.0035 | |
| A | 42 | 54 | 48 |
Values are for 14NO complexes with 96 μM YC-1 or 20 μM BAY 41-2272.
Two models of line broadening were employed, both consisting of Gaussian distributions of the g-values (g-strain) to reflect disorder. The first model is based on the simulations reported by Stone et al., where the broad feature in the sGC spectrum was modeled with a large amount of g-strain (21). The more resolved spectra were fit using a smaller amount of g-strain. Full width at half maximum (FWHM) values of the distributions of each principal g-value are given in Tables 1 and 2.
Preparation of RR samples
NO complexes of β1(1-194) and β2(1-217) were generated using acidified nitrite as described above. Samples were 30 μM and 25 μM for β1(1-194) and β2(1-217), respectively, in 50 mM HEPES, pH 7.4, 50 mM NaCl, 1 mM DTT.
RR spectroscopy
RR spectra were collected in backscattering geometry at 25 K with a cryogenic sample holder (26) connected to a Model CSA-202E Displex closed-cycle liquid He refrigerator (Air Products, Allentown, PA). Excitation wavelength at 400 nm was obtained by frequency doubling, using a non-linear lithium triborate crystal, of a Ti:sapphire laser (Photonics International TU-UV), which was pumped by the second harmonic of a Q-switched Nd:YLF laser (Photonics Industries International, GM-30-527). Laser power at the sample was kept minimum (4 mW) to avoid the photolysis of bound NO. The scattered light was dispersed by a triple spectrograph (Spex 1877) equipped with a CCD detector (Roper Scientific, Model 7375-0001) operating at −110 °C. Data acquisition times ranged from 60-90 min. Variations in the signal-to-noise ratios in the various spectra are the result of different acquisition times, and small changes in the alignment of the sample. Spectra were calibrated with dimethyl formamide and DMSO. The reported frequencies are accurate to ± 1 cm−1 and the resolution of the spectra is ~3 cm−1.
Results
EPR of sGC heme domains
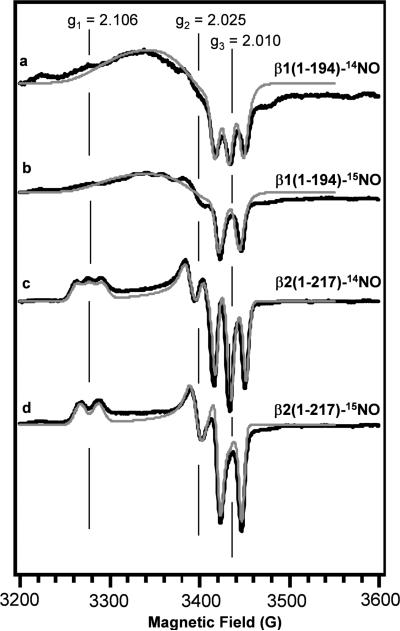
EPR was used to characterize the sGC truncations β1(1-194) and β2(1-217). The EPR spectrum of the full-length α1β1 sGC-NO complex (Figure 3a) is well characterized (21, 22). This spectrum has a broad feature with some subtle structure in the low field region of the spectrum, and a negative feature with sharp hyperfine splittings in the high field region, g3=2.01. The β1(1-194)-14NO complex (Figure 2a) has a spectrum similar to that of the full length protein. Compared to the β1(1-194)-14NO complex, the EPR spectrum of the β2(1-217)-14NO complex (Figure 2c) shows significant variation in the position of the g-values, as well as a better resolved g-tensor showing a clear rhombic powder pattern with resolved hyperfine splittings. A 2-line spectrum is observed with the 15NO complexes (15N, I = ½) and a 3-line spectrum for the 14NO complexes (14N, I=1) in both β1(1-194) and β2(1-217) (Figure 2) showing that the hyperfine interaction is between the unpaired electron spin and the nucleus of the nitrogen of the NO ligand. The g-values of the sGC-NO and the β2(1-217)-NO complexes did not change when the temperature was adjusted from 10-100 K (data not shown), suggesting the conformations of the FeII-NO species are independent of temperature in the studied range.
Figure 3.
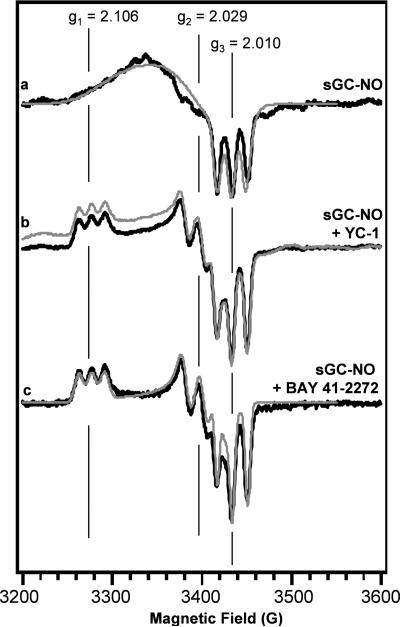
Electron paramagnetic resonance spectra of the sGC-NO complex in the absence and presence of YC-1 or BAY 41-2272. (a) sGC-14NO complex (5.9 μM), (b) sGC-14NO complex (5.9 μM) in the presence of YC-1 (150 μM), and (c) sGC-14NO (5.9 μM) in the presence of BAY 41-2272 (20 μM). Experimental (black line) and simulated (grey line) spectra are shown for all samples. Samples were in 50 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM MgCl2, 1 mM DTT. The spectra shown are the average of 12 scans, and are representative of experiments repeated 2-3 times.
Figure 2.
Electron paramagnetic resonance spectra of sGC β1(1-194)-NO and β2(1-217)-NO complexes at 25 K. (a) β1(1-194)-14NO complex (20 μM) and (b) β1(1-194)-15NO complex (20 μM). (c) β2(1-217)-14NO complex (22 μM) and (d) β2(1-217)-15NO complex (22 μM). Experimental (black line) and simulated (grey line) spectra are shown for all samples. Samples were in 50 mM HEPES, pH 7.4, 50 mM NaCl, 1 mM DTT. The spectra shown are representative of experiments repeated 2 times. Spectra are the average of 16 scans and were normalized to the intensity of the high field triplet (g3=2.010) for comparison.
Simulations of the spectra were performed for both β1(1-194)-NO and β2(1-217)-NO complexes (Figure 2a-d). Similar to the previously reported simulations of the sGC-NO complexes (21), large g-strain was needed to simulate the β1(1-194)-NO complexes. Table 1 shows that the β1(1-194)-14NO complex is similar to the sGC-14NO complex, but the β2(1-217)-14NO complex has shifted g1- and g2-values. Additionally, the β2(1-217)-14NO complex was simulated with lower g-strain, indicating there is less variability in the FeII-NO conformation when compared to the β1 constructs.
Allosteric modulators of sGC
YC-1 or BAY 41-2272 alone marginally activates sGC; however, they fully activate the enzyme when a ligand is bound to the ferrous heme moiety (15, 16). EPR was used to probe the effects of YC-1 and BAY 41-2272 on the sGC-NO complex. In agreement with previous reports, YC-1 induced a change in the sGC-NO spectrum (Figure 3b) (17, 22). The presence of BAY 41-2272 caused a similar change in the sGC-NO spectrum (Figure 3c). Both activators lead to greater resolution of the powder pattern, revealing the 3-line hyperfine splitting at each g-value. Simulations of the sGC-NO complexes show that the g1- and g2-values are shifted in the presence of BAY 41-2272 and YC-1, and that the spectra can be fit without invoking large g-strain (Table 2). The effects of YC-1 and GTP on the EPR spectra of the β1(1-385)-NO, β1(1-194)-NO, and β2(1-217)-NO complexes were also examined. The molecules did not change the EPR signal of these proteins (Figure S1), suggesting the small molecules do not bind to these domains, or they are unable to lead to the same conformational change as seen in the full-length protein.
Effects of excess NO and GTP on the sGC-NO spectrum
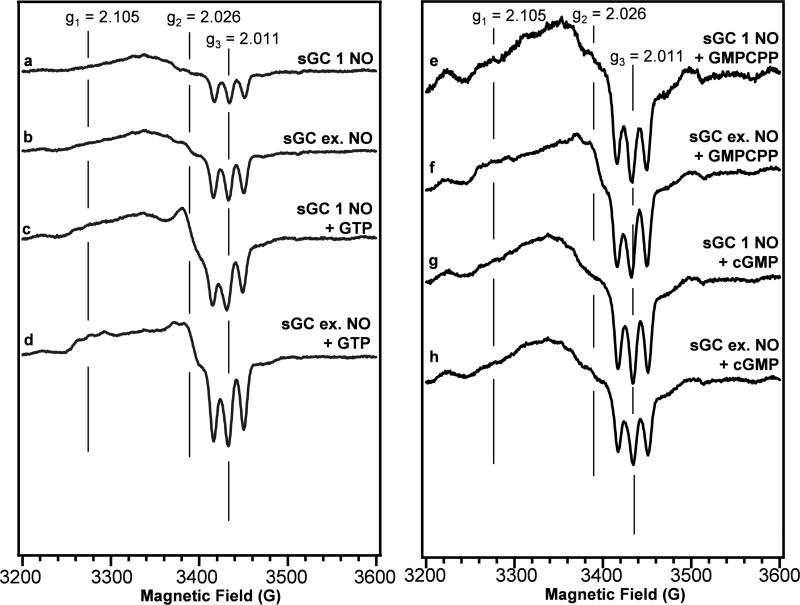
sGC activity is influenced by the presence of substrate and NO in excess of that bound to the sGC heme (5, 20). Therefore, the effects of GTP, the non-cyclizable GMPCPP, and cGMP on the sGC-NO EPR spectrum in the presence and absence of excess NO were investigated. sGC with 1 equivalent of heme-bound NO (1-NO) was made by treating the protein with excess DEA/NO followed by desalting into a NO-free buffer. After this procedure the protein was mostly NO-bound, but there was a small population of unligated protein (~5 %) (Figure S2). The presence of GTP, GMPCPP, or cGMP did not affect the peak position or intensity of the electronic absorption spectra, indicating the complex remained 5-coordinate. Subtle changes in the sGC-NO EPR spectrum were observed upon addition of excess NO (Figures 4a and 4b) around the g2-value. In the presence of GTP both the 1-NO and excess-NO sGC EPR spectrum (Figures 4c and 4d) shifted in a unique manner, indicating that substrate affects the heme environment differently depending on the amount of NO. With excess NO and GTP, the sGC-NO EPR spectrum shows increased hyperfine resolution at g1 and g2 similar to that observed in the presence of YC-1 or BAY 41-2272. The effects of GMPCPP and cGMP on the sGC-NO spectra were examined to determine if the observed effect of GTP was dependent on enzyme turnover or catalysis. In the presence of GMPCPP the sGC-NO spectrum with excess NO (Figure 4f) is similar to the spectrum with GTP and excess NO. Additionally, the spectra shifted depending on the amount of NO used to form the FeII-NO complex (Figure 4e and 4f). There was no significant change in the sGC-NO spectra in the presence of cGMP (Figures 4g and 4h).
Figure 4.
Electron paramagnetic resonance spectra of the sGC-NO complexes. (a) and (b) sGC-NO complexes with 1 NO and excess NO, respectively. (c) and (d) sGC-NO complexes with 1 NO and excess NO in the presence of 1 mM GTP. (e) and (f) sGC-NO complexes with 1 NO and excess NO in the presence of 0.9 mM GMPCPP. (g) and (h) sGC-NO complexes with 1 NO and excess NO in the presence of 1 mM cGMP. All samples were in 50 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM MgCl2, 1 mM DTT. Spectra are the average of 12 scans, and are representative of experiments repeated 2-3 times.
Low-temperature RR
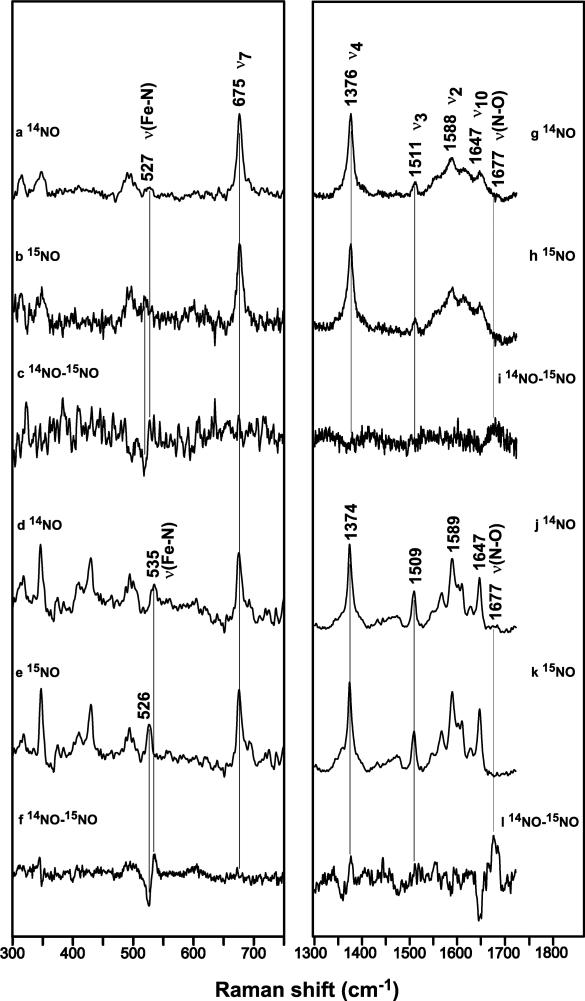
Significant changes are observed in the EPR spectra of sGC in the presence of allosteric activators. Interestingly, the sGC-NO EPR signal is similar to the β1(1-194)-NO signal, and this signal shifts to resemble the β2(1-217)-NO signal in the presence of YC-1, BAY 41-2272 or GTP with excess NO (Tables 1, 2 and Figures 2, 3). Low-temperature resonance Raman spectroscopy was performed on the sGC heme domains to further characterize this observation. The high-frequency spectra of both β1(1-194)-NO and β2(1-217)-NO complexes at 25 K (Figure 5g and 5j) exhibit heme skeletal marker bands that are similar to the bands observed at 298 K (Table 3). This indicates that both proteins remain 5-coordinate at 25 K. Isotope-sensitive bands are observed in the low-frequency spectra of the β1(1-194)-NO and β2(1-217)-NO complexes (Figure 5c and 5f) at 527 and 535 cm−1, respectively. These bands are assigned to the Fe-NO stretching vibration. At room temperature these bands were reported at 526 and 527 cm−1 for the β1(1-194)-NO and β2(1-217)-NO complexes, respectively (9). In the high-frequency spectra a band at 1677 cm−1 is sensitive to isotope substitution for both β1(1-194)-NO and β2(1-217)-NO complexes (Figure 5i and 5l). This band is assigned to the N-O stretching vibration, and is not significantly shifted from the N-O stretching vibration reported for the spectra acquired at room temperature (1676-1677 cm−1) (9). This indicates that the Fe-NO stretching vibration of the β2(1-217)-NO complex is most significantly affected by the reduced temperature (8 cm−1 shift), and confirms the EPR data which suggests that the β2(1-217)-NO complex adopts a 5-coordinate conformation that is distinct from the β1(1-194)-NO complex. We were not able to carry out low-temperature resonance Raman spectroscopy on the sGC-NO complex due to limitations in the amount of full-length protein available. However, since the EPR spectrum of the sGC-NO complex resembles that of the β2(1-217)-NO complex, we can infer that the sGC-NO complex in the presence of YC-1, BAY 41-2272, or GTP with excess NO also exhibits a similar shift in the Fe-NO stretching vibration.
Figure 5.
Resonance Raman spectra of FeII-NO complexes of sGC heme domains. β1(1-194)-14NO and 15NO complexes and their difference spectra in the low- (a-c) and high- (g-i) frequency regions, respectively; β2(1-217)- 14NO and 15NO complexes and their difference spectra in the low- (d-f) and high- (j-l) frequency regions, respectively. Spectra were obtained with 400 nm excitation at 25 K. Data acquisition times ranged from 60-90 min.
Table 3.
Resonance Raman frequencies and mode assignments for various heme proteinsa
| Temp. | Protein | Coord. | ν10 | ν2 | ν3 | ν4 | ν(Fe-NO) | ν(N-O) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 298 K | Mb (pH 7) | 6 | 1635 | 1583 | 1500 | NPb | 560 | 1613 | (27) |
| Mb (pH 4) | 5 | 1645 | 1584 | 1508 | NP | 524 | 1668 | (27) | |
| β2(1-217) | 5 | 1646 | 1586 | 1510 | 1376 | 527 | 1676 | (9) | |
| β1(1-194) | 5 | 1647 | 1586 | 1510 | 1376 | 526 | 1677 | (9) | |
| sGC | 5 | 1646 | 1584 | 1509 | 1375 | 525 | 1677 | (28) | |
| 25 K | Mb (pH 4) | 5 | 1648 | 1588 | 1511 | 1375 | 524 | 1665 | * |
| β2(1-217) | 5 | 1647 | 1589 | 1509 | 1374 | 535 | 1677 | * | |
| β1(1-194) | 5 | 1646 | 1588 | 1510 | 1375 | 527 | 1677 | * |
Vibrations in cm−1
Not presented
this work.
To test the effect of temperature on another 5-coordinate FeII-NO complex, the RR spectra of the model heme protein myoglobin was examined. At pH 7 myoglobin forms a 6-coordinate complex with NO, but at pH 4 the ferrous NO complex is 5-coordinate (27). For the Mb-NO complex (pH 4), the heme skeletal marker bands and the N-O stretching vibration are most significantly effected by temperature, not the Fe-NO stretching vibration (Table 3). This confirms that the change in the Fe-NO stretching vibration of the β2(1-217)-NO complex is not a general effect due to the change in temperature.
Discussion
Acquiring molecular details about NO binding to sGC is critical to elucidating the mechanism of enzyme activation. sGC and the isolated heme domains from the β1 and β2 isoforms form indistinguishable 5-coordinate complexes with NO, based on electronic absorption and resonance Raman spectroscopy done at room temperature (9, 28, 29). These methods provide details about the porphyrin environment, but subtle differences in NO coordination may not be resolved with these experiments. EPR can probe directly the environment around an unpaired electron making it a particularly useful method for studying Fe-NO complexes.
Comparison of the EPR spectra of the β1(1-194)-NO and β2(1-217)-NO complexes show that the β2(1-217)-NO spectrum is significantly different from both the previously published sGCNO spectrum (21) and the β1(1-194)-NO spectrum (Figure 2a). The β2(1-217)-NO complex is similar to certain 5-coordinate FeII-NO model compounds (30) and the sGC-NO complex that is seen in the presence of YC-1 (17, 22). Simulations of the β1(1-194)-NO and β2(1-217)-NO EPR spectra show that the proteins have significantly different g-values (Table 1) and that the β2(1-217)-NO EPR signal can be simulated with reduced g-strain relative to the EPR signal of the sGC-NO and β1(1-194)-NO complexes. The large g-strain needed to simulate the sGC-NO and β1(1-194)-NO complexes may be due to conformational inhomogeneity of the NO complexes, which would be consistent with NO dissociation experiments that suggest sGC-NO is a mixture of different 5-coordinate FeII-NO conformations (24).
The presence of YC-1 or BAY 41-2272 causes the sGC-NO EPR spectrum to shift to a 5-coordinate species characterized by g1 = 2.106, g2 = 2.029, g3 = 2.010 (Table 2). This shift in the sGC-NO EPR signal is also observed in the presence of excess NO and GTP (Figure 4d) (17), conditions where maximal enzyme activity is observed. This suggests a common mechanism of activation for this family of compounds, by which they shift the sGC FeII-NO conformation to a highly active form. Several binding site(s) have been proposed for YC-1 and BAY 41-2272, including within the heme binding pocket (31, 32) and a pseudosymmetric site within the catalytic domains (22, 33, 34). To examine if YC-1 or GTP can bind to the sGC heme domains, the EPR spectra of the β1(1-385)-NO, β1(1-194)-NO, and β2(1-217)-NO complexes were collected in the presence of YC-1 and GTP (Figure S1). The spectra of these heme domains were not affected by the small molecules, suggesting that these allosteric activators do not bind to these domains or the domains lack the means to transduce the signal to the heme binding pocket.
To further examine the correlation between the sGC-NO EPR signal and enzyme activity, the EPR spectrum of the sGC-NO complex was examined with a stoichiometric amount of NO and excess NO relative to the sGC heme. This experiment was critical in light of recent studies showing that a low activity 5-coordinate FeII-NO complex is isolated in the presence of stoichiometric amounts of NO (5, 20). We observed a change in the heme environment of the sGC-NO complex in the presence of excess NO and GTP (Figure 4), conditions where sGC exhibits high activity, that was not observed with the low-activity sGC-NO species that was formed in the presence of stoichiometric amounts of NO and GTP. The difference in the EPR signal suggests that NO binds to the sGC-NO complex, and this binding event increases the signal intensity and resolution of the hyperfine splitting at g1 = 2.106 and g2 = 2.029. Addition of the non-cyclizable substrate analogue, GMPCPP, also increases the differences between the 1-NO and excess-NO EPR spectra as the hyperfine splitting of the g1 and g2 values become more apparent in the sample with excess NO. This indicates that the GTP effect is independent of catalysis or the presence of product (Figure 4). These experiments support the hypothesis that the sGC-NO conformation that exhibits an EPR signal characterized by g1 = 2.106, g2 = 2.029, g3 = 2.010 represents a highly active sGC species. Additionally, this provides the first spectroscopic evidence that NO binds to the sGC-NO complex, and shows that this binding event leads to a change in the sGC heme environment.
The EPR signals observed with GTP and excess NO, BAY 41-2272 or YC-1 (g1 = 2.106, g2 = 2.029, g3 = 2.010) represent a highly active state of sGC-NO, and the similar spectrum of the β2(1-217)-NO complex suggests that it adopts this FeII-NO conformation without the presence of the activators. This makes the β2(1-217)-NO complex a good model for studying the conformational change in the sGC-NO complex induced upon activator binding. Low-temperature resonance Raman spectroscopy was carried out on the β1(1-194)-NO and β2(1-217)-NO complexes to acquire more details about the different FeII-NO conformations. This study confirmed that both proteins form 5-coordinate FeII-NO complexes at 25 K, and revealed that the Fe-NO stretching vibration was significantly shifted in the β2(1-217)-NO complex compared to the β1(1-194)-NO complex (Table 3). The different Fe-NO stretching vibrations and EPR signals observed for the FeII-NO complexes may be attributed to changes in the Fe-N-O angle or variation in the heme pocket conformation (35, 36). We predict that this change in the Fe-NO stretching vibration also occurs when YC-1, BAY 41-2272 or GTP with excess NO bind to sGC. Tomita and colleagues did not observe a change in the sGC Fe-NO stretch in the presence of GTP; their resonance Raman spectra were acquired at 293 K and in the absence of excess NO (37). However, since their work, the effect of excess NO on the activity of sGC has been well established (5, 20, 38).
The varying activities of the seemingly identical 5-coordinate sGC-NO conformations have now been tied to spectroscopically distinct EPR signals. Taken together with the low-temperature resonance Raman data, this suggests that the mechanism of sGC activation by excess NO involves a conformational change that affects the environment around the NO molecule bound at the heme. The allosteric activators YC-1 and BAY 41-2272 can induce a similar change within the distal heme pocket, suggesting that these molecules may activate sGC by mimicking a native state of the protein normally induced by the binding of GTP and excess NO, and facilitate a conformational change from a lower-activity 5-coordinate state to a higher-activity 5-coordinate state. Further experiments involving multifrequency EPR or x-ray absorption fine-structure (XAFS) spectroscopy could provide insight into the molecular details of this conformational change within the heme pocket that leads to sGC activation.
Supplementary Material
Acknowledgements
We would like to thank Dr. W. Robert Scheidt, Dr. J. Timothy Sage, and Dr. Jonathan Winger for helpful discussions, Sarah Deng for assistance with sample preparations, and members of the Marletta lab for critical reading of this manuscript.
Footnotes
Funding was provided by NIH grants GM077365 (M.A.M.), GM73789 (R.D.B.) and GM33576 (T.G.S.)
Abbreviations: sGC, soluble guanylate cyclase; NO, nitric oxide; GTP, guanosine 5′-triphosphate; cGMP, cyclic guanosine 3′, 5′-monophosphate; GMPCPP, guanosine-5'-[(α,β-methylene]triphosphate; EPR, electron paramagnetic resonance; RR, resonance Raman; Mb, myoglobin; Sf9, Spodoptera frugiperda; DEA/NO, diethylammonium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate; YC-1, 3-(5′-hydroxymethyl-3′-furyl)-1-benzylindazole; HEPES, 4-(2-hydroxyethyl)-1-piperazineethane sulfonic acid; DTT, dithiothreitol; DMSO, dimethyl sulfoxide; EIA, enzyme immunoassay.
sGC amino acid numbering is that of the rat enzyme unless otherwise noted.
Supporting Information Available
Figure S1. GTP and YC-1 do not change the EPR spectra of the β1(1-385)-NO and β2(1-217)-NO complexes. Figure S2. A representative electronic absorption spectrum of the sGC-NO complex with and without addition of excess NO. Both complexes have an absorbance maximum at 399 nm. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Buechler WA, Ivanova K, Wolfram G, Drummer C, Heim JM, Gerzer R. Soluble guanylyl cyclase and platelet function. Ann. NY Acad. Sci. 1994;714:151–7. doi: 10.1111/j.1749-6632.1994.tb12039.x. [DOI] [PubMed] [Google Scholar]
- 2.Denninger JW, Marletta MA. Guanylate cyclase and the NO/cGMP signaling pathway. BBA-Bioenergetics. 1999;1411:334–350. doi: 10.1016/s0005-2728(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 3.Munzel T, Feil R, Mulsch A, Lohmann SM, Hofmann F, Walter U. Physiology and pathophysiology of vascular signaling controlled by guanosine 3′,5′-cyclic monophosphate-dependent protein kinase [corrected]. Circulation. 2003;108:2172–83. doi: 10.1161/01.CIR.0000094403.78467.C3. [DOI] [PubMed] [Google Scholar]
- 4.Warner TD, Mitchell JA, Sheng H, Murad F. Effects of cyclic GMP on smooth muscle relaxation. Adv. Pharmacol. 1994;26:171–94. doi: 10.1016/s1054-3589(08)60054-x. [DOI] [PubMed] [Google Scholar]
- 5.Cary SP, Winger JA, Marletta MA. Tonic and acute nitric oxide signaling through soluble guanylate cyclase is mediated by nonheme nitric oxide, ATP, and GTP. P. Natl. Acad. Sci. USA. 2005;102:13064–9. doi: 10.1073/pnas.0506289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz-Stewart I, Tiyyagura SR, Lin JE, Kazerounian S, Pitari GM, Schulz S, Martin E, Murad F, Waldman SA. Guanylyl cyclase is an ATP sensor coupling nitric oxide signaling to cell metabolism. P. Natl. Acad. Sci. USA. 2004;101:37–42. doi: 10.1073/pnas.0305080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harteneck C, Wedel B, Koesling D, Malkewitz J, Bohme E, Schultz G. Molecular cloning and expression of a new alpha-subunit of soluble guanylyl cyclase. Interchangeability of the alpha-subunits of the enzyme. FEBS Lett. 1991;292:217–22. doi: 10.1016/0014-5793(91)80871-y. [DOI] [PubMed] [Google Scholar]
- 8.Yuen PS, Potter LR, Garbers DL. A new form of guanylyl cyclase is preferentially expressed in rat kidney. Biochemistry. 1990;29:10872–8. doi: 10.1021/bi00501a002. [DOI] [PubMed] [Google Scholar]
- 9.Karow DS, Pan D, Davis JH, Behrends S, Mathies RA, Marletta MA. Characterization of functional heme domains from soluble guanylate cyclase. Biochemistry. 2005;44:16266–16274. doi: 10.1021/bi051601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winger JA, Marletta MA. Expression and characterization of the catalytic domains of soluble guanylate cyclase: Interaction with the heme domain. Biochemistry. 2005;44:4083–90. doi: 10.1021/bi047601d. [DOI] [PubMed] [Google Scholar]
- 11.Koglin M, Vehse K, Budaeus L, Scholz H, Behrends S. Nitric oxide activates the beta 2 subunit of soluble guanylyl cyclase in the absence of a second subunit. J. Biol. Chem. 2001;276:30737–43. doi: 10.1074/jbc.M102549200. [DOI] [PubMed] [Google Scholar]
- 12.Stone JR, Marletta MA. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry. 1996;35:1093–9. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, Brandish PE, Ballou DP, Marletta MA. A molecular basis for nitric oxide sensing by soluble guanylate cyclase. P. Natl. Acad. Sci. USA. 1999;96:14753–8. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derbyshire ER, Tran R, Mathies RA, Marletta MA. Characterization of nitrosoalkane binding and activation of soluble guanylate cyclase. Biochemistry. 2005;44:16257–65. doi: 10.1021/bi0515671. [DOI] [PubMed] [Google Scholar]
- 15.Friebe A, Schultz G, Koesling D. Sensitizing soluble guanylyl cyclase to become a highly CO-sensitive enzyme. EMBO J. 1996;15:6863–8. [PMC free article] [PubMed] [Google Scholar]
- 16.Stasch JP, Becker EM, Alonso-Alija C, Apeler H, Dembowsky K, Feurer A, Gerzer R, Minuth T, Perzborn E, Pleiss U, Schroder H, Schroeder W, Stahl E, Steinke W, Straub A, Schramm M. NO-independent regulatory site on soluble guanylate cyclase. Nature. 2001;410:212–215. doi: 10.1038/35065611. [DOI] [PubMed] [Google Scholar]
- 17.Makino R, Obayashi E, Homma N, Shiro Y, Hori H. YC-1 facilitates release of the proximal His residue in the NO and CO complexes of soluble guanylate cyclase. J. Biol. Chem. 2003;278:11130–7. doi: 10.1074/jbc.M209026200. [DOI] [PubMed] [Google Scholar]
- 18.Martin E, Czarnecki K, Jayaraman V, Murad F, Kincaid J. Resonance Raman and infrared spectroscopic studies of high-output forms of human soluble guanylyl cyclase. J. Am. Chem. Soc. 2005;127:4625–31. doi: 10.1021/ja0440912. [DOI] [PubMed] [Google Scholar]
- 19.Pal B, Kitagawa T. Interactions of soluble guanylate cyclase with diatomics as probed by resonance Raman spectroscopy. J. Inorg. Biochem. 2005;99:267–279. doi: 10.1016/j.jinorgbio.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 20.Russwurm M, Koesling D. NO activation of guanylyl cyclase. EMBO J. 2004;23:4443–50. doi: 10.1038/sj.emboj.7600422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone JR, Sands RH, Dunham WR, Marletta MA. Electron paramagnetic resonance spectral evidence for the formation of a pentacoordinate nitrosylheme complex on soluble guanylate cyclase. Biochem. Bioph. Res. Co. 1995;207:572–577. doi: 10.1006/bbrc.1995.1226. [DOI] [PubMed] [Google Scholar]
- 22.Yazawa S, Tsuchiya H, Hori H, Makino R. Functional characterization of two nucleotide-binding sites in soluble guanylate cyclase. J. Biol. Chem. 2006;281:21763–70. doi: 10.1074/jbc.M508983200. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim M, Xu C, Spiro TG. Differential sensing of protein influences by NO and CO vibrations in heme adducts. J. Am. Chem. Soc. 2006;128:16834–45. doi: 10.1021/ja064859d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winger JA, Derbyshire ER, Marletta MA. Dissociation of nitric oxide from soluble guanylate cyclase and H-NOX domain constructs. J. Biol. Chem. 2006;292:897–907. doi: 10.1074/jbc.M606327200. [DOI] [PubMed] [Google Scholar]
- 25.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Czernuszewicz RS. Closed-cycle refrigerator solution and rotating solid sample cells for anaerobic resonance Raman spectroscopy. Appl. Spectrosc. 1986;40:571–573. [Google Scholar]
- 27.Tomita T, Hirota S, Ogura T, Olson JS, Kitagawa T. Resonance Raman investigation of Fe-N-O structure of nitrosylheme in myoglobin and its mutants. J. Phys. Chem. B. 1999;103:7044–7054. [Google Scholar]
- 28.Deinum G, Stone JR, Babcock GT, Marletta MA. Binding of nitric oxide and carbon monoxide to soluble guanylate cyclase as observed with resonance Raman spectroscopy. Biochemistry. 1996;35:1540–7. doi: 10.1021/bi952440m. [DOI] [PubMed] [Google Scholar]
- 29.Schelvis JP, Zhao Y, Marletta MA, Babcock GT. Resonance Raman characterization of the heme domain of soluble guanylate cyclase. Biochemistry. 1998;37:16289–97. doi: 10.1021/bi981547h. [DOI] [PubMed] [Google Scholar]
- 30.Wayland BB, Olson LW. Spectroscopic studies and bonding model for nitric oxide complexes of iron porphyrins. J. Am. Chem. Soc. 1974;96:6037–41. doi: 10.1021/ja00826a013. [DOI] [PubMed] [Google Scholar]
- 31.Denninger JW, Schelvis JPM, Brandish PE, Zhao Y, Babcock GT, Marletta MA. Interaction of soluble guanylate cyclase with YC-1: Kinetic and resonance Raman studies. Biochemistry. 2000;39:4191–4198. doi: 10.1021/bi992332q. [DOI] [PubMed] [Google Scholar]
- 32.Li ZQ, Pal B, Takenaka S, Tsuyama S, Kitagawa T. Resonance Raman evidence for the presence of two heme pocket conformations with varied activities in CO-bound bovine soluble guanylate cyclase and their conversion. Biochemistry. 2005;44:939–946. doi: 10.1021/bi0489208. [DOI] [PubMed] [Google Scholar]
- 33.Friebe A, Russwurm M, Mergia E, Koesling D. A point-mutated guanylyl cyclase with features of the YC-1-stimulated enzyme: implications for the YC-1 binding site? Biochemistry. 1999;38:15253–7. doi: 10.1021/bi9908944. [DOI] [PubMed] [Google Scholar]
- 34.Lamothe M, Chang FJ, Balashova N, Shirokov R, Beuve A. Functional characterization of nitric oxide and YC-1 activation of soluble guanylyl cyclase: structural implication for the YC-1 binding site? Biochemistry. 2004;43:3039–48. doi: 10.1021/bi0360051. [DOI] [PubMed] [Google Scholar]
- 35.More C, Belle V, Asso M, Fournel A, Roger G, Guigliarelli B, Bertrand P. EPR spectroscopy: a powerful technique for the structural and functional investigation of metalloproteins. Biospectroscopy. 1999;5:S3–18. doi: 10.1002/(SICI)1520-6343(1999)5:5+<S3::AID-BSPY2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 36.Ubbink M, Worrall JA, Canters GW, Groenen EJ, Huber M. Paramagnetic resonance of biological metal centers. Annu. Rev. Biophys. Biomol. Struct. 2002;31:393–422. doi: 10.1146/annurev.biophys.31.091701.171000. [DOI] [PubMed] [Google Scholar]
- 37.Tomita T, Ogura T, Tsuyama S, Imai Y, Kitagawa T. Effects of GTP on bound nitric oxide of soluble guanylate cyclase probed by resonance Raman spectroscopy. Biochemistry. 1997;36:10155–60. doi: 10.1021/bi9710131. [DOI] [PubMed] [Google Scholar]
- 38.Derbyshire ER, Marletta MA. Butyl isocyanide as a probe of the activation mechanism of soluble guanylate cyclase. Investigating the role of non-heme nitric oxide. J. Biol. Chem. 2007;282:35741–8. doi: 10.1074/jbc.M705557200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.