Abstract
Fluorescence tends to produce the lowest detection limits for most forms of capillary electrophoresis. Two issues have discouraged its use in capillary isoelectric focusing. The first issue is fluorescent labeling of proteins. Most labeling reagents react with lysine residues and convert the cationic residue to a neutral or anionic product. At best, these reagents perturb the isoelectric point of the protein. At worse, they convert each protein into hundreds of different fluorescent products that confound analysis. The second issue is the large background signal generated by impurities within commercial ampholytes. This background signal is particularly strong when excited in the blue portion of the spectrum, which is required by many common fluorescent labeling reagents. This paper addresses these issues. For labeling, we employ Chromeo P540, which is a fluorogenic reagent that converts cationic lysine residues to cationic fluorescent products. The reaction products are excited in the green, which reduces the background signal generated by impurities present within the ampholytes. To further reduce the background signal, we photobleach ampholytes with high-power photodiodes. Photobleaching reduced the noise in the ampholyte blank by an order of magnitude. Isoelectric focusing performed with photobleached pH 3–10 ampholytes produced concentration detection limits of 270 ± 25 fM and mass detection limits of 150 ± 15 zmol for Chromeo P540 labeled β-lactoglobulin. Concentration detection limits were 520 ± 40 fM and mass detection limits were 310 ± 30 zmol with pH 4–8 ampholytes. A homogenate was prepared from a Barrett’s esophagus cell line and separated by capillary isoelectric focusing, reproducibly generating dozens of peaks. The sample taken for the separation was equal to the labeled protein homogenate from three cells.
Isoelectric focusing has been a valuable tool for protein analysis, particularly because of the commercialization of gels with immobilized pH gradients.1 This technology offers outstanding resolution of proteins to 0.001 delta pI units and is widely employed in biotechnology. The separation mechanism is orthogonal to that produced by sieving electrophoresis, and isoelectric focusing is commonly coupled with SDS-PAGE for two-dimensional separations of complex protein samples.
As in other electrophoretic separations, performing isoelectric focusing in a capillary format has advantages compared with classic slab-gels. These advantages include the ability to employ autoinjectors for automated analyses, lower sample loadings, and faster separations. Hjertén was the first to report isoelectric focusing in a capillary format.2 The performance of capillary isoelectric focusing is limited by poor detection sensitivity due to the small dimension of the capillary. Perhaps the most impressive performance has been obtained with mass spectrometric detection, which has resolved hundreds of proteins from nanograms of cellular lysate.3 Such experiments are beyond the ability of most labs, which instead employ absorbance detection in the ultraviolet portion of the spectrum, and these measurements suffer from the short path length available for the Beer’s law measurement. Concentration detection limits are in the high picomolar range and mass detection limits are in the low femtomole range.
Improved detection limits for capillary isoelectric focusing would enable more applications. In general, the best detection limits for capillary electrophoresis methods are produced by laser-induced fluorescence. This detection technology is ubiquitous in DNA sequencing by capillary sieving electrophoresis,4 and single molecule detection limits have been obtained in favorable cases for multiply labeled oligonucleotides in capillary sieving electrophoresis and highly fluorescent proteins in capillary zone electrophoresis.5–6
There is a surprisingly small literature for laser-induced fluorescence detection in capillary isoelectric focusing of proteins. A large fraction of those reports employ the technique for affinity-based assays.7–12 In those experiments, the affinity reagent is labeled with a highly fluorescent dye. Those studies usually employed a point-detector near the distal end of the separation capillary; focused analyte must be mobilized and driven past the detector. Detection limits are in the picomolar concentration level and the attomole mass level.
Pawlyszin has explored the use of whole-column imaging with fluorescence detection.12–19 Whole column imaging avoids mobilization of proteins across the point detector and dramatically speeds up the analysis. Impressive results include concentration detection limits of 10−13 M and mass detection limits of 10−19 mole for the highly fluorescent proteins R-phycoerythrin and green fluorescent protein.20 Both proteins were detected based on their native fluorescence. Performance was roughly two-orders of magnitude poorer for labeled bovine serum albumin in the whole column imagine detector.16
These reports tend to be silent on two important limitations of laser-induced fluorescence as a detector for capillary isoelectric focusing. First, covalent protein labeling can result in a serious degradation in separation efficiency. Most fluorescent reagents react with the cationic ε-amine of lysine residues, converting them to neutral or anionic products and changing their isoelectric point. There are 2n-1 possible fluorescent products from the reaction with a protein that has n labeling sites, and these products can generate very complex peaks from a single protein.21–22 Second, commercial ampholytes tend to produce a very large background signal that can swamp protein fluorescence. This background signal was first reported by Righetti in 1975 and was interpreted as being due to nitrogen aromatic compounds produced as a byproduct of ampholyte synthesis.23
The change in pI upon labeling can be minimized if the label converts the cationic lysine residue to a cationic fluorescent product. The CY family of dyes has been used for this purpose.24 These highly fluorescent reagents must be used at high concentrations; unreacted reagent and trace level impurities can generate background signals that confound analysis of minute amounts of proteins. Alternatively, non-covalent reagents, such as Nanoorange, can also be used to label proteins, but tend to produce relatively modest detection limits.16 Wolfbeiss reported the Chromeo family of fluorogenic reagents; these reagents are weakly fluorescent but produce a relatively highly fluorescent cationic product.25–27 We have employed both Chromeo P465 and Chromeo P503 for protein analysis by capillary electrophoresis.27–30 The labeled proteins produced good separation behavior in both submicellar electrophoresis and isoelectric focusing. While labeled proteins produced low zeptomole detection limits in zone electrophoresis, their performance in isoelectric focusing has been three orders of magnitude poorer.30 Proteins labeled with these reagents require excitation in the blue portion of the spectrum; we employed the 473 nm solid-state laser for excitation for Chromeo P503 labeled proteins. Unfortunately, all ampholytes tested generated a large background signal when excited at this wavelength.30
There are several approaches to reducing the background signal in fluorescence experiments. In general, there is a dramatic decrease in background signal with longer wavelength excitation. Impurities can be removed by chemical means, such as oxidation, or by adsorption to activated charcoal or similar adsorbants.31–32 Finally, impurities can be removed by photobleaching.32–33
In this paper, we report two innovations that allow capillary isoelectric focusing to be performed on minute amounts of labeled proteins. We found that the use of the Chromeo P540 dye with excitation at 532 nm and the use of high-power photodiodes to photobleach ampholytes resulted in an extremely low background signal, producing femtomolar concentration detection limits, zeptomole mass detection limits, and the ability to analyze a protein homogenate corresponding to the content of two cells.
Materials and methods
Reagents and materials
Unless stated, all reagents were purchased from Sigma-Aldrich and used without further purification. Solutions were made with distilled deionized water and vacuum filtered through a 0.22 μm filter. Biolytes were purchased from Biorad; both Pharmalytes and Biolytes were treated as described below. Chromeo P540 was purchased from Active Motif. Fused-silica capillaries with a Guarant coating were purchased from Alcor Bioseparations; all others were from Polymicro Technologies. Ampholytes were photobleached using a bank of high-power green light emitting diodes (see Supporting Information for details).
Sample and ampholyte preparation
Protein samples were first dissolved in water at a concentration of 10 mg/mL, aliquoted, and stored at −20 °C. A new sample was taken each day from the freezer and thawed at room temperature. Proteins were then labeled with Chromeo P540 with the same procedure described previously for P503.29–30 Briefly, 5 μL of protein solution was added to 15 μL of borate buffer (10 mM) and 5 μL of Chromeo 540 dye. The mixture was incubated for 30 min at room temperature. The reaction was complete when the solution color changed from light purple to maroon. Once labeled, 475 μL of 1% Tween 20 (v/v) solution was added to quench the reaction.
The homogenate sample was prepared from an immortalized CP18821 cell line of premalignant epithelial esophageal cells, kindly donated by Peter Rabinovitch of the Department of Pathology, University of Washington. Cells were cultured with human recombinant epidermal growth factor (rEGF) and keratinocyte-serum free medium supplemented with bovine pituitary extract (BPE) (Invitrogen, Carlsbad, CA). The cells were exposed to 0.25% trypsin (v/v), and rinsed three times in PBS to remove the media from the cell suspension. Cells were lysed in 1% Triton X (v/v) (100 μL per 1 million cells), sonicated for 20 min, and 20 μL aliquots were stored at −80 °C. Labeling was performed the same as with standard proteins; a 5 μL aliquot, equivalent to the homogenate of 50,000 cells, was labeled and diluted to 500 μL after the reaction was complete.
For all studies, a 4% ampholyte solution was prepared containing either a mixture of 2% Pharmalytes (3–10), 1% Pharmalyte (4–6), and 1% Biolyte (5–8) in a solution of dd H2O and 1% Tween 20 (v/v) or a 1:1 ratio of Pharmalyte (4–6) and Biolyte (5–8). A protein/ampholyte solution was made by taking 5 μL of labeled protein, equivalent to a homogenate prepared from 500 cells, and adding it to 95 μL of the 4% ampholyte solution. Finally, 550 nL of this solution was used to fill the capillary for analysis, corresponding to the protein content of roughly 3 cells.
Instrumentation
Our laser-induced fluorescence instrument has been described in detail elsewhere.34–37 Briefly, a multipurpose injection block was connected to a CZE1000R high-voltage power supply (Spellman).38 Analytes were detected using a post-column sheath flow cuvette34–37, 39–40. Fluorescence was excited by an 8-mW 532 nm diode-pumped solid-state laser beam (Crystalaser), collected with an M-PLAN 60x, 0.7 NA microscope objective, and filtered with a 550–600 nm bandpass filter. Light was detected by an avalanche photodiode single-photon counting module. Voltage programming and fluorescence measurement were controlled by LabView software.
Capillary coating procedure
Separation was performed either on a Gaurant coated capillary that had a length of 24 cm and an ID/OD of 50/150 μM or on an acrylamide-coated capillary that had a length of 28 cm and an ID/OD of 50/150 μm.41–42 Before each experiment the Gaurant capillary was rinsed with citric acid (100 mM) for 5 min at 5 psi followed by dd H2O for 5 min at 5 psi.
Hjertén’s method was used to prepare acrylamide-coated capillaries. 42 The capillary was attached to a locally constructed gas-handling system that uses N2 gas to purge solutions at 30 psi. For the first step in the coating procedure, a capillary was rinsed with 1M NaOH for 1 hr. Distilled water was then rinsed through the capillary for 10 min, followed by 1% (w/v) acetic acid for 2 hrs. Next, a 1:1 ratio of 3-(trimethoxysilyl) propyl methacrylate (MAPS) and methanol was purged at 10 psi for 1 hr. Both ends of the capillary were placed in MAPS:methanol solution for the next 18 hrs. On day two, the capillary was rinsed with methanol for 1 hr at 10 psi. A freshly prepared solution containing 4% (w/v) acrylamide, 0.1% (v/v) TEMED, and 0.1% (w/v) of ammonium persulfate was flushed through the capillary for 10 min at 10 psi. Both ends of the capillary were left in the acrylamide solution for 18 hrs. On day three, the capillary was rinsed with distilled water for 10 min at 10 psi and stored in water until use. Between runs the acrylamide-coated capillary was rinsed with 3 M HCl for 5 min at 5 psi followed by distilled water for 5 min at 5 psi before use.43
cIEF procedure
The anode end of the capillary was placed in phosphoric acid (10 mM, pH 2) and the cathode end was placed in a cuvette where the sheath flow was sodium hydroxide (40 mM, pH 12). The capillary was filled with analyte in a 4% ampholyte solution by purging the solution through the capillary for 2 min at 5 psi. Focusing voltage was held constant at 833 V/cm for 7.5 min. Chemical mobilization at the cathode was used for migration of the proteins by changing the sheath flow buffer from sodium hydroxide to a zwitterionic solution (10 mM aspartic acid, pH = 2.5); the field strength remained at 833 V/cm during mobilization.
Laser Power study
To study the effect of laser power on the fluorescence intensity, samples were continually infused into the sheath flow cuvette detector. Laser power was adjusted by rotating a polarizing filter placed in the optical path.
Data processing
Data were collected at 10 Hz. The data were corrected for photodetector nonlinearity before further processing.44 The corrected data were treated with a 5-point median filter to remove spikes caused by the passage of particles through the laser beam and then smoothed by convolution with a Gaussian function that had either a forty-point (4.0 s) or sixty-point (6.0 s) standard deviation.
Results and discussion
Laser power optimization
The photostability of Chromeo P540 has not been reported previously. We continuously infused a solution of 1.3-nM labeled β-lactoglobulin and measured the fluorescence signal as a function of laser power, which was adjusted by means of a polarizing filter. We fit the photobleaching signal to a simple three-level saturation model, Supporting Information. The saturation behavior was similar to Chromeo P503 labeled proteins; 17 3-mW of laser power generated a signal one-half of the limiting signal.
Ampholyte photobleaching
Righetti has published extensively on ampholyte properties,45–51 and has reported that the best resolution comes from mixing ampholytes from different manufacturers and different pH ranges. For this study, we prepared two mixtures of ampholytes: the first covered a relatively narrow range pH 4–8 and was prepared by mixing Biolyte pH 5–8 ampholytes with Pharmalyte pH 4–6 ampholytes. The second mixture spanned a wider pH range and was prepared by mixing both of those ampholytes solutions with Pharmalyte pH 3–10 ampholytes. All ampholytes generated very strong background signals in our laser-induced fluorescence detector. This background was not associated with the labeling reagents; we observed no change in fluorescence when the ampholytes were treated with the reagent.
We investigated a number of methods to reduce the background signal from ampholytes, including treatment with oxidizing agents, use of activated charcoal to selectively remove impurities, and photobleaching with a UV hand lamp. None of these methods was particularly useful in reducing the background signal to acceptable levels, and some resulted in increased background fluorescence.
Background signals in fluorescence experiments tend to decrease at longer wavelengths. The use of 532-nm excitation for the Chromeo P540 labeled proteins reduced the background signal by an order of magnitude compared to our earlier results with Chromeo P504, which required excitation at 473 nm.30
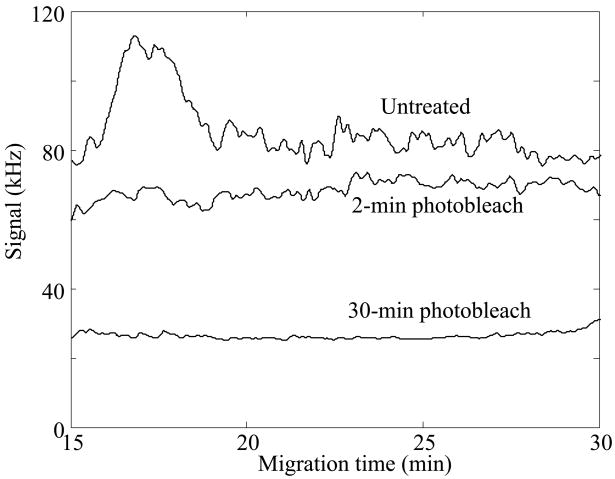
To photobleach the fluorescent impurities present within the ampholytes, we surrounded a 1-cm2 fluorescence cuvette with a bank of high-power green light emitting diodes. The spectra of these diodes peak near the 532 nm fluorescence excitation wavelength. Importantly, the diodes do not emit in the infrared portion of the spectrum and do not heat the solution. To determine the kinetics of photobleaching, a cuvette was filled with a mixture of ampholytes and placed in the ampholyte tanning booth. A 25-μL volume aliquot was removed periodically, diluted to 4%, and used to generate a capillary isoelectric focusing profile, Figure 1. The untreated ampholytes generate a strong background signal. Photobleaching reduced the background to 1/3 of its initial value and reduced the noise in the background signal by an order of magnitude. The narrow range ampholytes photobleached quickly; the background signal was reduced to negligible levels after 30 min. Addition of the Pharmalyte pH 3–10 ampholytes resulted in a much more photostable solution; five hours illumination was required to reduce the background to negligible levels, Supporting Information. Photobleaching produced a stable product; after 48 hrs in the dark, no fluorescent products returned. The background signal was similar to that produced by distilled water and presumably was due to weak Raman signal passing through our bandpass interference filter.
Figure 1.
Isoelectric focusing profile of pH 4–8 ampholyte blanks. The top curve is the untreated blank, the middle curve is after 2 minutes of photobleaching, and the bottom trace is after 30 minutes of photobleaching. Isoelectric focusing was performed in a Gaurant-coated capillary. Data not offset.
Limit of detection
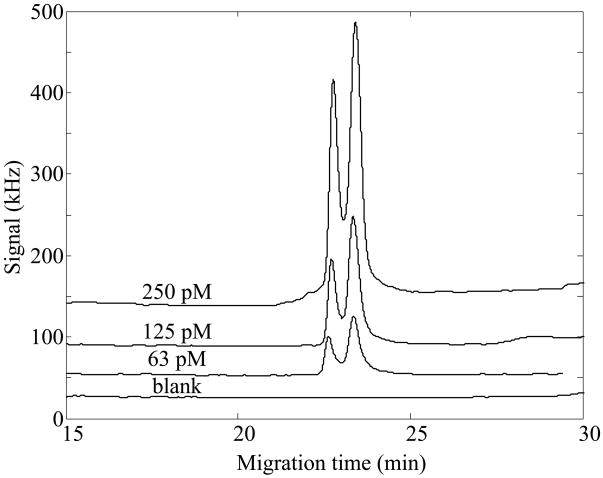
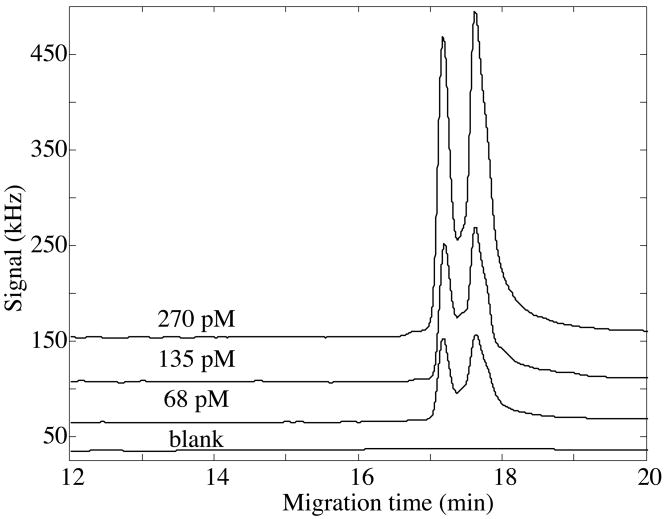
We characterized the performance of Chromeo P540-labeled β-lactoglobulin in isoelectric focusing with the two different ampholytes mixtures, Figures 2 and 3. In both cases, the ampholytes were photobleached to completion, producing a very low and featureless background signal. In both cases, the calibration curve was linear across the narrow concentration range tested (r>0.999).
Figure 2.
Capillary isoelectric focusing analysis of Chromeo p540-labeled β-lactoglobulin in photobleached pH 4–8 ampholytes. Isoelectric focusing was performed in a Gaurant coated capillary. Data offset for clarity.
Figure 3.
Capillary isoelectric focusing analysis of Chromeo p540-labeled β-lactoglobulin in photobleached pH 3–10 ampholytes. Isoelectric focusing was performed in an acrylamide-coated capillary. Data offset for clarity.
The calibration curve for the pH 4–8 range ampholytes produced concentration detection limits (3 s) of 520 ± 40 fM and mass detection limits of 310 ± 30 zmol. The calibration curve for the pH 3–10 range ampholytes produced concentration detection limits (3 s) of 325 ± 25 fM and mass detection limits of 180 ± 15 zmol. The wider range ampholytes had a steeper pH gradient, which resulted in sharper peaks and slightly improved detection limits compared to the narrow range ampholytes.
Separation of standards
A mixture of β-lactoglobulin and ovalbumin was subjected to isoelectric focusing with untreated and photobleached ampholytes, Supporting Information. The isoelectric focusing profiles for the proteins were identical; the background signal was much noisier for the untreated ampholytes.
Homogenate study
In our previous experiments, impurities in the ampholytes interfered with the analysis of dilute cellular homogenates. Relatively high concentration homogenates were required to generate a signal above the background, which led to problems with protein precipitation. These precipitates generated significant noise spikes.
The improved signal-to-noise produced by the use of Chromeo P540 with photobleached ampholytes allowed analysis of quite small amounts of cellular homogenate. In these experiments, the protein content of roughly 3 cells was aspirated into the capillary for analysis, with the exception of one experiment that used a more dilute solution, data below. Only a relatively few spikes were observed, typically in the 14–16 minute region.
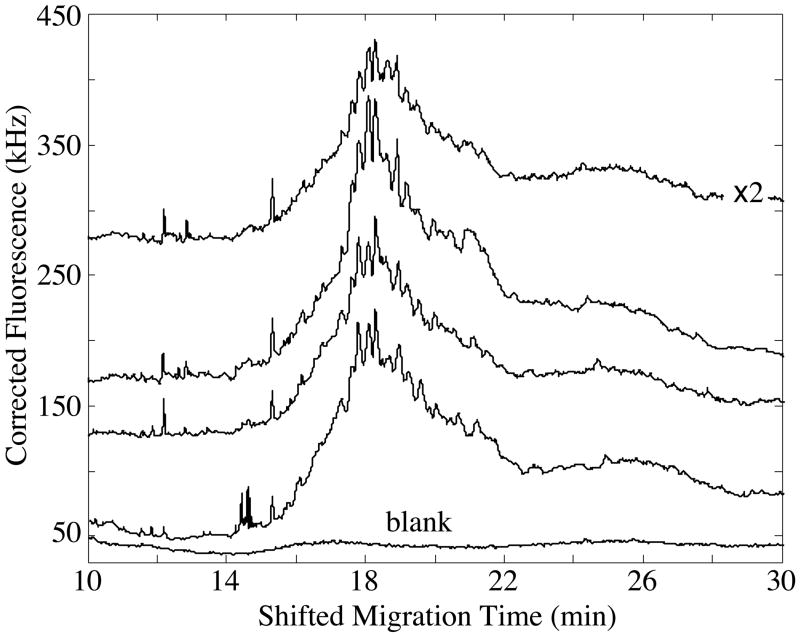
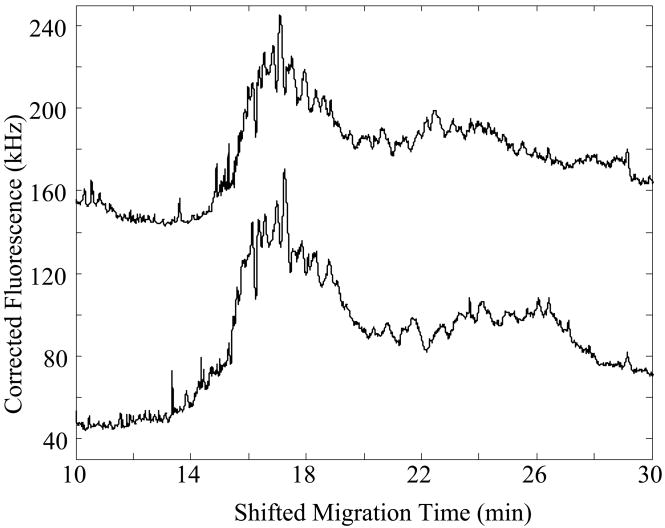
Figure 4 presents the capillary isoelectric focusing data generated from four samples in the pH 3–10 ampholyte mixture, generated over two days. In general, the focusing patterns show a broad envelope between 15–22 minutes, with a set of reproducible peaks, presumably caused by high-abundance components within the homogenate. Note that one sample was diluted by a factor of two; the protein content of that sample is slightly larger than the protein content of a single cell. The signal from that sample was multiplied by a factor of two to reflect the dilution.
Figure 4.
Capillary isoelectric focusing analysis of protein homogenate in pH 3–10 ampholytes. The amount of labeled protein taken for analysis corresponds to the content of ~3 cells, except for the top trace, which had been diluted by a factor of two; the data were multiplied by a factor of two to correct for the dilution. Separations performed in a Gaurant-coated capillary. Data offset for clarity.
Figure 5 presents two electropherograms generated with pH 4–8 ampholytes. Higher resolution is observed for these data than that of figure 4, as expected for the narrower pH range. Again, the electropherograms are reproducible. Perhaps 40 to 50 peaks can be observed in the electropherogram. Faster-migrating components with higher pI generate much sharper peaks than β-lactoglobulin, Supporting Information. 30
Figure 5.
Capillary isoelectric focusing analysis of protein homogenate in pH 4–8 ampholytes. The amount of labeled protein taken for analysis corresponds to the content of ~3 cells. Separations were performed in a Gaurant coated capillary. Data offset for clarity.
Conclusion
Isoelectric focusing with laser-induced fluorescence detection is shown to produce zeptomole mass detection limits for β-lactoglobulin and is capable of detecting the protein content from a few cells. However, significant effort will be required for single cell analysis. In the homogenate experiment, we employed ultrasonic disruption to lyse cells. In general, alternative methods are required for cellular disruption in single cell experiments, typically by treatment with surfactants.52,53 Our earlier experience has been with anionic surfactants for cell lysis; unfortunately, these surfactants are not compatible with isoelectric focusing. It will be necessary to employ nonionic or zwiterionic surfactants for lysis and protein solubilization.
There are several modifications that can be envisioned for the experiment. Fluorescently labeled peptides have been proposed as standards in capillary electrophoresis.54 These standards could be prepared with dyes whose emission differs from Chromeo P540, and a two-color fluorescence detection could be used to discriminate the signal from standards and samples.55
It obviously would be desirable to couple capillary isoelectric focusing with capillary sieving electrophoresis for two-dimensional protein separations.56–57 Also, it would be intriguing to replace the fluorescence detector with a high-sensitivity refractive index detector or native fluorescence detector to perform label-free analysis.58–60
Supplementary Material
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We acknowledge support from the National Institutes of Health grant R33CA122900. We are particularly grateful to Professor Robert Campbell of the University of Alberta, who suggested the use of green LEDs in the construction of the ampholyte tanning booth. We acknowledge Professors Paul Hopkins and Forest Michael of the chemistry department at the University of Washington for useful conversations on destruction of fluorescent impurities. We also acknowledge Roy Olund and Lon Buck for the design and construction of the ampholyte tanning booth.
Literature cited
- 1.Bjellqvist B, Ek K, Righetti PG, Gianazza E, Görg A, Westermeier R, Postel W. J Biochem Biophys Methods. 1982;6:317–339. doi: 10.1016/0165-022x(82)90013-6. [DOI] [PubMed] [Google Scholar]
- 2.Hjertén S, Elenbring K, Kilar F, Liao JL, Chen AJ, Siebert CJ, Zhu MD. J Chromatogr. 1987;403:47–61. doi: 10.1016/s0021-9673(00)96340-4. [DOI] [PubMed] [Google Scholar]
- 3.Jensen PK, Pasa-ToliV L, Anderson GA, Horner JA, Lipton MS, Bruce JE, Smith RD. Anal Chem. 1999;71:2076–2084. doi: 10.1021/ac990196p. [DOI] [PubMed] [Google Scholar]
- 4.Dovichi NJ, Zhang J. Angew Chem Int Ed Engl. 2000;39:4463–4468. doi: 10.1002/1521-3773(20001215)39:24<4463::aid-anie4463>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen DY, Dovichi NJ. Anal Chem. 1996;68:690–696. [Google Scholar]
- 6.Haab BB, Mathies RA. Anal Chem. 1995;67:3253–3260. doi: 10.1021/ac00114a023. [DOI] [PubMed] [Google Scholar]
- 7.Shimura K, Karger BL. Anal Chem. 1994;66:9–15. doi: 10.1021/ac00073a004. [DOI] [PubMed] [Google Scholar]
- 8.Shimura K, Hoshino M, Kamiya K, Katoh K, Hisada S, Matsumoto H, Kasai K. Electrophoresis. 2002;23:909–917. doi: 10.1002/1522-2683(200203)23:6<909::AID-ELPS909>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Bornemann C, Burggraef T, Heimbüchel G, Hanisch FG, Winkels S. Anal Bioanal Chem. 2003;376:1074–1080. doi: 10.1007/s00216-003-2038-3. [DOI] [PubMed] [Google Scholar]
- 10.Cunliffe JM, Liu Z, Pawliszyn J, Kennedy RT. Electrophoresis. 2004;25:2319–2325. doi: 10.1002/elps.200405953. [DOI] [PubMed] [Google Scholar]
- 11.Knittle JE, Roach D, Horn PB, Voss KO. Anal Chem. 2007;79:9478–9483. doi: 10.1021/ac071537z. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Drabovich AP, Krylov SN, Pawliszyn J. Anal Chem. 2007;79:1097–1100. doi: 10.1021/ac061876c. [DOI] [PubMed] [Google Scholar]
- 13.Wu XZ, Wu J, Pawliszyn J. Electrophoresis. 1995;16:1474–1478. doi: 10.1002/elps.11501601244. [DOI] [PubMed] [Google Scholar]
- 14.Fang X, Tragas C, Wu J, Mao Q, Pawliszyn J. Electrophoresis. 1998;19:2290–2295. doi: 10.1002/elps.1150191307. [DOI] [PubMed] [Google Scholar]
- 15.Mao Q, Pawliszyn J. J Biochem Biophys Methods. 1999;39:93–110. doi: 10.1016/s0165-022x(99)00006-8. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Pawliszyn J. Anal Chem. 2003;75:4887–4894. doi: 10.1021/ac034587m. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Pawliszyn J. J Proteome Res. 2004;3:567–571. doi: 10.1021/pr034114w. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Pawliszyn J. Anal Biochem. 2005;336:94–101. doi: 10.1016/j.ab.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Pawliszyn J. Anal Chem. 2005;77:165–171. doi: 10.1021/ac049229d. [DOI] [PubMed] [Google Scholar]
- 20.Huang T, Pawliszyn J. Analyst. 2000;125:1231–1233. [Google Scholar]
- 21.Zhao JY, Waldron KC, Miller J, Zhang JZ, Harke HR, Dovichi NJ. J Chromatogr. 1992;608:239–242. doi: 10.1016/0021-9673(92)87129-v. [DOI] [PubMed] [Google Scholar]
- 22.Richards D, Stathakis C, Polakowski R, Ahmadzadeh H, Dovichi NJ. J Chromatogr A. 1999;853:21–25. doi: 10.1016/s0021-9673(99)00687-1. [DOI] [PubMed] [Google Scholar]
- 23.Righetti PG, Righetti A, Galante E. Anal Biochem. 1975;63:423–432. doi: 10.1016/0003-2697(75)90365-6. [DOI] [PubMed] [Google Scholar]
- 24.Unlu M, Morgan M, Minden J. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 25.Wetzl BK, Yarmoluk SM, Craig DB, Wolfbeis OS. Angew Chem Int Ed Engl. 2004;43:5400–5402. doi: 10.1002/anie.200460508. [DOI] [PubMed] [Google Scholar]
- 26.Craig DB, Wetzl BK, Duerkop A, Wolfbeis OS. Electrophoresis. 2005;26:2208–2213. doi: 10.1002/elps.200410332. [DOI] [PubMed] [Google Scholar]
- 27.Wojcik R, Swearingen KE, Dickerson JA, Turner EH, Ramsay LM, Dovichi NJ. J Chromatogr A. 2008;1194:243–248. doi: 10.1016/j.chroma.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swearingen KE, Dickerson JA, Turner EH, Ramsay LM, Wojcik R, Dovichi NJ. J Chromatogr A. 2008;1194:249–252. doi: 10.1016/j.chroma.2008.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner EH, Dickerson JA, Ramsay LM, Swearingen KE, Wojcik R, Dovichi NJ. J Chromatogr A. 2008;1194:253–256. doi: 10.1016/j.chroma.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsay L, Dickerson J, Dovichi NJ. Electrophoresis. 2008 doi: 10.1002/elps.200800498. in press. [DOI] [PubMed] [Google Scholar]
- 31.Stephan J, Dorre K, Brakmann S, Winkler T, Wetzel T, Lapczyna M, Stuke M, Angerer B, Ankenbauer W, Foldes-Papp Z, Rigler R, Eigen M. J Biotechnol. 2001;86:255–267. doi: 10.1016/s0168-1656(00)00417-x. [DOI] [PubMed] [Google Scholar]
- 32.Billinton N, Knight AW. Anal Biochem. 2001;291:175–197. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- 33.Affleck RL, Ambrose WP, Demas JN, Goodwin PM, Schecker JA, Wu JM, Keller RA. Anal Chem. 1996;68:2270–2276. doi: 10.1021/ac9512517. [DOI] [PubMed] [Google Scholar]
- 34.Cheng YF, Dovichi NJ. Science. 1988;242:562–564. doi: 10.1126/science.3140381. [DOI] [PubMed] [Google Scholar]
- 35.Wu S, Dovichi NJ. J Chromatogr. 1989;480:141–155. doi: 10.1016/s0021-9673(01)84284-9. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Wu J, Baker GB, Parent M, Dovichi NJ. J Chromatogr A. 2001;914:293–298. doi: 10.1016/s0021-9673(01)00539-8. [DOI] [PubMed] [Google Scholar]
- 37.Kraly JR, Jones MR, Gomez DG, Dickerson JA, Harwood MM, Eggertson M, Paulson TG, Sanchez CA, Odze R, Feng Z, Reid BJ, Dovichi NJ. Anal Chem. 2006;78:5977–5986. doi: 10.1021/ac061029+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krylov SN, Starke DA, Arriaga EA, Zhang Z, Chan NW, Palcic MM, Dovichi NJ. Anal Chem. 2000;72:872–877. doi: 10.1021/ac991096m. [DOI] [PubMed] [Google Scholar]
- 39.Dovichi NJ, Martin JC, Jett JH, Keller RA. Science. 1983;219:845–847. doi: 10.1126/science.6823553. [DOI] [PubMed] [Google Scholar]
- 40.Dovichi NJ, Martin JC, Jett JH, Trkula M, Keller RA. Anal Chem. 1984;56:348–534. doi: 10.1021/ac00267a010. [DOI] [PubMed] [Google Scholar]
- 41.Dolnik V, Gurske WA, Padua A. Electrophoresis. 2001;22:707–719. doi: 10.1002/1522-2683(200102)22:4<707::AID-ELPS707>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 42.Hjertén S, Liao JL, Yao KQ. J Chromatogr. 1987;387:127–138. doi: 10.1016/s0021-9673(01)94519-4. [DOI] [PubMed] [Google Scholar]
- 43.Suratman A, Watzig H. J Sep Sci. 2008;31:1834–1840. doi: 10.1002/jssc.200700678. [DOI] [PubMed] [Google Scholar]
- 44.Turner EH, Lauterbach K, Pugsley HR, Palmer VR, Dovichi NJ. Anal Chem. 2007;79:778–781. doi: 10.1021/ac061778r. [DOI] [PubMed] [Google Scholar]
- 45.Righetti PG. Isoelectric focusing: Theory, Methodology, and Applications. Elsevier Biomdeical Press; Amsterdam: 1989. [Google Scholar]
- 46.Sebastiano R, Simó C, Mendieta ME, Antonioli P, Citterio A, Cifuentes A, Peltre G, Righetti PG. Electrophoresis. 2006;27:3919–3934. doi: 10.1002/elps.200600170. [DOI] [PubMed] [Google Scholar]
- 47.Simó C, Mendieta ME, Antonioli P, Sebastiano R, Citterio A, Cifuentes A, Peltre G, Righetti PG. Electrophoresis. 2006;27:4849–4858. doi: 10.1002/elps.200600344. [DOI] [PubMed] [Google Scholar]
- 48.Simó C, Mendieta ME, Antonioli P, Sebastiano R, Citterio A, Cifuentes A, Righetti PG. Electrophoresis. 2007;28:715–723. doi: 10.1002/elps.200600499. [DOI] [PubMed] [Google Scholar]
- 49.Simó C, Citterio A, Righetti PG. Electrophoresis. 2007;28:1488–1494. doi: 10.1002/elps.200600853. [DOI] [PubMed] [Google Scholar]
- 50.Simó C, Citterio A, Righetti PG. Electrophoresis. 2007;28:3156–3162. doi: 10.1002/elps.200700123. [DOI] [PubMed] [Google Scholar]
- 51.Righetti PG, Simó C, Sebastiano R, Citterio A. Electrophoresis. 2007;28:3799–3810. doi: 10.1002/elps.200700232. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Krylov S, Arriaga EA, Polakowski R, Dovichi NJ. Anal Chem. 2000;72:318–322. doi: 10.1021/ac990694y. [DOI] [PubMed] [Google Scholar]
- 53.Hu S, Michels DA, Fazal MA, Ratisoontorn C, Cunningham ML, Dovichi NJ. Anal Chem. 2004;76:4044–4049. doi: 10.1021/ac0498314. [DOI] [PubMed] [Google Scholar]
- 54.Shimura K, Kasai K. Electrophoresis. 1995;16:1479–1484. doi: 10.1002/elps.11501601245. [DOI] [PubMed] [Google Scholar]
- 55.Chen D, Harke HR, Dovichi NJ. Nucleic Acids Res. 1992;20:4873–4880. doi: 10.1093/nar/20.18.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michels DA, Hu S, Schoenherr RM, Eggertson MJ, Dovichi NJ. Mol Cell Proteomics. 2002;1:69–74. doi: 10.1074/mcp.t100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 57.Michels DA, Hu S, Dambrowitz KA, Eggertson MJ, Lauterbach K, Dovichi NJ. Electrophoresis. 2004;25:3098–3105. doi: 10.1002/elps.200405939. [DOI] [PubMed] [Google Scholar]
- 58.Bornhop DJ, Dovichi NJ. Anal Chem. 1986;58:504–505. [Google Scholar]
- 59.Bornhop DJ, Latham JC, Kussrow A, Markov DA, Jones RD, Sørensen HS. Science. 2007;317:1732–1736. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- 60.Lillard SJ, Yeung ES. J Chromatogr B. 1996;687:363–369. doi: 10.1016/s0378-4347(96)00253-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.