Abstract
Current approaches to protein site-directed mutagenesis require an independent user operation for each mutation. This can impede large-scale scanning mutagenesis projects such as mapping protein interaction surfaces, active sites, or epitopes. It also prevents the creation of protein libraries of defined complexity for directed evolution purposes. Here we present a simple, fast, and effective way to perform scanning codon mutagenesis throughout a protein sequence. The process allows the researcher to define the new codon change therefore any amino acid mutation can be achieved. We demonstrate this approach by creating a library of proteins that contain single unnatural amino acid mutations encoded by the amber stop codon, TAG. The mutant proteins generated by this method can be expressed and assayed individually, or used together as a mixed population of “rationally diversified” protein sequences.
Keywords: scanning mutagenesis, amino acid, unnatural, transposon, photo-crosslinking
Whether through chemical synthesis or genetic mutagenesis, the ability to rationally change the amino acid sequence of proteins has had a profound impact upon the understanding of structure and function. Diversity-oriented approaches to protein mutagenesis such as error-prone PCR(1), DNA shuffling(2), or cassette mutagenesis(3) have expanded on this and can easily create mixtures of complexity that exceed protein screening capabilities. Scanning mutagenesis utilizes a different approach by making small changes, such as single alanine replacements(4), so that the contribution of individual side chains can be deciphered and later exploited. This generates mixtures of modest, defined complexity and is well suited for mapping protein-protein interactions, solvent accessibility, or active sites using low to medium-throughput assays. The limitation, however, is the time and effort spent creating the mutations via PCR-based methods. For example there are a clear, defined number of single alanine mutants that could be made of a protein 500 amino acids long, but generating those mutants with conventional methods is expensive, time consuming and most likely the limiting factor when considering such a project.
We faced this conceptual challenge when seeking to investigate protein surfaces using photo-crosslinking unnatural amino acids that are genetically encoded by the amber stop codon, TAG(5-7). If one is attempting to map multiple binding sites on a protein, it is important to create mutations that are close, but not integral to binding interfaces. Ideally, one would want to sample many different sites - perhaps every site - but making hundreds of amber codon mutations is not practical. Further, it's not possible to randomly introduce these codons via single nucleotide exchanges incorporated with error-prone PCR because of the redundancy and composition of the genetic code. For example the probability of randomly changing a proline codon (CCC) to an amber codon (TAG) by three successive nucleotide mutations is effectively zero. Here we present a rational approach to randomly replace single codons within an open reading frame with TAG such that every site is sampled. This collection of gene mutants then gives rise to the independent mutation of every amino acid residue in the protein with an unnatural amino acid. In contrast to other methods that are used to perform scanning mutagenesis (8, 9), these library members are not redundant or silent and are enriched with members containing only a single mutation. Moreover, we have insured that that mutation is in the correct reading frame of the protein being studied. This results in a “cleaner” library and allows every possible mutant to be represented in fairly small collection of clones particularly when compared to other methods. This convergent process, consisting of relatively simple DNA manipulations, can be applied to any open reading frame encoding a protein.
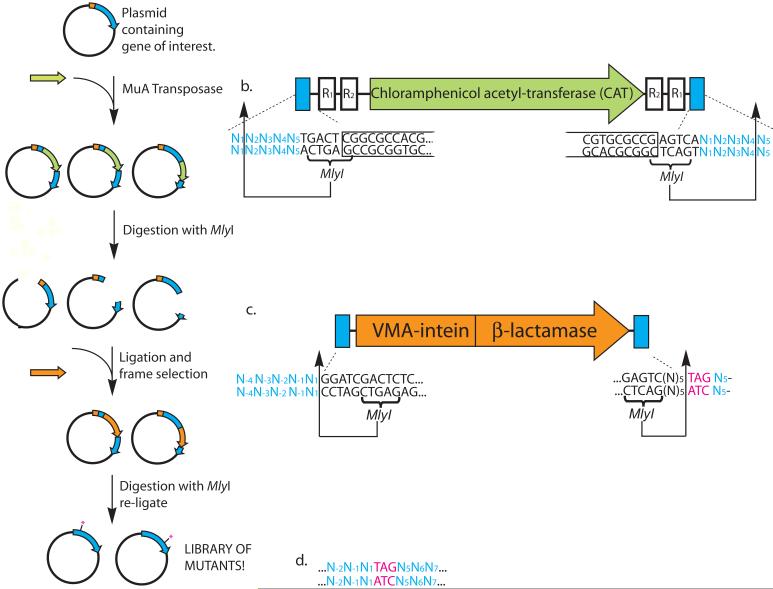
Previously, transposon methods have been used to map protein domains via randomly distributed amino acid linker insertions (10-12). Recently Jones and co-workers described a transposon-based system for removal of triplet nucleotides from plasmid DNA using TypeIIS asymmetric restriction enzymes(13) and have gone on replace those deletions with new sequence in an approach referred to as Tri-NEX(14). While this method replaces nucleotides, our goal was to perform single unnatural amino acid mutations in a protein and not affect the sequence that flanks the mutation. We adopted a similar strategy in which we performed nucleotide substitutions consisting of an amber stop codon and then ensure that this replacement is in-frame with the original coding sequence by genetic selection. The process consists of i) a transposon reaction to randomly insert two unique MlyI restriction sites, ii) digestion with MlyI, which randomly breaks a coding DNA sequence and removes three nucleotides, iii) replacement and re-assembly with three new nucleotides consisting of the desired mutation, and iv) simultaneous verification that this replacement occurred in the correct reading frame of the gene (Figure 1).
Figure 1.
Schematic diagram of Codon Scanning Mutagensis. a. A plasmid containing the gene (blue) to be mutated is first targeted with a modified Mu transposon (green) that is inserted in vitro by the action of MuA transposase, resulting in a library of plasmids that are recovered in E. coli. This insertion event results in the duplication of five nucleotides, N1N2N3N4N5, at the site of insertion (shown in b). The library can be purified to only contain members where the transposon was inserted into the gene of interest by restriction digest. b. The transposon fragment with recognition sequences, R1 and R2, is modified to have MlyI restirciton sites on either end. Three base pairs, N2N3N4 of the original plasmid sequence is then removed by digestion with MlyI, and (c.) a new frame selectable TAG linker segment of DNA (orange) is ligated into this digestion site, resulting in a library of plasmids that are selected for correct reading frame in E. coli. The basis for the reading frame selection is assembly of a functional β-lactamase-intein chimera which splices to yield colonies resistant to ampicillin. This library is then subjected to a second digestion and re-ligated, thus resulting in (d.) selective removal of the linker and a net replacement of a new codon.
As a proof of principle, we performed this mutagenesis reaction on a plasmid containing a target gene encoding glutathione-S-transferase (GST) from Schistosoma japonicum. The plasmid (pIT-GST) we used is a derivative of pInSALect which expresses proteins as N-terminal fusions to a β-lactamase-VMA intein fusion protein. This is a system that has been used in the past for isolating intact open reading frames(15). Using a modified Mu transposon, a library of this plasmid was generated and recovered in E. coli resulting in >104 independent clones, sufficient to accommodate all possible insertion sites assuming a random event distribution. Previous studies have shown that when performed upon naked plasmid DNA this event is random, (16, 17). The mutant plasmids were verified by restriction mapping of 10 individual clones which were chosen randomly. To ensure that mutations would only occur in the target gene, GST, the library was digested with BamHI and SalI removing GST and the transposon and then the fragment ligated with the original plasmid backbone. The total pool was then combined and digested with MlyI to remove the transposon and in doing so, generate a “library of vectors” all of which are exactly the same size and have been randomly linearized with concomitant removal of three base pairs and have ends ready for ligation. To remove those clones which have been linearized out-of-frame with the target GST gene, we created a DNA linker segment containing the mutant codon TAG, that is frame-selectable and compatible with the original plasmid pIT-GST. Importantly, this fragment consists of a fusion gene which must be ligated in-frame with the partner on the target plasmid to confer resistance to ampicillin (Figure 1). Upon successful in-frame ligation with the fusion gene on the original plasmid, constructs are generated that express a fusion protein where two halves of β-lactamase resistance marker are separated by a self-splicing intein, and the remnant piece of the coded target protein. This precursor protein is rapidly spliced in vivo by the intein(15), generating functional β-lactamase. Out-of-frame ligations do not allow for this pre-protein to be expressed correctly and are therefore removed. As expected, when we re-plated this library containing the linker replacement on media containing ampicillin, approximately 90% of all library members were removed (as judged by colony count comparison to non-selective plates), consistent with removal of the 5/6th of the library derived from out-of-frame and reversed ligations. Moreover, DNA sequence analysis of the plasmids derived from the surviving colonies verified that all clones enriched in this step consisted of only in-frame fusion genes that were randomly distributed.
This frame-selected collection of clones was pooled and the plasmid DNA extracted. The pooled library DNA was then again digested with MlyI, to remove the linker segment (Figure 1), re-ligated, and re-transformed into E. coli. Simultaneously, the plasmid is re-circularized, resulting in no net addition or subtraction of nucleotides and a seamless replacement of the original codon with a new amber stop codon, TAG. To verify that each of these steps proceeded as expected, we isolated 48 individual clones as representatives of the library and re-sequenced the ligation sites, one of which is shown in Figure 2. Out of the 48 clones, we observed 15 different clones (>30%) that were entirely correct sequences containing in-frame mutations. In the other clones we did observe unexpected deformations in the coding sequence such as the addition or removal of single nucleotides at the ligation sites (supporting info). We believe that this is due to the fidelity of the MlyI restriction digests in the final step of library assembly. Nevertheless, the transposon insertions and the resulting mutations are distributed across the protein coding sequence, consistent with previous reports of the very limited specificity of Mu transposomes(17). Further, the step-wise nature of this process allows one to recover and amplify the plasmid DNA in cells and therefore count colonies to ensure that the modest, yet statistically sampled diversity (~103 clones) is more than maintained throughout the process.
Figure 2.
Sequence of a single clone proceeding through the process. a. Sequence of wild-type glutathione S-transferase, the highlighted ACA is the codon removed by MlyI digestion of the transposon. b. The linker is inserted in-frame resulting in ampicillin resistance, the MlyI restriction sites are highlighted and red arrows indicate the cut sites. Upon digestion with MlyI and subsequent ligation the codon TAG (shown in yellow c.) replaces ACA.
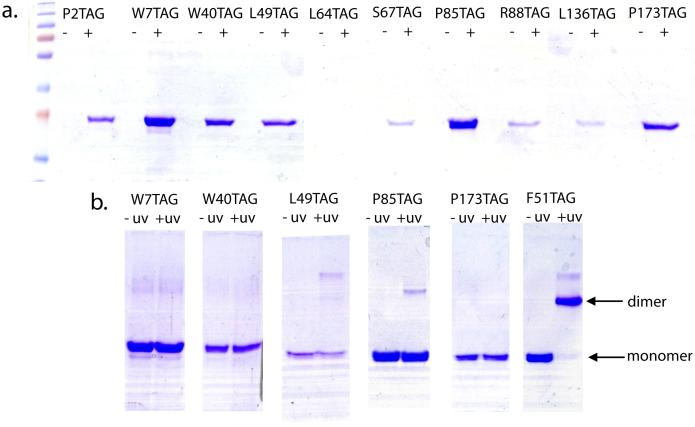
After library construction, ten plasmids containing GST with in-frame TAG mutations were chosen based on sequencing results to be used for unnatural amino acid incorporation. The ten mutant clones obtained from the library were sub-cloned into an overexpression vector and then used to transform E. coli cells which harbor the translational machinery capable of inserting the unnatural photo-crosslinking amino acid p-benzoylphenylalanine (pBpa)(6). The resulting double transformants were then grown as individual cultures and assayed for protein expression in the presence and absence of pBpa. As can be seen in Figure 3 these clones only express protein in the presence of the unnatural amino acid. The protein expression yields of the mutants was variable and indeed, one clone (L64TAG) failed to produce at all. We believe this variability is based on the context effects amber suppression (18), and in the case of no expression, the stability of the mutant protein. Just as with standard site-directed mutagenesis, one would not expect all mutants to be equally stable. In order to probe the photoreactivity of those mutants which showed good expression while displaying pBpa (W7, W40, L49, P85, and P173), we irradiated them at 365nm and analyzed for the formation of covalent cross-links across the dimeric GST interface (Figure 3). Of the five mutants we assayed, only P85 showed an indication of successful cross-link formation, albeit minor when compared to a mutant that is known to cross-link very well (F51)(6). This is consistent with the expectation that only those mutations in or near the dimeric interface (such as P85 and F51) would be capable of capturing the interface. More importantly however, these individual protein expression results can be viewed as members of the larger population of clones that exists in the library mixture, each expressing a unique protein mutant containing a single unnatural amino acid mutation at a random position. That is, overexpression of the pool of clones created using this approach would generate a mixture of proteins (in this case GST) all of which are the same apart from a single, random unnatural amino acid mutation.
Figure 3.
a. SDS/PAGE analysis of expression of 10 amber codon mutants in the presence (+) and absence (-) of pBpa. Amino acid residues mutated to TAG codons are shown. One mutant, L65TAG, did not express for unknown reasons. b. Photo-activity of high expression mutants derived from scanning compared to a mutant known to cross-link (F51).
In summary, we have created an alternative, non-PCR-based approach for creating mixtures of (all possible) single residue, unnatural protein mutants to facilitate the initiation of large-scale scanning mutagenesis projects. The requirement for the application of this method is the lack of the unique and rare restriction sites used in the cloning process. If present in a candidate open reading frame, these sites can be removed easily by traditional site-directed silent mutagenesis or total gene synthesis(19, 20). In this case, we chose to express mutant proteins bearing photo-reactive groups at the site of mutation. One could envision other uses for scanning unnatural amino acid mutagenesis including strategies for bioconjugation, isotope or fluorophore labeling, and post-translational modifications. There is also evidence that enzymes bearing unnatural amino acids can exhibit improved catalytic activity, above and beyond that available using the 20 genetically encoded amino acids (21). In addition to amber codon mutations, an analogous approach (using a different codon linker segment) could be taken to perform alanine scanning to map protein epitopes(4), cysteine scanning to map protein surfaces(22), or lysine or aspartic acid scanning to “super-charge” proteins(23). Indeed, we have now created the full complement of the required 20 reading-frame-selectable linkers, such that a combined approach in which multiple linkers coding for custom mixtures of amino acids (or all 20) could be used as a method for creating defined molecular diversity in protein libraries. This represents an alternative to error prone mutagenesis that is not limited by the redundancy of the genetic code. This allows a much more efficient sampling of protein sequence space. Finally, because the approach is convergent, it could be iterated to generate libraries of proteins that contain two, three, or four mutations, etc, or collections of single mutants recombined in vitro (24, 25) to create a distribution of these possibilities. Rational protein diversification methods such as these could generate protein libraries that are much more likely to contain new or improved protein function.
Methods
Scanned library construction
To create a codon-scanned library the target plasmid (in this case pIT-GST which is described in the supporting info) was used in an in vitro transposon mutagenesis reaction. In a 20 μL reaction, 400 ng of target plasmid was mixed with a 1.3 molar excess of the transposon, which is a BglII digested PCR product. To the DNA mixture, 2 μL of 10X HyperMu reaction buffer (1.5 M potassium acetate, 0.5 M Tris-acetate (pH 7.5), 0.1 M magnesium acetate and 40 mM spermidine) and 1 μL of Mu transposase (Epicentre) was added and the reaction incubated at 30 °C for 4 hours. The reaction was then stopped by adding SDS to a final concentration of 0.1%(w/v) and heating to 75 °C for 10 minutes. The reaction was then placed on ice and 4 μL was transformed into 200 μL of chemically competent E. coli, recovered in 1 mL SOC for 1hr, and then plated on LB agar supplemented with 50 μg/mL kanamycin and 10 μg/mL chloramphenicol and grown at 37 °C. This resulted in ~17,000 resistant colonies and a transposition efficiency of ~2.5%, when compared to control plates containing only kanamycin. Ten of these individual colonies were picked and analyzed by restriction to insure random digestion. The library was then pooled and plasmid DNA isolated. Because the transposon inserts randomly into the target plasmid, we chose to further purify the library to isolate only those insertions in the GST gene. The library was digested with BamHI and SalI resulting in four bands; pIT backbone + transposon, pIT backbone, GST + transposon, and GST. The bands for the pIT backbone and GST + transposon were isolated by gel electrophoresis, re-ligated and then used to transform chemically competent E. coli, recovered in 4 mL SOC for 1 hour then plated on LB agar supplemented with 50 μg/mL kanamycin and 10 μg/mL chloramphenicol and grown at 37 °C. The ligation reaction resulted in ~30,000 resistant colonies, of which ten were picked and verified with restriction digest. All colonies were then pooled and plasmid DNA isolated. The purified library was then digested with MlyI to remove the transposon and gel purified. The randomly linearized library was then ligated overnight with the frame selectable TAG linker in a 1:5 library to linker ratio. The ligation was transformed by adding 800 μL of chemical competent E. coli, heat-shocked and recovered in 4 mL SOC then plated on LB agar supplemented with 50 μg/mL kanamycin and 40 μg/mL ampicillin. At this stage the plates were grown at 30 °C which is a critical step for correct intein mediated splicing(15). The selected library resulted in 1635 kanamycin and ampicillin resistant colonies. The library was then digested with MlyI to remove the linker and generate the TAG codon scar. The digested library was gel extracted and ligated in dilute solution at room temperature for 2 hours. The mutant library ligation was then transformed by mixing 5 μL of the ligation reaction with 200 μL of chemically competent E. coli, recovered in 1 mL SOC for one hour and plated on LB agar supplemented with 50 μg/mL kanamycin and grown at 37 °C, which resulted in > 40,000 individual colonies.
Expression of mutant proteins containing unnatural amino acids and photo-crosslinking
Of the 48 clones we isolated, ten were chosen as representative examples to transfer into the pBADmycHisA expression vector for production with the unnatural amino acid p-benzoylphenylalanine. The TAG mutants were subcloned with BamHI-SalI into the BglII-SalI sites of pBADmycHisA. The ligation was transformed into chemically competent E. coli harboring the plasmid pSUP-pBpa(26). All expressions were done on a 50 mL scale in LB media 50 μg/mL ampicillin and 30 μg/mL chloramphenicol supplemented with 20 mM p-benzoylphenylalanine (racemic). All individual clones were expressed in both the presence and absence of amino acid. Cultures were grown to OD600 = ~0.5-0.6 then induced with 0.04% arabinose and grown for an additional 6 hours at 37 °C. All cultures were centrifuged to pellet cells and resuspended in 2 mL native binding buffer (100 mM HEPES, 10 mM imidazole, 1 mM PMSF). Cells were lysed by sonication (3 minutes, 10 seconds on, 10 seconds off) then centrifuged at 13,000 rpm for 20 minutes to clear the lysate and 100 μL of a 50% slurry mix of Promega HisLink resin was added. The resin and lysate were incubated on ice for 30 minutes with mixing. The resin was washed twice with 1.5 mL wash buffer (100 mM HEPES, 50 mM imidazole) followed by elution with 200 μL elution buffer (100 mM HEPES, 500 mM imidazole). Photo-crosslinking was performed in vitro, by irradiating the proteins in elution buffer at 365nm (handheld-UV) for 15 minutes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the University of Maryland and NIH (GM084396) for financial support. KAD is a recipient of a GAANN fellowship (P200A060238). We thank Prof. Stefan Lutz (Emory University) for the plasmid pInSALect, and Prof. Peter G. Schultz for the plasmid pSUP-BP. We are grateful to Prof. Steve Rokita for helpful comments on the manuscript.
Footnotes
Supporting Information Available Construction and sequences of plasmids used in the study. This material is available free of charge via the Internet.
REFERENCES
- (1).Caldwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Applic. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- (2).Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–91. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- (3).Wells JA, Vasser M, Powers DB. Cassette mutagenesis: an efficient method for generation of multiple mutations at defined sites. Gene. 1985;34:315–23. doi: 10.1016/0378-1119(85)90140-4. [DOI] [PubMed] [Google Scholar]
- (4).Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–5. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- (5).Cropp TA, Schultz PG. An expanding genetic code. Trends Genet. 2004;20:625–30. doi: 10.1016/j.tig.2004.09.013. [DOI] [PubMed] [Google Scholar]
- (6).Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci U S A. 2002;99:11020–4. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wilkins BJ, Daggett KA, Cropp TA. Peptide mass fingerprinting using isotopically encoded photo-crosslinking amino acids. Mol Biosyst. 2008;4:934–6. doi: 10.1039/b801512k. [DOI] [PubMed] [Google Scholar]
- (8).Sidhu SS, Fellouse FA. Synthetic therapeutic antibodies. Nat Chem Bio. 2006;l2:682–688. doi: 10.1038/nchembio843. [DOI] [PubMed] [Google Scholar]
- (9).Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97:8950–4. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hallet B, Sherratt DJ, Hayes F. Pentapeptide scanning mutagenesis: random insertion of a variable five amino acid cassette in a target protein. Nucleic Acids Res. 1997;25:1866–7. doi: 10.1093/nar/25.9.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Petyuk V, McDermott J, Cook M, Sauer B. Functional mapping of Cre recombinase by pentapeptide insertional mutagenesis. J Biol Chem. 2004;279:37040–8. doi: 10.1074/jbc.M406042200. [DOI] [PubMed] [Google Scholar]
- (12).Pajunen M, Turakainen H, Poussu E, Peranen J, Vihinen M, Savilahti H. High-precision mapping of protein protein interfaces: an integrated genetic strategy combining en masse mutagenesis and DNA-level parallel analysis on a yeast two-hybrid platform. Nucleic Acids Res. 2007;35:e103. doi: 10.1093/nar/gkm563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jones DD. Triplet nucleotide removal at random positions in a target gene: the tolerance of TEM-1 beta-lactamase to an amino acid deletion. Nucleic Acids Res. 2005;33:e80. doi: 10.1093/nar/gni077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Baldwin AJ, Busse K, Simm AM, Jones DD. Expanded molecular diversity generation during directed evolution by trinucleotide exchange (TriNEx) Nucleic Acids Res. 2008;36:e77. doi: 10.1093/nar/gkn358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gerth ML, Patrick WM, Lutz S. A second-generation system for unbiased reading frame selection. Protein Eng Des Sel. 2004;17:595–602. doi: 10.1093/protein/gzh068. [DOI] [PubMed] [Google Scholar]
- (16).Haapa-Paananen S, Rita H, Savilahti H. DNA transposition of bacteriophage Mu. A quantitative analysis of target site selection in vitro. J Biol Chem. 2002;277:2843–51. doi: 10.1074/jbc.M108044200. [DOI] [PubMed] [Google Scholar]
- (17).Poussu E, Vihinen M, Paulin L, Savilahti H. Probing the alpha-complementing domain of E. coli beta-galactosidase with use of an insertional pentapeptide mutagenesis strategy based on Mu in vitro DNA transposition. Proteins. 2004;54:681–92. doi: 10.1002/prot.10467. [DOI] [PubMed] [Google Scholar]
- (18).Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J Mol Biol. 1983;164:73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- (19).Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat Methods. 2008;5:37–9. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- (20).Richmond KE, Li MH, Rodesch MJ, Patel M, Lowe AM, Kim C, Chu LL, Venkataramaian N, Flickinger SF, Kaysen J, Belshaw PJ, Sussman MR, Cerrina F. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 2004;32:5011–8. doi: 10.1093/nar/gkh793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jackson JC, Duffy SP, Hess KR, Mehl RA. Improving nature's enzyme active site with genetically encoded unnatural amino acids. J Am Chem Soc. 2006;128:11124–7. doi: 10.1021/ja061099y. [DOI] [PubMed] [Google Scholar]
- (22).Silverman JA, Harbury PB. Rapid mapping of protein structure, interactions, and ligand binding by misincorporation proton-alkyl exchange. J Biol Chem. 2002;277:30968–75. doi: 10.1074/jbc.M203172200. [DOI] [PubMed] [Google Scholar]
- (23).Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129:10110–2. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci U S A. 1994;91:10747–51. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechno. 1998;l16:258–61. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- (26).Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–5. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.