Abstract
Site-directed mutagenesis of Pseudomonas aeruginosa azurin C112D at the M121 position has afforded a series of proteins with elevated Cu(II/I) reduction potentials relative to the Cu(II) aquo ion. The high potential and low axial hyperfine splitting [Cu(II) EPR A‖] of the C112D/M121L protein are remarkably similar to features normally associated with type 1 copper centers.
The capacity for precise tuning of active site reduction potentials in folded polypeptide environments has afforded nature the freedom to access myriad chemistries despite its limited repertoire of incorporated elements.1-3 The reduction potential of Cu(II) in Pseudomonas aeruginosa azurin has long been known to be sensitive to substitutions within both the inner and outer type 1 copper coordination spheres. 4-12 This sensitivity makes it possible to tune the potential over 0.5 V higher than the aqueous redox couple.8
We have exploited this sensitivity to obtain robust azurins with relatively high reduction potentials in the absence of soft ligands. Here we report work on the C112D protein (Figure 1), where the C to D substitution is made to avoid the unwanted irreversible oxidation of the cysteine thiol group.13-15 We have generated mutants possessing charged axial ligands as well as one lacking axial coordination from position 121 to evaluate the potential range of these robust scaffolds.
Figure 1.
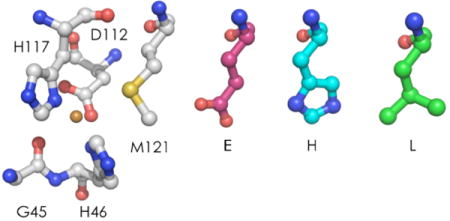
PyMol rendering of the active site of C112D azurin (PDBID: 1AG0).13
We constructed plasmids encoding azurins C112D, C112D/M121E, C112D/M121L, and C112D/M121H via site-directed mutagenesis of a T7-controlled plasmid encoding the wild-type protein. We expressed all four proteins in E. coli BL21(DE3). Periplasmic fractions were passed through Q-Sepharose media in 50 mM Tris pH 7.8 containing 100 mM NaCl to remove anionic contaminants. The proteins were desalted into 10 mM CHES pH 9.0 and purified to homogeneity on a MonoQ column. Purity was assessed by silver-stained SDS-PAGE and ESI-MS. Apoazurin concentrations were determined using ε280 = 8800 cm -1M-1. For well-resolved spectra and for electrochemistry, a slight excess of CoCl2 or CuSO4 was added to apoprotein solution, which was subsequently desalted to remove surface-bound metal ions.
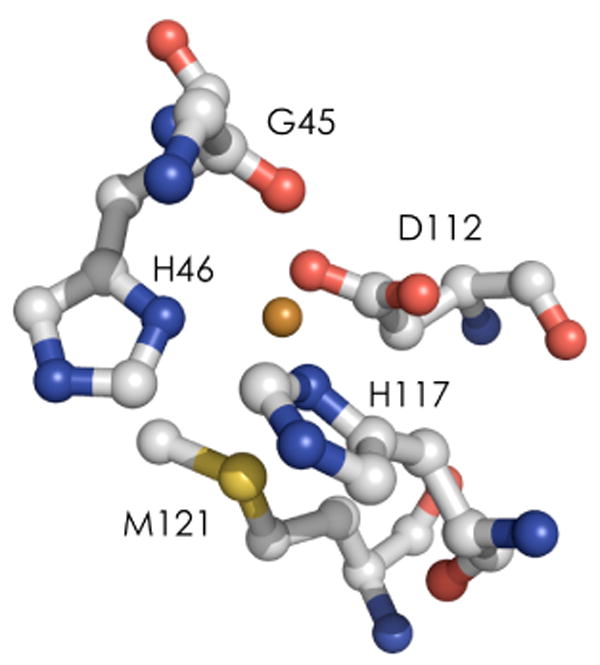
Electronic absorption spectra of the Cu(II) and Co(II) C112D/M121X (X = M, E, H, L) azurins in pH 7 aqueous solution are shown in Figure 2 (data are set out in Table 1). The M121L substitution results in a red shift of the Cu(II) ligand field (LF) absorption to 800 nm; consistent with this decrease in LF splitting, there also is a small red shift of the imidazole to Cu(II) ligand-to-metal charge-transfer (LMCT) band. The Cu(II) LF band of C112D/M121L Cu(II) azurin is much sharper than that of C112D. This could indicate decreased site reorganization in the case of a single transition comprising the d-d band, though further characterization will be required to dissect this spectroscopic feature. The Co(II) C112D/M121L LF system near 600 nm is virtually identical with that of the single mutant, confirming that the inner-sphere electronic interactions involve mainly the equatorial ligands.
Figure 2.
Electronic absorption spectra of Cu(II) (A) and Co(II) (B) C112DM121X azurins in 10 mM sodium phosphate pH 7.0. X = M (black), L (green), E (red), H (blue).
Table 1.
| X | Cu(II) λmax (nm) |
Co(II) λmax (nm) |
gx | gy | gz | A⊥ (10-4 cm-1) |
A‖ (10-4 cm-1) |
Cu (II/I) E°1/2 (V vs NHE), pH 7 |
|---|---|---|---|---|---|---|---|---|
| M | 754 (100) | 518 (210), 560 (200), 610 (280), 630 (650, sh) | 2.07(1) | 2.05(1) | 2.31(1) | 1.58(4) | 151(1) | 180 |
| E | 875 (70) | 569 (270), 531 (330), 600 (330) | 2.06(3) | 2.070(1) | 2.32(3) | 2.15(1) | 151(1) | 270 |
| H | 657 (70) | 549 (210, sh), 577 (240), 608 (280), 630 (170) | 2.064(2) | 2.06(5) | 2.30(3) | 4(2) | 165(1) | 305 |
| L | 798 (100) | 519 (170), 559 (170), 609 (220), 628 (170, sh) | 2.11(1) | 2.05(1) | 2.385(1) | 15.07(3) | 101(1) | 280 |
Parenthetical values represent molar extinction coefficient (M-1cm-1) calculated by titration of apoazurin with CuSO4 or CoCl2 These values are correct to within 5% uncertainty.
EPR parameters were simulated using SpinCount. Parenthetical values represent uncertainty due to g or A strain. See ref 17.
The Cu(II) LF band in the spectrum of C112D/M121E azurin is red-shifted to 875 nm. The Co(II) LF absorption system exhibits structure characteristic of tetrahedral coordination, indicating that the E121 carboxylate is ligated to the metal and that this interaction forces the metal out of the plane of the H46, D112, and H117 donor atoms.16 The Cu(II) LF band in the spectrum of the C112D/M121H protein is blueshifted to 680 nm. Based on the profile of its 600-nm absorption system, the Co(II) geometry appears intermediate between five- and four-coordinate, raising the possibility that the H121 imidazole does not interact strongly with the metal center.
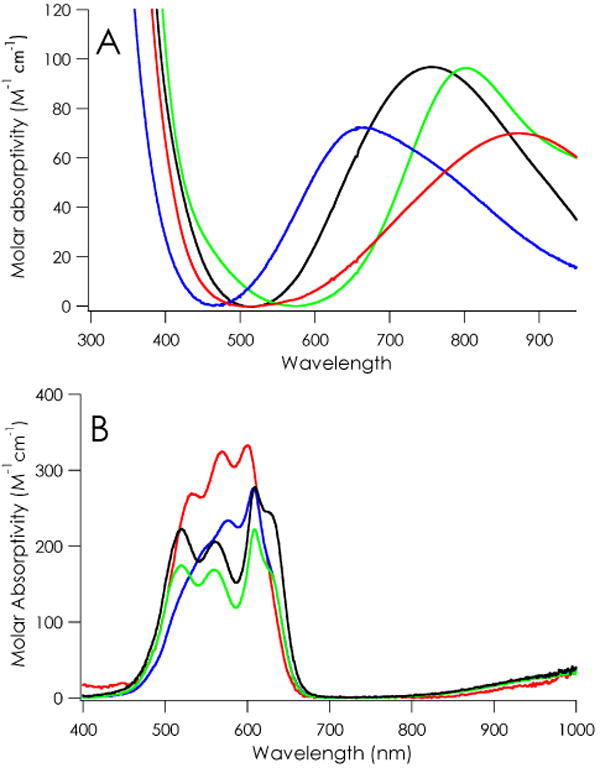
The coordination geometries that can be inferred from examination of the Co(II) spectra of C112D/M121X (X = M, E, H) are supported by 77 K X-band EPR data of Cu(II) analogues (Table 1, Figure 3).17 The C112D/M121E and C112D/M121H proteins possess axial hyperfine splittings lack of significant anisotropy in equatorial g-tensors, these parameters suggest similar axial coordination environment to that of the C112D mutant in which the X121 side-chain weakly coordinates the metal.
Figure 3.
77 K X-band EPR spectra of Cu(II) C112DM121X azurins in 10 mM sodium phosphate pH 7.0. X = M (black), L (green), E (red), H (blue).
The C112D/M121L EPR spectrum is particularly interesting. The spin Hamiltonian parameters represent a departure from the other azurins, displaying a substantial increase in gz and decrease in A‖. Normally this would imply a shift toward tetrahedral site geometry.18 Furthermore, there is considerable g-tensor anisotropy that may be attributed to dz2 mixing into a dxy ground state.19,20 Together these features suggest that C112D/M121L Cu(II) is in a unique electronic environment.
Cyclic voltammograms (CVs) of Cu(II) C112D/M121X (X = M, L) azurins adsorbed onto [CH3(CH2)8SH and HO(CH2)8SH] mixed self-assembled-monolayer (SAM) modified gold electrodes21,22 are shown in Figure 4, with comparable to that of the single mutant. Concomitant with the midpoint potentials appearing in Table 1. Azurin C112D/M121E is coupled poorly to the electrode, as indicated by weak CV signals (Figure S1); however, square wave voltammetry (SWV) provides sufficient current for midpoint potential determination. The C112D/M121L and C112D/M121E Cu(II/I) midpoint potentials are both higher than C112D, likely a result of weaker overall ligand fields. From the position of the LF band in the C112D/M121E spectrum, we would have predicted an even larger upshift were it not for the negative charge of the carboxylate. Earlier work has shown that a carboxylate interaction with Cu(II) in azurin leads to a ∼100 mV decrease in reduction potential.8
Figure 4.
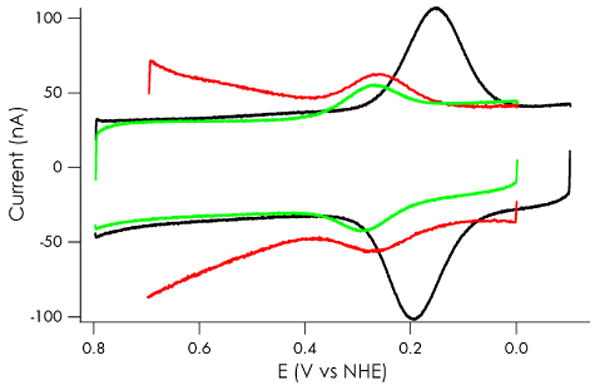
Electrochemistry of C112D/M121X azurins on SAM-modified Au electrodes. C112D (black) and C112D/M121L (green) potentials were measured by CV in 10 mM sodium phosphate pH 7.0 at a scan rate of 50 mV/s referenced to Ag/AgCl in saturated KCl (197 mV vs NHE). Azurin C112D/M121E (red) was measured by SWV in 10 mM NaPi with a square wave frequency of 8 Hz.
We were not able to determine the reduction potential of C112D/M121H azurin from electrochemical experiments, as the CV was very broad and asymmetric (Figure S2). Based on a redox titration with P. aeruginosa cytochrome c551 (Figure S3), we estimate a reduction potential near 305 mV. An optical absorption pH titration suggests that the pKa of the H121 imidazole is ∼8 (Figure S4), so the high potential is likely attributable to relative destabilization of Cu(II) by the nearby protonated side chain. The elevated pKa suggests that there is a favorable electrostatic interaction between the D112 carboxylate and protonated H121.
Our work on C112D/M121L azurin demonstrates that type 2 copper can acquire type 1 character, even in the absence of sulfur ligation. Properties such as a relatively small A‖ value and high reduction potentials (relative to C112D, and by extension, Cu(II) aquo ion) until now have been associated almost exclusively with blue copper centers.10,11,23,24 We suggest that electron donation from the equatorial ligands will reduce the positive charge on Cu(II) in the hydrophobic axial environment, effectively mimicking the covalency attributable mainly to cysteine thiolate ligation in a blue protein.
Supplementary Material
pH dependent UV/VIS, CV and redox titration of C112D/M121H azurin, CV of C112D/M121E azurin. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Yuling Sheng and Matthew R. Hartings for assistance, and Israel Pecht for useful discussions. This work was supported by the NSF (CHE-0553150 to HBG, JRW; graduate fellowship to KML), GCEP (Stanford), and NIH (DK19038 to HBG).
References
- 1.Gray HB. Proc Natl Acad Sci USA. 2003;100:3563–3568. doi: 10.1073/pnas.0730378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zong CH, Wilson CJ, Shen TY, Wittung-Stafshede P, Mayo SL, Wolynes PG. Proc Natl Acad Sci USA. 2007;104:3159–3164. doi: 10.1073/pnas.0611149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malmstrom BG, Wittung-Stafshede P. Coord Chem Rev. 1999;186:127–140. [Google Scholar]
- 4.Garner DK, Vaughan MD, Hwang HJ, Savelieff MG, Berry SM, Honek JF, Lu Y. J Am Chem Soc. 2006;128:15608–17. doi: 10.1021/ja062732i. [DOI] [PubMed] [Google Scholar]
- 5.Ralle M, Berry SM, Nilges MJ, Gieselman MD, van der Donk WA, Lu Y, Blackburn NJ. J Am Chem Soc. 2004;126:7244–56. doi: 10.1021/ja031821h. [DOI] [PubMed] [Google Scholar]
- 6.Berry SM, Gieselman MD, Nilges MJ, van Der Donk WA, Lu Y. J Am Chem Soc. 2002;124:2084–5. doi: 10.1021/ja0169163. [DOI] [PubMed] [Google Scholar]
- 7.Berry SM, Ralle M, Low DW, Blackburn NJ, Lu Y. J Am Chem Soc. 2003;125:8760–8. doi: 10.1021/ja029699u. [DOI] [PubMed] [Google Scholar]
- 8.Pascher T, Karlsson BG, Nordling M, Malmstrom BG, Vanngard T. Eur J Biochem. 1993;212:289–96. doi: 10.1111/j.1432-1033.1993.tb17661.x. [DOI] [PubMed] [Google Scholar]
- 9.Chang TK, Iverson SA, Rodrigues CG, Kiser CN, Lew AY, Germanas JP, Richards JH. Proc Natl Acad Sci USA. 1991;88:1325–9. doi: 10.1073/pnas.88.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canters GW, Gilardi G. FEBS Lett. 1993;325:39–48. doi: 10.1016/0014-5793(93)81410-2. [DOI] [PubMed] [Google Scholar]
- 11.Gray HB, Malmstrom BG, Williams RJP. J Biol Inorg Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 12.Gray HB, Malmstrom BG. Comments Inorg Chem. 1983;2:203–209. [Google Scholar]
- 13.Faham S, Mizoguchi TJ, Adman ET, Gray HB, Richards JH, Rees DC. J Biol Inorg Chem. 1997;2:464–469. [Google Scholar]
- 14.Mizoguchi TJ, Di Bilio Angel J, Gray Harry B, Richards John H. J Am Chem Soc. 1992;114:10076–10078. [Google Scholar]
- 15.Mizoguchi TJ. PhD Thesis. California Institute of Technology; 1996. [Google Scholar]
- 16.Cotton FA, Goodgame M, Goodgame DM. J Am Chem Soc. 1961;83:4690–&. [Google Scholar]
- 17.Golombek AP, Hendrich MP. J Mag Res. 2003;165:33–48. doi: 10.1016/j.jmr.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Bencini A, Gatteschi D, Zanchini C. J Am Chem Soc. 1980;102:5234–5237. [Google Scholar]
- 19.Gewirth AA, Cohen SL, Schugar HJ, Solomon EI. Inorg Chem. 1987;26:1133–1146. [Google Scholar]
- 20.Fittipaldi M, Steiner RA, Matsushita M, Dijkstra BW, Groenen EJJ, Huber M. Biophys J. 2003;85:4047–4054. doi: 10.1016/S0006-3495(03)74818-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita K, Nakamura N, Ohno H, Leigh BS, Niki K, Gray HB, Richards JH. J Am Chem Soc. 2004;126:13954–13961. doi: 10.1021/ja047875o. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama K, Leigh BS, Sheng Y, Niki K, Nakamura N, Ohno H, Winkler JR, Gray HB, Richards JH. Inorg Chim Acta. 2008;361:1095–1099. doi: 10.1016/j.ica.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solomon EI, Szilagyi RK, George SD, Basumallick L. Chem Rev. 2004;104:419–458. doi: 10.1021/cr0206317. [DOI] [PubMed] [Google Scholar]
- 24.Solomon EI. Inorg Chem. 2006;45:8012–8025. doi: 10.1021/ic060450d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
pH dependent UV/VIS, CV and redox titration of C112D/M121H azurin, CV of C112D/M121E azurin. This material is available free of charge via the Internet at http://pubs.acs.org.


