Abstract
Electron capture dissociation (ECD) and electron transfer dissociation (ETD) of doubly protonated electron affinity (EA)-tuned peptides were studied to further illuminate the mechanism of these processes. The model peptide FQpSEEQQQTEDELQDK, containing a phosphoserine residue, was converted to EA-tuned peptides via β-elimination and Michael addition of various thiol compounds. These include propanyl, benzyl, 4-cyanobenzyl, perfluorobenzyl, 3,5-dicyanobenzyl, 3-nitrobenzyl and 3,5-dinitrobenzyl structural moieties, having a range of EA from -1.15 to 1.65 eV, excluding the propanyl group. Typical ECD or ETD backbone fragmentations are completely inhibited in peptides with substituent tags having EA over 1.00 eV, which are referred to as electron predators in this work. Nearly identical rates of electron capture by the dications substituted by the benzyl (EA = -1.15 eV) and 3-nitrobenzyl (EA = 1.00 eV) moieties are observed, which indicates the similarity of electron capture cross sections for the two derivatized peptides. This observation leads to the inference that electron capture kinetics are governed by the long range electron-dication interaction and are not affected by side chain derivatives with positive EA. Once an electron is captured to high-n Rydberg states, however, through-space or through-bond electron transfer to the EA-tuning tags or low-n Rydberg states via potential curve crossing occurs in competition with transfer to the amide π* orbital. The energetics of these processes are evaluated using time-dependent density functional theory with a series of reduced model systems. The intramolecular electron transfer process is modulated by structure-dependent hydrogen bonds and is heavily affected by the presence and type of electron withdrawing groups in the EA-tuning tag. The anion radicals formed by electron predators have high proton affinities (approximately 1400 kJ/mol for the 3-nitrobenzyl anion radical) in comparison to other basic sites in the model peptide dication, facilitating exothermic proton transfer from one of the two sites of protonation. This interrupts the normal sequence of events in ECD or ETD leading to backbone fragmentation by forming a stable radical intermediate. The implications which these results have for previously proposed ECD and ETD mechanisms are discussed.
Introduction
Following the development of electron capture dissociation (ECD) of multiply protonated peptide or protein ions,1 numerous studies have been carried out to investigate the mechanism of this process and to explore its broad applicability to mass spectrometry (MS)-based structural studies of peptides and proteins.2-5 Unlike collision-induced dissociation (CID)6 or infrared multiphoton dissociation (IRMPD),7 ECD and its analogue, electron transfer dissociation (ETD),8 generate abundant sequence ions and the sites of peptide backbone cleavage are relatively less discriminated by the side-chains of nearby amino acids. These methods also preserve labile side-chains with post-translational modifications (PTMs), allowing easier identification and localization of PTMs compared with CID or IRMPD.9 While ECD and ETD preferentially cleave a disulfide bond, thermal activation methods (CID and IRMPD) do not generate abundant C-S or S-S bond cleavage fragments unless peptides are cationized by metal ions.10 This makes ECD and ETD methods of choice for characterizing phosphorylation,11 glycosylation,12 methylation13 and disulfide linkage14 of proteins to elucidate important biological processes such as cell signaling and cell differentiation and proliferation. Owing to recent instrumental developments, ECD and ETD have been successfully implemented to various mass analyzers such as the linear ion trap,8 hybrid quadrupole-Time-Of-Flight (QqTOF),15 Fourier transform ion cyclotron resonance (FTICR)16 and, most recently, orbitrap17 instruments. These developments satisfy the varying requirements of a wide range of applications where resolution, sensitivity, dynamic range and compatibility with various chromatographic methodologies are important parameters to consider for the mass spectrometric analyses of biological samples of ever increasing complexity.
Since its conception, however, ECD has elicited lively discussions in the mass spectrometry community with regard to its mechanism. Initial electron capture to high-n Rydberg states was first proposed by McLafferty and co-workers.1,2,14,18 In this model, the protonation sites (i.e. protonated amine, guanidine or imidazole residues) of a peptide ion are believed to be internally solvated by amide oxygens via one or more hydrogen bonds. Electron localization occurs to one of the positively charged sites, which subsequently forms a hypervalent radical in the ground electronic state via internal conversion, with the energy released in this process contributing to the overall vibrational excitation of the ion. Subsequent transfer of a hydrogen atom to an amide oxygen facilitates β-cleavage of the adjacent N-Cα bond through an aminoketyl radical intermediate. The resulting fragments are the residues of the peptide N-terminus and C-terminus, denoted as c and z• ions, respectively. This process, referred to as the Cornell mechanism,19 was initially suggested to be a non-ergodic reaction.3 The preservation of non-covalent interactions along with backbone cleavages was demonstrated as a proof of non-ergodicity in ECD.20 Supportive theoretical and experimental observations for the Cornell mechanism were subsequently reported elsewhere.21
Even though the Cornell mechanism provided a reasonable picture for ECD, some backbone fragmentations were not easily explained.22 The characteristic ECD fragmentation processes are still observed in some peptide cations where electron capture does not yield a mobile hydrogen atom. These include peptides cationized by metal ion attachment23 or fixed charge derivatives (i.e. quaternary ammonium or phosphonium groups).24,25 In addition, the guanidinium groups in peptides are poorly solvated by amide oxygens and hydrogen atom transfer from an arginine radical to an amide carbonyl is endothermic.19,26 With either of these circumstances, c or z type ions are still prominent in ECD spectra.19
The Utah-Washington mechanism19 (UW mechanism), recently proposed independently by Simons and co-workers27-29 and Turecek and co-workers,19,24,26,30-35 provides an alternative view of the mechanism explaining the relatively indiscriminate distribution of N-Cα bond cleavage processes observed in ECD and ETD. Coulomb stabilization by positively charged groups allows the amide π* orbital to possess a positive electron affinity (EA).36 Electron attachment to Coulomb stabilized amide π* orbitals makes the amide group an exceptionally strong base with a proton affinity (PA) in the range 1100-1400 kJ/mol.32 The amide anion radical is able to abstract a proton in an energetically favorable process via conformational changes, even from relatively distant proton donors. The resulting intermediate is identical to the aminoketyl cation radical proposed in the Cornell mechanism and can undergo the same N-Cα bond cleavage. This process does not require invoking either the mobile “hot” hydrogen atom hypothesis or non-ergodicity of dissociation. ECD of multiply cationized ions where the charge carriers are metal ions or fixed charge derivatives can also be explained by ion-dipole interactions and the intramolecular electron transfer between the charge-stabilized amide π* orbital and the N-Cα σ* orbital, followed by N-Cα bond cleavage. The UW mechanism is supported by recent theoretical and experimental investigations.31,37
Despite many efforts of the past decade, there is still much to be learned about the mechanistic details of ECD and ETD. The sizes of peptides or proteins are too large to accurately quantify the energetics of these processes based on high level ab initio or density functional calculations. Recently, Williams and coworkers quantified the energetics of the ECD process involving a hydrated gaseous peptide dication by examining the extent of water evaporation resulting from electron capture.38 The conformational dynamics of multiply protonated peptides and proteins also contributes to uncertainties in identification of a particular charged site associated with the capture dynamics of an electron in high-n Rydberg states and the specification of the eventual site of electron localization in the cation radical. To circumvent these problems, relatively simple model systems have been investigated with high level quantum mechanical calculations.35,39 The amide-I vibration (C=O stretching mode) dynamics was also examined as a simple model of the vibrational energy propagation in α-helix fragmentation upon ECD and ETD.40
To constrain the charged or radical site, recent studies have shown the effect of incorporation of permanent charged tags in peptides on backbone24,25,41 and disulfide cleavage.42 Improved sequence coverage of glycosylated and phosphorylated peptides has also been demonstrated using permanently charged tags.43 Tags comprising strongly basic sites of proton localization as well as radical traps have been incorporated to study their effect on typical ECD fragmentations.34,44 However, electron traps with a range of EAs have not been considered.
Turecek and coworkers used 2-(4′-carboxypyrid-2′-yl)-4-carboxamide (pepy) group34 which has much higher gas phase basicity (923 kJ/mol) compared to other basic groups in the peptide with the expectation that it is always protonated in the peptide dication. Thus it actually functions in the same manner as permanently charged tags such as quaternary ammonium or phosphonium groups by trapping an electron at the site of protonation because of its higher recombination energy. The resulting radical is also stable and does not contribute a labile hydrogen atom that might be transferred to an amide carbonyl and lead to backbone cleavage. As a result, they observed the termination of N-Cα backbone cleavage in analogy with many other permanent tag experiments.
O'Connor and coworkers used the coumarin tag44 which has a relatively low electron affinity (<0.6 eV), and hence, based on the experiments described in this work, cannot terminate peptide backbone cleavage solely by operation as an electron trap. Instead, the courmarin group acts as a free radical (hydrogen atom) scavenger to terminate the ECD process. In the experiments of O'Connor and coworkers, it is likely that initial electron capture and subsequent relaxation of the charge reduced cation radical initially forms the aminoketyl intermediate, which in turn transfers the labile hydrogen atom to the coumarin substituent before cleavage of the peptide backbone can occur.
In the present work, we synthesized a series of EA-tuned peptides, which were generated from phosphopeptides, by attaching thiol groups having EAs ranging from -1.15 eV to 1.652 eV in their precursor forms. The model peptide, FQpSEEQQQTEDELQDK, was chosen because it has a C-terminal lysine residue, thus simulating a typical tryptic peptide, and also has a phosphoserine residue for inserting the EA-tuning tags between the N-terminal amine and the C-terminal lysine. For the synthesis of the EA-tuned peptides, a dehydroalanine residue is prepared by eliminating a phosphate group under basic conditions, followed by Michael addition of thiols to generate various benzylic cysteine residues. The derivatized peptide dications generated by electrospray are analyzed by ECD and ETD to investigate the effect of the EA-tuning tags. We observe that, with sufficiently high EA, the tag leads to inhibition of the backbone dissociation process normally observed in ECD and ETD experiments. We propose that this results from relaxation processes involving through-space or through-bond electron transfer from an initially formed high-n Rydberg state to the tag, followed by proton transfer to the resulting radical anion moiety. The implications of these results for previously proposed mechanisms of electron capture and electron transfer dissociation are discussed. In addition, the present experiments allow for interpretation of matrix-assisted laser desorption/ionization (MALDI) in-source decay processes45 resulting from MALDI plume chemistry involving electrons and multiply protonated ions and have important implications for the study of peptides possessing nitrated tyrosine as a PTM.46
Experimental Section
Materials
Monophosphopeptide from β-casein (FQpSEEQQQTEDELQDK) was obtained from Anaspec (San Jose, CA). Thioacetic acid (HSAc), 0.3 N saturated barium hydroxide (Ba(OH)2) solution, propanethiol (PT), benzyl bromide, 4-cyanobenzyl bromide, perfluorobenzyl bromide, 2-nitrobenzyl bromide, 4-nitrobenzyl bromide, 3,5-dinitrobenzyl chloride, 3-nitrobenzylthiol (3NBT), 1,3-dibromobenzaldehyde, sodium borohydride, mesyl chloride and α-cyano-4-hydroxycinnamic acid (CHCA) were acquired from Sigma-Aldrich (St. Louis, MO). Hydrochloric acid in methanol (~1.25 M) and 1-fluoro-3,5-dinitrobenzene were purchased from Fluka (Buchs, Switzerland). Methanol (MeOH), ethanol (EtOH), anhydrous N,N-dimethylformamide (DMF), anhydrous dichloromethane (DCM), dimethylether, acetonitrile (ACN), tetrahydrofuran (THF), ethyl acetate (EtOAc), anhydrous potassium carbonate (K2CO3) and OmniSolv™ high purity water were provided by EMD (Darmstadt, Germany). Dimethylsulfoxide (DMSO), formic acid (FA), and trifluoroacetic acid (TFA) were supplied by Mallinckrodt Inc. (Phillpsburg, NJ). All chemicals mentioned above were used as received without further purification. For desalting, OMIX™-100 μL size C-18 tips were purchased from Varian Inc. (Palo Alto, CA).
Synthesis of the EA-tuning tags and Derivatized Peptides
The EA-tuning tags (benzyl thiols) were prepared from the corresponding benzyl halides. The literature procedure was followed with minor modification for better yield.47 To synthesize thioesters, each benzyl halide (5 mmol) was dissolved in 15 mL of THF with 6 mmol of HSAc and 6 mmol of anhydrous K2CO3 in an air-free flask. The mixture was stirred at room temperature under a steady stream of N2. The reaction time for each precursor varied from 1 to 1.5 h and the completion of reactions was monitored by thin-layer chromatography (TLC). The crude thioacetate obtained after standard aqueous work-up was sufficiently pure to use directly in the next step. The deacetylation reaction was carried out by adding 3mL of hydrochloric acid in methanol to a solution of the crude thioacetate in methanol and stirring at ~55-60 °C for 15~18 h. The thiol products were purified by flash chromatography on silica (1:20 EtOAc/hexane eluent) and identified by 1H NMR (Supporting Information). Solid products such as 2-nitrobenzyl thiol, 4-nitrobenzyl thiol and 3,5-dinitrobenzyl thiol were dissolved in DMF at ~3-4 M concentration. All products were stored in sealed vials at 4 °C up to 6 months without any noticeable degradation.
Reactions involving formation of a dehydroalanine by β-elimination followed by Michael addition were used to attach the EA-tuning tags to our model phosphopeptide. A 20 μg portion of monophosphopeptide (FQpSEEQQQTEDELQDK) was dissolved in 40 μL of 4:3:1 mixture of H2O/DMSO/EtOH (Solvent A) or 40 μL of 20% ACN (Solvent B), which proved optimal after extensive screening of solvent systems. In particular, these solvent systems provide enhanced solubility of thiols as described elsewhere.48 Whereas solvent A generally worked well with all of the thiol compounds, solvent B proved better suited for perfluorobenzyl thiol. However, solvent B gave poor product recovery for nitrobenzyl thiols. An aliquot of 10 μL of 0.3 N (saturated) Ba(OH)2 solution was added and allowed to react at room temperature for 1h. One μL of each thiol either in its liquid form or DMF solution was then added to the peptide solution, and the mixture was allowed to react at 37 °C for 3h. The extended reaction time (~4-6 h) is required for less nucleophilic thiols such as 3,5-dinitrobenzyl thiols to improve the yield. Heating the mixture over 6 h at higher temperature results in poorer product recovery. The reaction was terminated by adding 1 μL of FA. The product mixture was voltexed and spun down by centrifugation. Supernatant was subjected to desalting using an OMIX™-100 μL size C-18 tip following the standard procedure. Identities of final products, eluted in 0.1% TFA, 50% ACN, 50% H2O for MALDI or 0.1% FA, 50% MeOH, 50% H2O for electrospray ionization (ESI), were confirmed by MS and directly used for ECD and ETD experiments. MALDI-MS spectra of the derivatized peptides were further investigated to seek the presence of prompt in-source decay backbone fragments (i.e. c and z ions). The synthetic procedures above and the EAs of precursors49 are summarized in Scheme 1 and Table 1, respectively. The details for synthesis of 3,5-dicyanobenzyl thiol are available in Supporting Information. 1-Fluoro-3,5-dinitrobenzene (Sanger's reagent)50 was conjugated to the N-terminal amine to be compared with 3,5-dinitrobenzylcysteine containing peptides synthesized by β-elimination and Michael addition reaction. The procedure described in the literature51 with reaction conditions optimized for the selective N-terminal amine derivatization was used without any modification.
Scheme 1.
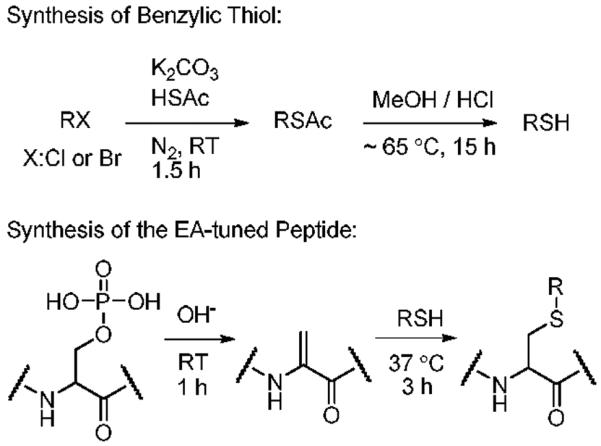
Table 1.
Electron Affinities of Thiol Precursors
| Name of Benzyl Group | Benzyl- | 4-Cyanobenzyl- | Perfluorobenzyl |
|---|---|---|---|
| Structure |  |
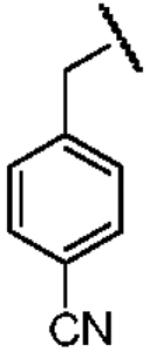 |
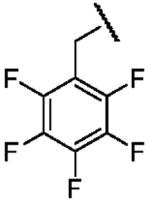 |
| Electron Affinity (eV) | -1.15 ± 0.05a | 0.258 ± 0.018b or 0.26 ± 0.1c | 0.434 ± 0.081e or 0.730 ± 0.080f |
| Name of Benzyl Group | 3,5-Dicyanobenzyl- | 3-Nitrobenzyl- | 3,5-Dinitrobenzyl- |
| Structure | 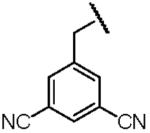 |
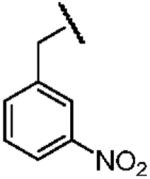 |
 |
| Electron Affinity (eV) | 0.91±0.1g | 1.00 ± 0.010h | 1.652 ± 0.048j |
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Electron affinities are quoted from Ref. 49, respectively. For Figure 7, c and e were chosen for each compound due to the consistency of experimental methods.
Mass Spectrometry
All ECD and IRMPD spectra were recorded using a 7-Tesla linear ion trap-Fourier transform (LTQ-FT) mass spectrometer (Thermo Scientific, San Jose, CA) with a nanoelectrospray ion source.52 The flow rate was ~50 nL/min and spray voltage was varied from 1.0 to 1.5 kV by monitoring ion signals. Other critical parameters were capillary temperature 200 °C, capillary voltage 30 V, and tube lens offset 200 V for maximal ion intensity. Other instrumental parameters were varied to optimize the intensities of the target ions in the linear ion trap prior to injection into the ICR cell. In ECD experiments electron irradiation occurred for 100 ms at ~5-7% of full energy scale, approximately corresponding to electron energy less than 1 eV and ~30 milliamp. Supplemental activation was accomplished by multiphoton excitation using a continuous 20 W CO2 infrared laser for 100 ms at ~45-90% of full energy scale, approximately corresponding to 5 J/cm2. The resolving power of FT MS scans was selected at 100,000 FWHM. For both ECD and IRMPD/ECD experiments, 100 scans were recorded.
ETD experiments were performed on a Thermo LTQ XL linear ion trap mass spectrometer (Thermo Scientific) modified for ETD. The eluted sample from the desalting step was directly infused into the microspray source at a flow rate of 2.0 μL/min. Spray conditions for maximizing ion counts included spray voltage 5.0 kV, capillary temperature 275 °C, capillary voltage 36 V, and tube lens offset 70 V. The electron transfer reagent generated from the chemical ionization (CI) source was introduced to the linear ion trap from the rear of the instrument and allowed to react with isolated ions. Fluoranthene (EA ~0.7 eV)53 was used for the CI reagent. The pressure of fluoranthene was 1×10-5 torr with a maximum injection time of 50 ms. Alternatively, isolated cations were collisionally activated for 200 ms prior to ETD in order to compare with IRMPD/ECD spectra.54 ETD spectra were accumulated for ~1 min (ca. 50 scans) to accumulate a reasonable signal-to-noise ratio.
MALDI TOF spectra were acquired using a Voyager DE-PRO mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a 20 Hz nitrogen laser (337 nm). All spectra were recorded in reflectron mode with 20 kV acceleration voltage, 150 ns delay extraction time and 75% grid voltage. 0.3 μL of the derivatized peptide solution was mixed with 0.3 μL of 10 mg/mL of CHCA matrix solution in 0.1% TFA, 50% ACN, 50% H2O and spotted on a stainless steel MALDI sample plate. Well-crystallized spots by the standard dried droplet method55 were introduced into the mass spectrometer for analysis. Usually 100 laser shots were averaged. Recorded spectra were analyzed using Xcalibur (Thermo Electron, San Jose, CA) for ECD and ETD and Data Explorer (Applied Biosystems, Foster City, CA) for MALDI. Fragment ion masses were calculated using MS-Product of Protein Prospector.56
Quantum Mechanical Calculation
The PC GAMESS57 (version 7.10) under Windows XP environment was used for the energetics of dicyanobenzene. To compare with previous work done by Polasek and Turecek, we used the same level of calculation and basis sets reported elsewhere.58 The geometries were optimized using Becke's general gradient exchange functional59 with Lee, Yang and Parr's correlation functional60 (B3LYP) with the 6-31+G(d,p) basis set for dicyanobenzene (DCB), protonated dicyanobenzene (DCBH+), dicyanobenzene anion radical (DCB-•) and hydrogen attached dicyanobenzene radical (DCBH•). For all open-shell systems, the spin-unrestricted method (UB3LYP) was used. Observed spin contamination in UB3LYP was small enough to be ignored (<S2> expectation values were 0.75-0.77). Optimized structures were further characterized by calculating vibrational frequencies and thermodynamic values using the same level of theory at 298.15K and 1.0 atm. To further refine the electronic energy of the system, electronic energies from UB3LYP/6-311+G(2df,p) and spin-restricted MP2 (ROMP2) with the same basis set were averaged (B3-ROMP2 energy).58 Spin contamination in spin-unrestricted MP2 (UMP2) for open-shell systems was significant with an <S2> expectation value ~1.6. Therefore, the UMP2 method was not used for this work.
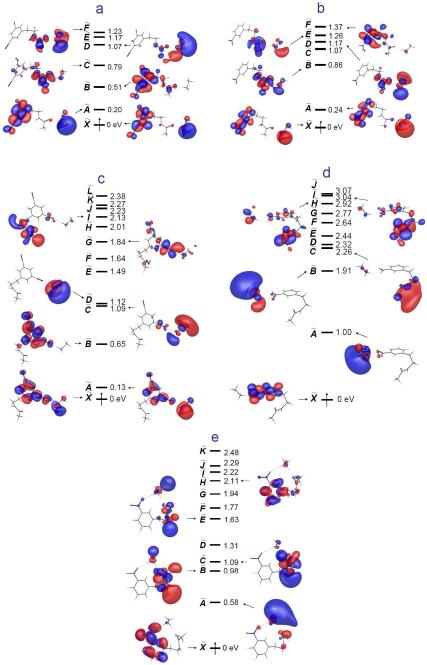
All other quantum mechanical calculations, including time-dependent density functional excited states analyses, were performed by GAMESS-US61 (version April 11, 2008 R1) under linux environment. The geometries of the model systems (Figure 8) were optimized at the B3LYP/6-31++G(d,p) level. All vertical electron affinities and recombination energies of the model systems were calculated without geometry relaxation. Further energy refinement was performed at the same level of theories described above for dicyanobenzene with the 6-311++G(2df,p) basis set. The M06 density functional62 with the same basis sets was also used to estimate the energetics of the electron capture process. Calculations of the energetics of vertical electron capture with excited states were performed using time-dependent density functional theory (TDDFT) at the UB3LYP/6-31++G(d,p) and 6-311++G(2df,p) level as implemented in GAMESS for open shell systems. Molecular orbitals (MOs) of excited states were prepared by linear combination of virtual orbitals with given coefficients from TDDFT calculations. Generated MOs were plotted using MacMolPlt63.
Figure 8.
Structures of the model compounds for quantum mechanical calculations. These are prepared by a combination of each aromatic functional group (1-6) with either acetamide (A) or methyl ammonium acetamide complex by a hydrogen bond to the amide carbonyl (B). Some methyl ammonium complexes having 3,5-dicyanophenyl (4) and 3-nitrophenyl ring (5) form hydrogen bonds with the cyano and the nitro group (C4 and C5) and both the amide carbonyl and nitro group, simultaneously (D5).
All geometries of optimized structures from quantum mechanical calculations with electronic, zero-point energy, enthalpy corrections and excited state energies are available in Supporting Information.
Results
ECD of the EA-tuned Peptides
Each derivatized peptide was confirmed by electrospray ionization to form mainly doubly protonated ions. These ions are respectively denoted as [P+2H]2+, [B+2H]2+, [4CB+2H]2+, [PFB+2H]2+, [35DCB+2H]2+, [3NB+2H]2+ and [35DNB+2H]2+ for the model peptides FQX*EEQQQTEDELQDK, where X* is propanylcysteine, benzylcysteine, 4-cyanobenzylcysteine, perfluorobenzylcysteine, 3,5-dicyanobenzylcysteine, 3-nitrobenzylcysteine and 3,5-dinitrobenzylcysteine. To examine the effect of substitution position in the nitrobenzyl moiety, 2-nitrobenzyl and 4-nitrobenzylcysteine containing peptides were studied. The peptides derivatized with 2NBT and 4NBT gave ECD and ETD spectra essentially identical to those of 3NBT (Supporting Information). Therefore, only spectra of [3NB+2H]2+ are discussed in this paper. To investigate the effect of the location of the EA-tuning tags in the peptide, the 3,5-dinitrophenyl group was attached to the N-terminal amine of the model peptide using 1-fluoro-3,5-dinitrobenzene and the resulting peptide was subject to ECD experiments. 3,5-Dicyanobenzyl thiol (35DCBT) derivatized peptides were studied to compare different types of functional groups for tags having EA near 1.00 eV. The spectra acquired from the 2NBT, 4NBT and Nα-3,5-dinitrophenyl derivatized peptides are available in Supporting Information.
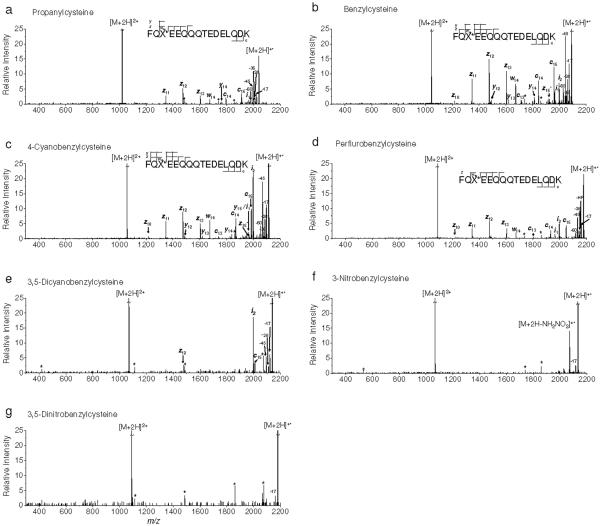
Figure 1 depicts ECD spectra of the derivatized peptides. Except Figure 1a, the spectra are presented in order of increasing EA of the benzyl substituents. The fragment ions induced by subsequent β-fission of a zn• ion and side chain losses (-R• or -RS•; R is a substituent side-chain) of [M+2H]+• are denoted as wn, i1 and i2, respectively. The C-terminal ions (z10 to z15) and the N-terminal ions (c13 to c15) were detected in most of the spectra. Some of the C-N amide bond cleavages (y ions) were also observed in ECD of [B+2H]2+ and [4CB+2H]2+ (Figures 1b-c).
Figure 1.
ECD of doubly protonated model peptides. a) propanylcysteine, b) benzylcysteine, c) 4-cyanobenzylcysteine, d) perfluorobenzylcysteine, e) 3,5-dicyanobenzylcysteine, f) 3-nitrobenzylcysteine and g) 3,5-dinitrobenzylcysteine containing peptides, respectively. An asterisk indicates instrumental noise.
The most prominent peak among ECD type ions is z12 as discussed by Savitski et al.64 Note that -1 or +1 Da shift from c or z• ions by the abstraction of a Cα hydrogen were observed as reported by O'Connor et al.5 and Savitski et al.65 We label these as c• and z ions which match with c-1 and z•+1 ions. In some cases, both c•/c ions and z•/z ions are identified simultaneously. Predominant z ions from z• ions are believed to be formed by the abstraction of the Cα hydrogen in the derivatized cysteine residues which contain a methionine-like thioether bond and a strong electron withdrawing group at benzylic side-chains, resulting in a more reactive Cα-H bond. The z14 ion was not observed in any ECD spectrum. Related to this, the presence of w14 indicates the facile side-chain loss reaction pathway for the EA-tuning tags compared to side-chain losses from the remaining amino acids in the model peptides (Figure 1).66
The ECD spectrum of [P+2H]2+ shown in Figure 1a exhibits a pattern of ECD backbone fragmentation typical of that observed tryptic peptide dications.64 Our model peptides have flexible gas phase structures, allowing frequent interactions between protonated sites and backbone amide carbonyls. Considering the effects of Coulomb stabilization and hydrogen bonded carbonyls, both the Cornell mechanism and the UW mechanism are expected to be operational in this case and it remains unknown which one is more dominant for elucidating ECD spectrum of [P+2H]2+.
As the EA of the tag is increased, the relative abundance of ECD type ions diminishes (Figure 1). Relative yields of typical ECD backbone fragment ions, which all ECD peaks in each spectrum are normalized for comparison, are summarized in Figure 2. The peaks from ECD of [PFB+2H]2+ deviate from the observed trend that the peak abundance is decreasing as EA of the tag is increased. The unusually high abundance of i2 in ECD of [4CB+2H]2+ and [35DCB+2H]2+ (Figures 1c and e) can be attributed to the stability of the RS• radical.
Figure 2.
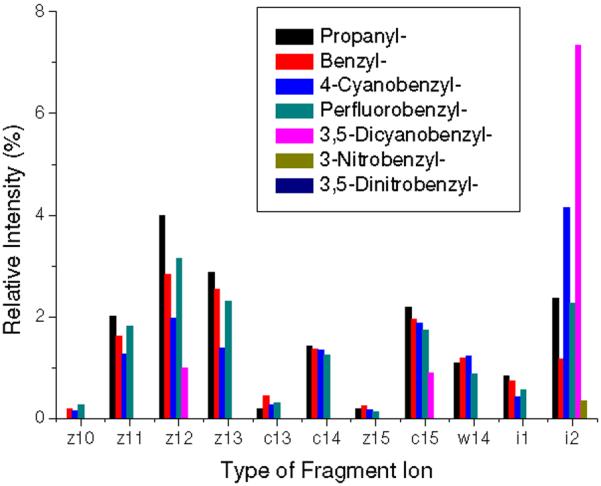
Relative intensities of ECD fragment ions. Intensities are taken from ECD spectra and reported as a total percent of the sum of the intensities of backbone fragments, side-chain losses and the charge reduced cation radical. The intensities attributed by -1 or +1 Da shift from c or z• ions by the abstraction of a Cα hydrogen are summed up to those of c or z• ions.
Remarkably, ECD spectra of [35DCB+2H]2+, [3NB+2H]2+ and [35DNB+2H]2+ exhibit essentially very small or no backbone fragmentation (Figures 1e-g). The loss of 17 Da from [3NB+2H]+• and [35DNB+2H]+• at m/z 2116.875 and 2161.862 turns out to be hydroxyl radical rather than ammonia by the comparison of measured and calibrated exact masses (Figures 1f-g). The mass deviation from the loss of hydroxyl radical in ECD of [3NB+2H]2+ and [35DNB+2H]2+ is 0.73 and 0.06 ppm while that of ammonia is 10.51 and 11.07 ppm, respectively. Polasek and Turecek previously reported loss of hydroxyl radical from the phenylnitronic radical and characterized the energetics of this process.58 More details about hydroxyl radical loss will be discussed in the following section. The loss of 17 Da from the remaining cation radicals is determined as ammonia.
In addition, ECD of [PFB+2H]2+ contains a product involving HF neutral loss (-20 Da) at m/z 2158.843, indicating possible formation of the perfluorobenzyl anion radical group, followed by proton transfer and loss of HF (Figure 1d). A similar process has been reported for gaseous perfluorobenzylalkylammonium ions forming the zwitterionic neutral radical by electron transfer and subsequent intramolecular proton transfer.67
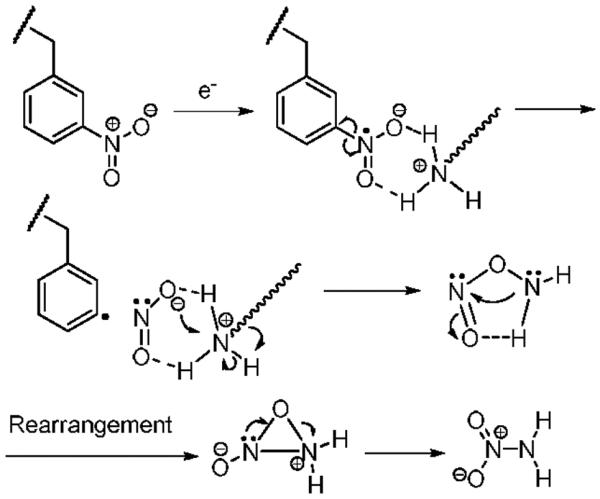
A neutral loss of 62 Da from [3NB+2H]2+ was observed at m/z 2071.882 as a main fragment (Figure 1f). Considering the specific Coulomb interaction between positively charged groups (i.e. the N-terminal amine and the ε-amine of lysine) and the nitrobenzylic cysteine anion radical formed by electron attachment, NH2NO2 is proposed as a reasonable candidate for this loss. However, it is not straightforward to propose a mechanism for NH2NO2 neutral loss. We tentatively suggest the process for NH2NO2 loss shown in Scheme 2. In the ECD spectrum of [35DNB+2H]2+, a cation radical, [35DNB+2H]+•, is the most abundant product ion (Figure 1g). After NH2NO2 loss from [35DNB+2H]+•, the resulting product is less stable in comparison to that of [3NB+2H]+•. Therefore, the product involving 62 Da loss in the ECD spectrum of [35DNB+2H]2+ is not significant.
Scheme 2.
The ECD spectrum of doubly protonated Nα-3,5-dinitrophenyl derivatized peptide was also investigated to demonstrate the effect of the position of 3,5-dinitrophenyl group and its connectivity (from thioether to secondary amine) in the model peptide. No ECD type backbone fragmentation is observed while most of the prominent side-chain losses remain as unknown peaks (Supporting Information). This observation is consistent with ECD of [35DNB+2H]2+. It also clearly demonstrates that the presence of the 3,5-dinitrophenyl group in the model peptide is responsible for inhibition of ECD and ETD backbone cleavage processes rather than its location or chemical connectivity.
IRMPD/ECD of the EA-tuned Peptides
To further examine the stability of cation radicals considered in this study, IRMPD with ECD (IRMPD/ECD) was performed. Precursor ions were heated by infrared photons to just below the onset of backbone cleavage. Electrons were simultaneously injected into the ICR cell without isolation of heated precursor ions. It is reasonable to assume that the IRMPD/ECD spectra contain not only ECD fragments of heated precursor ions but some direct IRMPD fragments. Delayed electron injection (100 ms) into the ICR cell for reaction with ions preheated by infrared photons did not generate spectra significantly different from those obtained with simultaneous activation. Therefore, only simultaneous excitation by infrared photons and electrons (IRMPD/ECD) is discussed in this work.
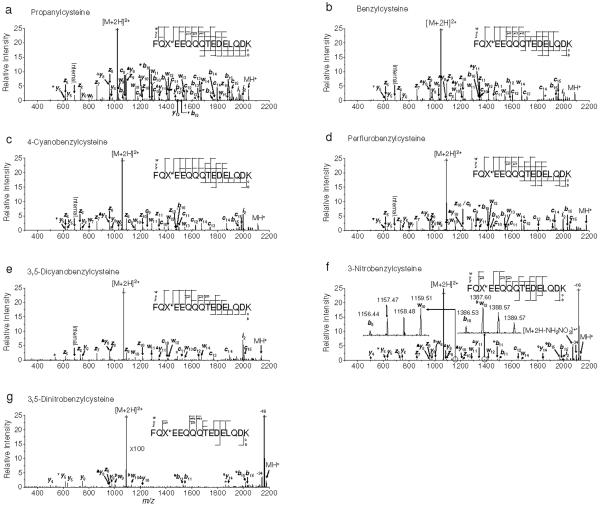
The IRMPD/ECD spectra are shown in Figure 3. In comparison with ECD-only spectra, many of the C-N bond cleavages (b, y ions) from cation radicals were detected.68 Hydrogen atom loss from the charge reduced cation radical, [M+2H]+•, is predominant in every IRMPD/ECD spectra, yielding [M+H]+. The loss of 17 and 18 Da from b and y ions in IRMPD/ECD spectra are assigned as ammonia and water, respectively. It is worth noting that abundant ECD type fragments (c, z and w ions) are observed in IRMPD/ECD spectra of [P+2H]2+, [B+2H]2+, [4CB+2H]2+ and [PFB+2H]2+ while those of [3NB+2H]2+ and [35DNB+2H]2+ exhibit a lower yield of these fragments (Figure 3). The IRMPD/ECD of [35DCB+2H]2+ presents slightly reduced but still prominent peak intensities (Figure 3e). The existence of abundant w ions is attributed to the higher level of vibrational excitation provided by infrared photons. Unusual w-C2H4 ions are observed in Figures 3f-g, which are also believed to be induced by additional vibrational excitation.
Figure 3.
IRMPD/ECD of doubly protonated model peptides. a) propanylcysteine, b) benzylcysteine, c) 4-cyanobenzylcysteine, d) perfluorobenzylcysteine, e) 3,5-dicyanobenzylcysteine, f) 3-nitrobenzylcysteine and g) 3,5-dinitrobenzylcysteine containing peptides, respectively. Precursor ions were heated by infrared photons to just below the onset of backbone cleavage. Electron irradiation was applied simultaneously with infrared excitation without isolation of heated precursor ions. Symbolic superscript appendixes °, Δ, ▼ and # indicates loss of hydroxyl radical from [b+1]+• and [y+1]+• ions, and ammonia, water and ethylene from either b and y or w ions, respectively. An asterisk indicates instrumental noise.
The isotope distributions of b ions in the IRMPD/ECD spectra were investigated for the presence of [b+1]+• ions formed by addition of a hydrogen atom to a typical b ion (Figure 3). The b8 and b10 ions have abundant peaks 1 Da higher than their calculated monoisotopic masses. Mass deviations from the theoretical masses were, however, large enough not to assign those peaks as [b+1]+• ions unlike a previous report.34 The most dominant b ions (b11 and b15) are observed at the C-terminus of aspartic acid residues and likely result from a salt-bridge mechanism (Figure 3).69 However, no significant yield of [b+1]+• ions from b11 and b15 ions was found, suggesting that the origin of b11 and b15 ions is the consequence of the direct IRMPD (data not shown). IRMPD/ECD spectra of the model peptides (Figure 3) were carefully examined for the presence of [y+1]+• ions but none was detected.
ETD of the EA-tuned Peptides
In a separate set of experiments, ETD spectra of the derivatized peptides were obtained to investigate possible differences between ECD and ETD. Without supplemental activation by collision prior to the electron transfer reaction, significant yields of c or z fragment ions were not observed in any ETD spectra. Hence, only spectra from the ETD of collisionaly activated ions (ETcaD)54 are discussed in this work.
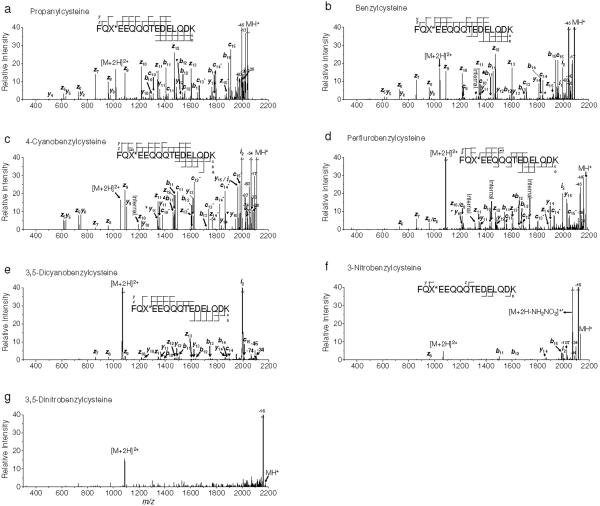
ETcaD spectra of the derivatized peptides are shown in Figure 4. While peptide dications are the most abundant peaks in ECD and IRMPD/ECD spectra, hydrogen atom loss (Figures 4a-e) or hydroxyl radical loss (Figures 4f-g) from [M+2H]+• is dominant in the ETcaD spectra. The relative intensities of precursor peptide dications and charge reduced cation radicals observed in ECD and ETcaD spectra indicate that ETcaD has a higher dissociation product yield than ECD (Figures 1 and 4). ECD-like side chain losses such as -17, -28, -36, -45 and -60 Da for ETcaD of [P+2H]2+, [B+2H]2+, [4CB+2H]2+ and [PFB+2H]2+ (Figures 4a-d) were identified. Loss of hydroxyl radical and NH2NO2 from [3NB+2H]+• and hydroxyl radical from [35DNB+2H]+• (Figures 4f-g) were observed. With ETD the coverage of sequence ions is generally better than that observed in ECD spectra. The ETcaD spectrum of [P+2H]2+ (Figure 4a) includes 6 out of 15 possible c ions (c8 to c15) and 10 out of 15 possible z ions (z5 to z13 and z15) while that of the ECD spectrum spans 2 out of 15 possible c ions (c14 and c15) and 4 out of 15 possible z ions (z10 to z13) (Figure 1a). The pattern of hydrogen abstraction forming c•/c ions and z•/z ions becomes more complex in comparison to the ECD data (Figures 1 and 4). No evidence was found for the presence of [b+1]+• and [y+1]+• ions in an examination of the isotope distributions of b and y ions. Therefore, all b and y ions are believed to be induced by direct action of vibrational excitation prior to ion/ion reaction.
Figure 4.
ETD of doubly protonated model peptides. a) propanylcysteine, b) benzylcysteine, c) 4-cyanobenzylcysteine, d) perfluorobenzylcysteine, e) 3,5-dicyanobenzylcysteine, f) 3-nitrobenzylcysteine and g) 3,5-dinitrobenzylcysteine containing peptides, respectively. Supplemental activation was performed prior to reaction with fluoranthene anion. Symbolic superscript appendixes °, Δ and ▼ indicates loss of hydroxyl radical from [b+1]+• and [y+1]+• ions, and ammonia, water from b and y ions, respectively.
Despite differences between ECD and ETD (i.e. electron capture/transfer cross section, exothermicity from electron transfer reaction depending on the electron affinity of the electron carrier reagent, and time scale of reaction or ion detection), typical backbone fragmentation is almost completely inhibited in ETcaD spectra of [35DCB+2H]2+, [3NB+2H]2+ and [35DNB+2H]2+ (Figures 4e-g). This observation reinforces the validity of the electron predator model for both ETD and ECD.
The presence of cleaved but hydrogen-bonded c, z fragment complexes were hypothesized in a previous study.5 This possibility can be explored using a high level of vibrational excitation in the peptide cation radicals. As seen in Figures 3f-g and 4f-g, this fails to yield significant abundances of ECD or ETD type backbone fragments. This supports the conjecture that stable peptide cation radicals are formed rather than hydrogen bonded c and z fragment complexes. However, the IRMPD/ECD of [35DCB+2H]2+ exhibits slightly more abundant fragment yields compared to the corresponding ECD and ETcaD spectra (Figures 1e, 3e and 4e). This also indicates that the nascent [35DCB+2H]+• cation radical is less stable compared to [3NB+2H]+• and [35DNB+2H]+• under the higher level of vibrational excitation.
Hydroxyl Radical Loss and Ion Formation Mechanism in MALDI plumes
As seen in Figures 1f-g, hydroxyl radical loss occurs from [3NB+2H]+• and [35DNB+2H]+•. In IRMPD/ECD, several peaks are observed 16 Da less than some b and y ions, indicating loss of hydroxyl radical from intermediately formed [b+1]+• and [y+1]+• ions (Figures 3f-g). Relevant to hydroxyl radical and related losses, formation of the phenylnitronic radical and its dissociation energetics were investigated in detail by Polasek and Turecek.58 The phenylnitronic radical is quite stable on the microsecond life time58 and does not appear to initiate significant backbone cleavages or other side chain losses in ECD of [3NB+2H]2+ and [35DNB+2H]2+. However, the phenylnitronic radical group easily undergoes a direct homolytic cleavage leading to hydroxyl radical loss and this process, which has an extremely low reverse reaction barrier (ca. ~0 kJ/mol),58 is especially prominent with higher levels of vibrational excitation (Figures 3f-g and 4f-g). The loss of HONO is calculated to be less energetically favorable,58 consistent with our observation that this is a less prominent dissociation pathway (Figures 1f-g, 3f-g and 4f-g). These theoretical calculations and experimental observations clearly support the formation of nitrobenzyl anion radical group and intramolecular proton transfer to it in ECD, IRMPD/ECD and ETcaD spectra of the nitrobenzylcysteine containing peptides.
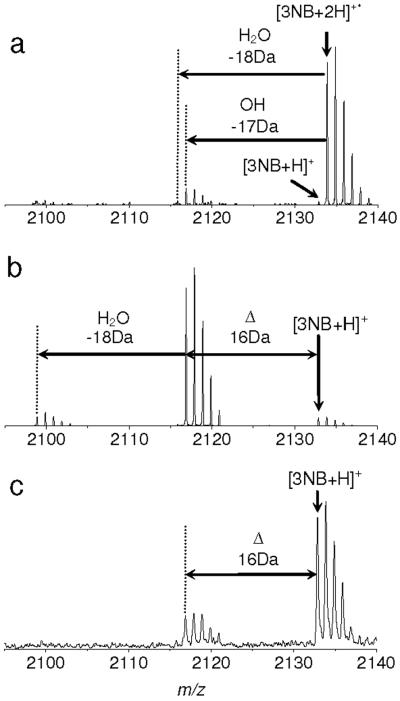
Hydroxyl radical loss also provides an explanation for the product appearing 16 Da less than [3NB+H]+ in the MALDI MS (Figure 5c). A similar loss from the 3-nitrotyrosine residue in MALDI MS of peptides has been reported previously.70 In the MALDI plume, a number of free electrons exist and may react with desorbed primary ions and neutrals.71 Protons can also be provided by numerous matrix molecules. From these observations, we suggest that ion yields in MALDI may in part result from charge neutralization process by electron capture of multiply protonated ions. This has also been discussed in several papers.72 However, prompt in-source decay backbone fragments (i.e. c and z ions) from the derivatized peptides were not observed in this work (data not shown).
Figure 5.
Hydroxyl loss from the charge reduced cation radical of 3-nitrobenzylcysteine containing peptide. a) ECD, b) IRMPD/ECD and c) MALDI-TOF MS of 3-nitrobenzylcysteine containing peptide. The peak 16 Da less than [3NB+H]+ is loss of hydroxyl radical from the charge reduced cation radical, [3NB+2H]+• based on our discussion. a) and b) are magnified Figures 1f and 3f, respectively, in m/z region between 2050 and 2140 around [3NB+2H]+• ion. c) was recorded using a time of flight mass spectrometer equipped with a 337nm N2 laser in the reflector mode. 100 shots were averaged. 10 mg/ml CHCA was used for matrix.
Kinetics of Electron Capture
At the inception of this study, we speculated that the tags having positive electron affinities might increase the overall efficiency of electron capture. This would be the case if, following the initial electron capture event, electron autodetachment competes with further relaxation of the nascent radical cation to yield ECD products. To investigate this possibility, ECD spectra of simultaneously isolated [B+2H]2+ and [3NB+2H]2+ ions were recorded. Similar initial ion signal intensities of peptide dications in the FT MS spectrum ([B+2H]2+/[3NB+2H]2+ = ~0.95) were established, and electron irradiation time was sequentially increased from 75 to 250 ms in order to monitor the relative electron capture kinetics. Assuming a constant electron flux during the irradiation period, the rate of electron capture can be expressed as in eq 1,
| (1) |
where [(M+2H)2+] and [e-]s are the number of the precursor ions and electrons, and kobs is the observed rate constant of the electron capture process. Eq 1 yields first order kinetics for the doubly charged ions, demonstrated by the data in Figure 6, where the logarithm of the [B+2H]2+ and [3NB+2H]2+ ion intensities versus electron irradiation time in the ICR cell are plotted. The nearly identical slopes indicates similar electron capture rates for [B+2H]2+ and [3NB+2H]2+. No change is observed that can be attributed to the higher EA tag. This is consistent with earlier studies which conclude that electron capture rates into high-n diffuse Rydberg states possess probabilities that vary as the square of the total charge of the ion.2,3 The eventual site at which the electron becomes localized is determined by through-space and through-bond electron transfer processes subsequent to the initial capture.29
Figure 6.
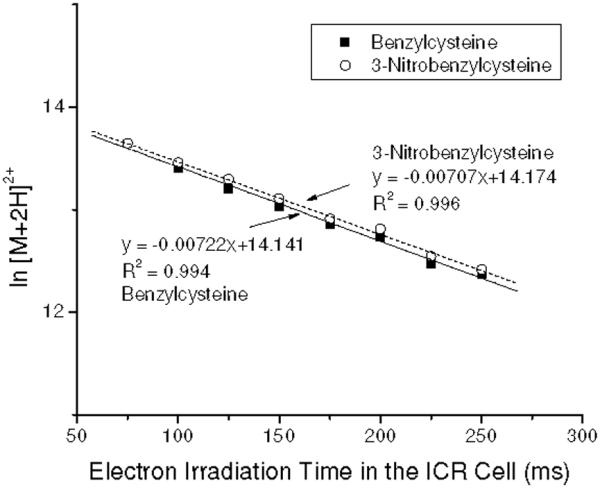
Variation in the natural logarithm of [B+2H]2+ and [3NB+2H]2+ with electron irradiation time in the ICR cell. Both precursor ions were simultaneously isolated for ECD with similar ion intensities. Slopes indicate that the electron predator has no effect on the rate of electron capture.
Discussion
Effect of EA-tuning Tags on Nascent Cation Radicals
The percent yield of each ECD fragmentation channel is depicted as a function of EA of tags in Figure 7. Equations 2-5 are used to calculate relative yield of different ECD processes, where a = charge reduced radical cations ([M+2H]+•), b = Σ [ci + zi + wi ions], c = Σ [side-chain loss] and d = Σ [other backbone fragments (b and y ions) and subsequent loss of H2O or NH3]. For each term, background noise was subtracted and isotopic contributions of each ion were summed up.
| (2) |
| (3) |
| (4) |
| (5) |
As seen in Figure 7, yield of c and z type backbone fragmentation generally diminishes with increasing EA of tags in the model peptides. Typical ECD type backbone fragments start to disappear when EA of the tag exceeds ~1.0 eV, independent of the functionality of the tag. It should be also noted that abundant side-chain losses in ECD of [35DCB+2H]2+, [3NB+2H]2+ and [35DNB+2H]2+ are mostly contributed by tag-related peaks such as RS• (i2), •OH and NH2NO2 losses, and not by other amino acids in the peptides.
Figure 7.
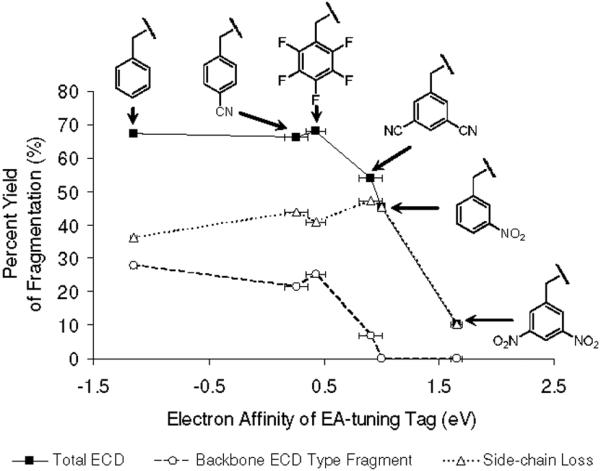
Relationship between the electron affinities of the tags and percent yields of various ECD fragmentation channels, including total ECD (solid line), backbone ECD type fragment (dash line) and side-chain loss yield (dotted line), respectively. The horizontal error bars are taken from references for the electron affinities of tags in Ref. 49. Each isotope distribution of ions is summed and normalized followed by eq. 2-5.
Different electron relaxation processes have different exothermicities, but they also lead to final states with dissociation pathways having very disparate activation energies. Therefore, it is important to consider the factors related to the stability of nascent peptide cation radicals formed in the electron capture and relaxation process. Figure 7 clearly demonstrates that EA of the tag is the most important parameter relating to stability of the cation radicals. A secondary factor appears to be the PAs of different intermediate anion radicals. Namely, if two tags have similar positive EA with different PAs of the corresponding anion radicals, ECD type backbone fragmentation of the tag with lower PA is more prominent. This idea is supported by calculated energetics of dicyanobenzene and nitrobenzene (Table 2) and by observed ECD spectra (Figures 1e-f). It is obvious that the most stable cation radical is [35DNB+2H]+• which exists mostly as a nascent cation radical with minimal fragmentation. To summarize, exceptional stability of nascent cation radicals is conferred by the generation of a stable radical center by electron capture followed by intramolecular proton transfer.
Table 2.
Enthalp from Quantum Mechanical Calculations on 1,3-dicyanobenzene. Enthalpies of Protonated, Electron and Hydrogen Attached Species of 1,3-dicyanobenzene were calculated and Compared with Those of Nitrobenzene from Polasek and Turecek.
| 1,3-Dicyanobenzene | |||||
|---|---|---|---|---|---|
| Level of Quantum Mechanical Calculation | Proton Affinitya | Electron Affinityb | Proton Affinity of Aniona | Hydrogen Affinitya | |
| Theoretical | B3LYP/6-31+G(d,p) | 787.5 | 1.073 | 1312.4 | 108.7 |
| B3LYP/6-311+G(2df,p) | 787.7 | 1.096 | 1307.0 | 100.5 | |
| ROMP2/6-311+G(2df,p) | 756.1 | 0.780 | 1269.9 | 39.1 | |
| B3-ROMP2c | 771.9 | 0.938 | 1288.5 | 68.9 | |
| Experimental | 779.3 | 0.91 | N/A | N/A | |
| Nitrobenzene | |||||
|---|---|---|---|---|---|
| Level of Quantum Mechanical Calculation | Proton Affinitya | Electron Affinityb | Proton Affinity of Aniona | Hydrogen Affinitya | |
| Theoretical | B3LYP/6-31+G(d,p) | 806.5 | 1.288 | 1384.8 | 201.8 |
| B3LYP/6-311+G(2df,p) | 806.6 | 1.252 | 1386.9 | 195.5 | |
| ROMP2/6-311+G(2df,p) | 776.5 | 0.718 | 1385.5 | 148.7 | |
| B3-ROMP2c | 791.6 | 0.985 | 1386.2 | 172.1 | |
| Experimental | 800.3 | 1.00 | N/A | N/A | |
unit of kJ/mol
unit of eV
1/2 (B3LYP + ROMP2).
The present investigation also leads to the conclusion that ECD and ETD may not generate abundant backbone cleavages in characterization of tyrosine nitration, which is widely observed in proteins as a post-translational modification.46
Quantum Mechanical Calculations
To further investigate the energetics and mechanism of electron capture in the presence of our tags, we performed several quantum mechanical calculations using a series of model compounds. First, the energetics of adding an electron, proton and hydrogen atom to the electron predators were evaluated to illuminate the stability and reactivity of model nascent cation radicals. Dicyanobenzene and nitrobenzene were chosen as model compounds to represent electron predators. The energetics of each process for nitrobenzene are derived from a previous study58 and are used here. Second, time-dependent density functional calculations of a series of reduced model peptide systems (Figure 8) were performed to estimate the relative energies among the excited states of cation radicals. These model systems comprise a series of N-(substituted-phenyl)acetamides with (B1-B6) or without (A1-A6) methyl ammonium, which forms a strong hydrogen bond to the amide carbonyl. For N-(3-nitrophenyl)acetamide and N-(3,5-dicyanophenyl)acetamide, the structures having strong hydrogen bonds to the substituted moieties such as the nitro or cyano groups are considered (C4 and C5). In particular, for N-(3-nitrophenyl)acetamide, the very stable structure formed with strong hydrogen bonds to both amide carbonyl and nitro oxygen (D5) is investigated. The vertical electron affinities and recombination energies were also calculated to provide vertical electronic energies of the lowest electronic states of each model species. This facilitates evaluation of the relative exothermicities of different electron relaxation processes to specific orbitals related to different reaction pathways (i.e. forming a stable radical intermediate or forming precursors that can lead to typical ECD backbone fragmentation processes).
Before discussing the electron capture process, it is appropriate to consider the sites of protonation in our model peptide cations. Unlike the 2-(4′-carboxypyrid-2′-yl)-4-carboxamide group studied by the Turecek group34 as a radical trap, our electron predators, a term used to describe the superior electron trapping abilities of 3,5-dicyanobenzyl, 3-nitrobenzyl and 3,5-dinitrobenzyl groups, are not stronger gasphase bases (PA[1,3-Dicyanobenzene] = 779.3 kJ/mol, PA[Nitrobenzene] = 800.3 kJ/mol)73 than other possible protonation sites such as the N-terminal amine (PA[Glycine] = 866.5 kJ/mol)73 or the ε-amine of lysine (PA[Lysine] = 966.0 kJ/mol).73 Therefore, peptide dications are not likely to be protonated at the site of the EA-tuning tags. The probable sites of protonation in the model peptide chosen for this study are the N-terminal amine and lysine amine.
Table 2 summarizes all calculated energies related to dicyanobenzene and nitrobenzene. The protonation sites of 1,3-dicyanobenzene (DCB) and nitrobenzene are the nitrogen of one of the cyano groups and the oxygen of the nitro group, respectively.58,74 The full sets of optimized structures and electronic energies, zero-point energy corrections and enthalpies of DCB, DCBH+, DCB-• and DCBH• are available in Supplemental Information. The enthalpy of each species is compared with that of Polasek and Turecek's report for nitrobenzene.58 The adiabatic electron affinity of 1,3-dicyanobenzene calculated at the B3-ROMP2/6-311+G(2df,p)//B3LYP/6-31+G(d,p) level in this work is 0.937 eV, in good agreement with the experimental value of 0.91 eV.
An important observation from these calculations is the difference of hydrogen affinity of 1,3-dicyanobenzene (69.8 kJ/mol) and nitrobenzene (172.1 kJ/mol), which contrasts with their similar EAs (EA[1,3-dicyanobenzene] = 0.91 eV, EA[nitrobenzene] = 1.00 eV). The ~2.5 times higher hydrogen affinity of nitrobenzene compared to that of 1,3-dicyanobenzene may in part be responsible for the absence of any significant ECD type backbone fragment from the 3-nitrobenzyl derivatized peptide (Figure 1f) while the 3,5-dicyanobenzyl derivatized peptide exhibits small yields of c and z ions (Figure 1e). It is also noteworthy that both tags have higher hydrogen affinity than the amide carbonyls (21-41 kJ/mol).30
To estimate the overall energy released by the electron capture process, we calculated the vertical electron affinity of the neutrals and the vertical recombination energy of the cation-neutral complexes by adding an electron to each system without geometry optimization (Table 3). The general trend observed in Table 3 is reasonable in comparison with the electron affinities of the tags listed in Table 2, regardless of the presence of Coulomb stabilization conferred by the methyl ammonium ion. Notably, electron affinities of A3 and B3 were estimated as slightly negative values regardless of the calculation methods, in contrast to the experimentally reported values in Table 1. However, Frazier et al. reported negative electron affinities of the π* orbitals of perfluorobenzene,75 which lends support to the validity of the calculated negative vertical electron affinities. ROMP2 vertical electron affinities for A2 through A5 seem to be erroneous showing all negative values. This manifest error may be caused by the limitation of the restricted spin calculation. It should be stressed that recombination energies of methyl ammonium complexes are highly dependent on their particular hydrogen bond acceptors. Also, although B6 has two nitro groups on the phenyl ring, C5 undergoes the most exothermic recombination process.
Table 3.
The Vertical Electron Affinities and Vertical Recombination Energies of the Model Compounds Described in Figure 8 at Various Levels of Theories. All Energies are in Units of Electron Volt.
| Species | UB3LYP | UM06 | UB3LYP | UM06 | ROMP2 | B3-ROMP2a |
|---|---|---|---|---|---|---|
| 6-31++G(d,p) | 6-311++G(2df,p) | |||||
| A1 | -0.424 | -0.605 | -0.394 | -0.519 | -0.589 | -0.491 |
| A2 | 0.074 | 0.171 | 0.136 | 0.275 | -0.390 | -0.127 |
| A3 | -0.025 | -0.194 | -0.028 | -0.141 | -0.418 | -0.223 |
| A4 | 0.801 | 0.947 | 0.875 | 1.040 | -0.262 | 0.306 |
| A5 | 0.819 | 0.962 | 0.852 | 1.019 | -0.393 | 0.230 |
| A6 | 1.584 | 1.725 | 1.602 | 1.753 | N/Ab | N/Ab |
| B1 | 2.950 | 2.698 | 2.946 | 2.739 | 2.740 | 2.843 |
| B2 | 3.184 | 2.969 | 3.196 | 3.033 | 2.872 | 3.034 |
| B3 | 3.120 | 2.890 | 3.112 | 2.930 | 2.858 | 2.985 |
| B4 | 3.494 | 3.348 | 3.523 | 3.425 | 2.947 | 3.235 |
| B5 | 3.521 | 3.403 | 3.530 | 3.455 | 2.834 | 3.182 |
| C4 | 3.569 | 3.567 | 3.612 | 3.657 | 2.919 | 3.266 |
| C5 | 4.668 | 4.798 | 4.700 | 4.853 | 4.305 | 4.502 |
| D5 | 4.060 | 4.209 | 4.095 | 4.264 | 3.599 | 3.847 |
| B6 | 3.966 | 4.000 | 3.974 | 4.031 | 2.991 | 3.482 |
1/2 (UB3LYP + ROMP2) without zero point energy correction.
Unrestricted open-shell SCF was not converge.
To further investigate the relative energetics of excited states during the relaxation of a captured electron, we performed time-dependent density functional calculations on the model systems shown in Figure 8. Excited state orbitals of charge neutralized B4, B5, C4, C5 and D5 radicals generated by TDDFT calculations are depicted in Figure 9.76 These excited MOs clearly reveal the effects of different hydrogen bonding partners. As seen in Figures 9a and 9b, a hydrogen bond to the amide carbonyl lowers the energy of the amide π* orbital, while the nitrophenyl π* orbital mixed with the ground Rydberg orbital of the methyl ammonium ion give rise to nearly degenerate lowest states (X and A states). The relative energy gaps among orbitals in which we are interested are quite similar in both B4 and B5 (Figures 9a-b). If the methyl ammonium ion directly interacts with an oxygen of the nitro group as in C5, it significantly stabilizes the nitrophenyl π* orbital, pushing the ground Rydberg orbital (A state) and the amide π* orbital (H and I states) to higher levels (Figure 9d). This effect is diminished by having another hydrogen bond with the amide carbonyl simultaneously with the nitro group (Figure 9e). However, this reordering of orbitals is not observed in the case of C4 despite the presence of the similar hydrogen bond with the cyano group (Figure 9c). As seen in excited state MOs of B4 and B5, the first two excited states of C4 are constituted from the dicyanophenyl π* orbitals mixed with the ground Rydberg orbitals of the methyl ammonium ion, being nearly degenerate.
Figure 9.
Excited state molecular orbitals obtained from time-dependent density functional calculations of a) B4 b) B5 c) C4 d) C5 and e) D5 at the UB3LYP/6-311++G(2df,p) level. See Figure 8 for the structure of each species.
In summary, these theoretical calculations and experimental observations lead to two conclusions. First, the inhibition of typical ECD backbone fragmentation requires a certain level of intrinsic positive electron affinity of the tag. The efficiency of the electron trap is further augmented by structure-dependent hydrogen bonds to the derivatized functional groups. In particular, the higher proton affinity of the nitro group compared to the cyano group (Table 2) facilitates more stable hydrogen bond formation with the N-terminal amine or lysine ε-amine. This results in higher populations of structural conformations which stabilize the nitrophenyl π* orbital and push other orbitals to higher levels. It is thus a reasonable prediction that the nascent [35DCB+2H]+• cation radical would be less stable than [3NB+2H]+• and [35DNB+2H]+•. This prediction is consistent with our observations of small fractions of typical ECD backbone fragmentation in ECD, IRMPD/ECD and ETcaD of [35DCB+2H]2+ (Figures 1e, 3e and 4e). Therefore, we conclude that the electron relaxation process after the initial electron capture to high lying Rydberg states is modulated by the presence of tags with positive EAs and their structure-dependent hydrogen bonds.
Second, the formation of a stable and regiospecific radical center68,77 on the nitrophenyl tags raises a question regarding the operation of the UW mechanism for ECD type backbone fragmentation in the EA-tuned peptides. This mechanism invokes the engagement of Coulomb stabilized amide π* orbitals in the electron relaxation and subsequent backbone cleavage processes. Although this process is energetically exothermic and has a lower barrier than the Cornell mechanism,19,24,28,32 backbone fragmentation was not observed in the presence of electron predators. In addition, the proton affinity of the amide carbonyl group (PA[CH3CONHCH3] = 888.5 kJ/mol, the protonation site being the carbonyl oxygen)73,78 is higher than those of the cyanophenyl and nitrophenyl groups (Table 2). This suggests that the amide carbonyl groups would more frequently participate in strong hydrogen bond formation than either the cyanophenyl or nitrophenyl group. Thus, more populated conformations that could induce the formation of the aminoketyl intermediate should contribute to the probability leading to typical ECD cleavage processes. However, backbone fragmentation is inhibited in the presence of the electron predator. This contradiction leads to the implication that, even with the assistance of Coulomb stabilization, the amide π* orbital cannot capture an electron to form a stable bound state that in turn would be expected to result in backbone fragmentation processes. However, it is possible that the presence of the electron predator could modulate the probability of intramolecular electron transfer from a hign-n Rydberg orbital to the amide π* orbital by intercepting and trapping the electron. This may prevail even when transient conformations of the peptide render electron capture by the amide π* orbital energetically more favorable.
Comparison of ECD, ETD and the Effect of Augmented Vibrational Excitation
The ECD and ETD experimental methodologies have several different aspects. The electron capture/transfer cross sections are different due to different electron transfer media (i.e. free electron for ECD and anion radical for ETD). Both methods also have dissimilar recombination energies, modified by the EA of the electron transfer reagent. In addition, the time scales associated with different instruments or instrumental parameters during the electron capture/transfer process, followed by dissociation, are different.
Inelastic scattering as well as electron transfer during energetic collisions between electron transfer reagent anions and peptide dications could result in higher internal energies of the resulting peptide cation radicals. Similarly, in the case of ECD, recombination involving energetic electrons as well as inelastic electron-peptide cation collisions may yield peptide cation radicals with excess internal energy. As a result, it is difficult to assess the internal energy distribution of peptide cation radicals formed by electron capture or transfer reactions. Therefore, we only discuss the recombination energy gained by the electron capture and transfer processes.
In the present work, we used fluoranthene with EA ~0.7 eV for the electron transfer reagent. Therefore, the overall recombination energy of ETD is smaller than that of ECD by ~0.7 eV, and fragmentation yields may be reduced in ETD relative to ECD. As noted above, supplemental activation by collision is required to acquire abundant backbone fragments. However, as seen in Figures 1, 3 and 4, the general dissociation patterns in ECD, IRMPD/ECD and ETcaD spectra are not significantly different, including the absence of ECD or ETD type fragmentation of [3NB+2H]2+ and [35DNB+2H]2+. This similarity leads to the conclusion that the overall recombination energy gained by either electron capture or transfer does not affect subsequent fragmentation processes. Excess vibrational excitation, provided either by IR photon absorption or by collisions with an inert gas, also does not produce any significant difference, which also indicates that the levels of vibrational excitation for dissociating ion populations in each case are similar.
Conclusion
We have elucidated some key aspects of the mechanism of electron capture dissociation and electron transfer dissociation of doubly protonated peptides. The twenty common amino acids, in the absence of post-translational modifications, do not have positive electron affinities. Using the model peptide FQpSEEQQQTEDELQDK, we have modified the phosphoserine residue to incorporate a range of functional groups of widely varying electron affinity, include propanyl, benzyl, 4-cyanobenzyl, perfluorobenzyl, 3,5-dicyanobenzyl, 3-nitrobenzyl and 3,5-dinitrobenzyl structural moieties, having a range of EA from -1.15 to 1.65 eV, excluding the propanyl group. Typical ECD or ETD backbone fragmentations are completely inhibited in peptides with substituent tags having EA over 1.00 eV, which we refer to as electron predators. The kinetics of the initial electron capture are not modified by the presence of the electron predators, consistent with the expectation that electron capture kinetics are governed by the long range electron-dication interaction. Once an electron is captured to high-n Rydberg states, however, we propose that through-space or through-bond electron transfer to the EA-tuning tags or low-n Rydberg states via potential curve crossing occurs in competition with transfer to the amide π* orbital. This conjecture is supported by time-dependent density functional theory applied to a series of reduced model systems. The intramolecular electron transfer process is modulated by structure-dependent hydrogen bonds and is heavily affected by the presence and type of electron withdrawing groups in the EA-tuning tag. The anion radicals formed by electron predators have high proton affinities (approximately 1400 kJ/mol for the 3-nitrobenzyl anion radical) in comparison to other basic sites in the model peptide dication, facilitating exothermic proton transfer from one of the two sites of protonation. This forms a stable radical intermediate and interrupts the normal sequence of events in ECD or ETD leading to backbone fragmentation through the intermediacy of an aminoketyl radical which fragments by β-cleavage of the adjacent N-Cα bond. Even in the presence of Coulomb stabilization from nearby charges it does not appear that one can infer that the amide π* orbital can compete with the electron predators, with electron affinities in excess of 1.0 eV, as the eventual site of localization of the captured electron.
The phenynitronic group formed by sequential electron and proton transfer to a nitrophenyl group in a peptide undergoes a facile hydroxyl loss. This process provides an explanation for the unusual peak observed in MALDI MS of peptides containing a nitrophenyl group, 16 Da less than [M+H]+. It indicates the role of electrons in charge reduction processes converting multiply charged peptides and proteins to the more usual singly charged ions observed in MALDI MS. Nitration of tyrosine is an important post-translational modification associated with cell signaling pathways and oxidative inflammatory responses.46 Interestingly, this process introduces an electron predator that exhibits behavior similar to what we observe with our derivatized peptides.79 We are exploring the possibility that this can be exploited to facilitate the detection of trace peptides where this PTM is present.
Supplementary Material
Acknowledgement
This work was supported by the National Science Foundation through grant CHE-0416381 and the Beckman Institute at California Institute of Technology. The computational resource was kindly provided by the Materials and Process Simulation Center at California Institute of Technology. C. H. S. acknowledges a fellowship from the Kwanjeong Educational Foundation. P. R. acknowledges support from the NIH/NIDCR UCLA Research Training Program (T32 DE007296). The NIH/NCRR High-End Instrumentation Program supported the acquisition of the LTQ-FT mass spectrometer (grant S10 RR023045 to J.A.L.). The authors thank Professor Jack Simons for discussions regarding the electron capture process, Professor Woon-Seok Yeo for help with synthesis, Professor Francis Turecek, Dr. Yousung Jung and Dr. Jiyoung Heo for assistance with the computational analysis, and Dr. Hugh I. Kim for discussions of reaction mechanisms.
Footnotes
Supporting Information Available: 1H-NMR peaks of thiol compounds, synthesis of 3,5-dicyanobenzyl thiol, ECD and IRMPD/ECD spectra of 2-nitrobenzyl, 4-nitrobenzyl and Nα-3,5-dinitrophenyl derivatized peptides, geometries, energetics and molecular orbitals of the model species from quantum mechanical calculations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- (2).Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Anal. Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- (3).Zubarev RA, Haselmann KF, Budnik B, Kjeldsen F, Jensen F. Eur. J. Mass Spectrom. 2002;8:337–349. [Google Scholar]
- (4).Zubarev RA. Mass Spectrom. Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]; Cooper HJ, Hakansson K, Marshall AG. Mass Spectrom. Rev. 2005;24:201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]; Leymarie N, Costello CE, O'Connor PB. J. Am. Chem. Soc. 2003;125:8949–8958. doi: 10.1021/ja028831n. [DOI] [PubMed] [Google Scholar]; Lin C, O'Connor PB, Cournoyer JJ. J. Am. Soc. Mass Spectrom. 2006;17:1605–1615. doi: 10.1016/j.jasms.2006.07.007. [DOI] [PubMed] [Google Scholar]
- (5).O'Connor PB, Lin C, Cournoyer JJ, Pittman JL, Belyayev M, Budnik BA. J. Am. Soc. Mass Spectrom. 2006;17:576–585. doi: 10.1016/j.jasms.2005.12.015. [DOI] [PubMed] [Google Scholar]
- (6).Senko MW, Speir JP, McLafferty FW. Anal. Chem. 1994;66:2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]; Laskin J, Futrell JH. Mass Spectrom. Rev. 2003;22:158–181. doi: 10.1002/mas.10041. [DOI] [PubMed] [Google Scholar]; Medzihradszky KF, Campbell JM, Baldwin MA, Falick AM, Juhasz P, Vestal ML, Burlingame AL. Anal. Chem. 2000;72:552–558. doi: 10.1021/ac990809y. [DOI] [PubMed] [Google Scholar]
- (7).Woodin RL, Bomse DS, Beauchamp JL. J. Am. Chem. Soc. 1978;100:3248–3250. [Google Scholar]; Little DP, Speir JP, Senko MW, Oconnor PB, McLafferty FW. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- (8).Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Domon B, Aebersold R. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]; Siuti N, Kelleher NL. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kim HI, Beauchamp JL. J. Am. Chem. Soc. 2008;130:1245–1257. doi: 10.1021/ja075698w. [DOI] [PubMed] [Google Scholar]; Kim HI, Beauchamp JL. J. Am. Soc. Mass Spectrom. 2009;20:157–166. doi: 10.1016/j.jasms.2008.10.003. [DOI] [PubMed] [Google Scholar]
- (11).Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Rapid Commun. Mass Spectrom. 2000;14:1793–1800. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]; Shi SDH, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Anal. Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]; Sweet SMM, Cooper HJ. Expert Rev. Proteomics. 2007;4:149–159. doi: 10.1586/14789450.4.2.149. [DOI] [PubMed] [Google Scholar]; Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mirgorodskaya E, Roepstorff P, Zubarev RA. Anal. Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]; Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Anal. Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]; Zaia J. Mass Spectrom. Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]; Mormann M, Paulsen H, Peter-Katalinic J. Eur. J. Mass Spectrom. 2005;11:497–511. doi: 10.1255/ejms.738. [DOI] [PubMed] [Google Scholar]; Morelle W, Canis K, Chirat F, Faid V, Michalski JC. Proteomics. 2006;6:3993–4015. doi: 10.1002/pmic.200600129. [DOI] [PubMed] [Google Scholar]; Adamson JT, Hakansson K. Anal. Chem. 2007;79:2901–2910. doi: 10.1021/ac0621423. [DOI] [PubMed] [Google Scholar]; Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Nat. Chem. Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- (13).Simon MD, Chu FX, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. J. Am. Chem. Soc. 1999;121:2857–2862. [Google Scholar]
- (15).Xia Y, Chrisman PA, Erickson DE, Liu J, Liang XR, Londry FA, Yang MJ, McLuckey SA. Anal. Chem. 2006;78:4146–4154. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liang XR, Xia Y, McLuckey SA. Anal. Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Marshall AG, Hendrickson CL, Jackson GS. Mass Spectrom. Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- (17).Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. J. Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]; McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Anal. Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]; McAlister GC, Berggren WT, Griep-Raming J, Horning S, Makarov A, Phanstiel D, Stafford G, Swaney DL, Syka JEP, Zabrouskov V, Coon JJ. J. Proteome Res. 2008;7:3127–3136. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).McLafferty FW, Horn DM, Breuker K, Ge Y, Lewis MA, Cerda B, Zubarev RA, Carpenter BK. J. Am. Soc. Mass Spectrom. 2001;12:245–249. doi: 10.1016/S1044-0305(00)00223-3. [DOI] [PubMed] [Google Scholar]
- (19).Chen XH, Turecek F. J. Am. Chem. Soc. 2006;128:12520–12530. doi: 10.1021/ja063676o. [DOI] [PubMed] [Google Scholar]
- (20).Haselmann KF, Budnik BA, Olsen JV, Nielsen ML, Reis CA, Clausen H, Johnsen AH, Zubarev RA. Anal. Chem. 2001;73:2998–3005. doi: 10.1021/ac0015523. [DOI] [PubMed] [Google Scholar]; Jackson SN, Dutta S, Woods AS. J. Am. Soc. Mass Spectrom. In Press, doi:10.1016/j.jasms.2008.08.021. [Google Scholar]
- (21).Breuker K, Oh HB, Lin C, Carpenter BK, McLafferty FW. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14011–14016. doi: 10.1073/pnas.0406095101. [DOI] [PMC free article] [PubMed] [Google Scholar]; Patriksson A, Adams C, Kjeldsen F, Raber J, van der Spoel D, Zubarev RA. Int. J. Mass Spectrom. 2006;248:124–135. [Google Scholar]; Rand KD, Adams CM, Zubarev RA, Jorgensen TJD. J. Am. Chem. Soc. 2008;130:1341–1349. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- (22).Hudgins RR, Kleinnijenhuis AJ, Quinn JP, Hendrickson CL, Marto JA. Proceedings of the 50th ASMS Conference on Mass Spectrometry and Allied Topics; Orlando, FL. June.2002. [Google Scholar]
- (23).Iavarone AT, Paech K, Williams ER. Anal. Chem. 2004;76:2231–2238. doi: 10.1021/ac035431p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chamot-Rooke J, Malosse C, Frison G, Turecek F. J. Am. Soc. Mass Spectrom. 2007;18:2146–2161. doi: 10.1016/j.jasms.2007.09.009. [DOI] [PubMed] [Google Scholar]
- (25).Xia Y, Gunawardena HP, Erickson DE, McLuckey SA. J. Am. Chem. Soc. 2007;129:12232–12243. doi: 10.1021/ja0736764. [DOI] [PubMed] [Google Scholar]
- (26).Hayakawa S, Matsubara H, Panja S, Hvelplund P, Nielsen SB, Chen XH, Turecek F. J. Am. Chem. Soc. 2008;130:7645–7654. doi: 10.1021/ja800207x. [DOI] [PubMed] [Google Scholar]
- (27).Sawicka A, Skurski P, Hudgins RR, Simons J. J. Phys. Chem. B. 2003;107:13505–13511. [Google Scholar]; Sobczyk M, Anusiewicz W, Berdys-Kochanska J, Sawicka A, Skurski P, Simons J. J. Phys. Chem. A. 2005;109:250–258. doi: 10.1021/jp0463114. [DOI] [PubMed] [Google Scholar]; Anusiewicz W, Berdys-Kochanska J, Simons J. J. Phys. Chem. A. 2005;109:5801–5813. doi: 10.1021/jp050218d. [DOI] [PubMed] [Google Scholar]; Sobczyk M, Simons J. J. Phys. Chem. B. 2006;110:7519–7527. doi: 10.1021/jp0604701. [DOI] [PubMed] [Google Scholar]; Sobczyk M, Simons J. Int. J. Mass Spectrom. 2006;253:274–280. [Google Scholar]; Skurski P, Sobczyk M, Jakowski J, Simons J. Int. J. Mass Spectrom. 2007;265:197–212. [Google Scholar]; Neff D, Sobczyk M, Simons J. Int. J. Mass Spectrom. 2008;276:91–101. [Google Scholar]
- (28).Anusiewicz I, Berdys-Kochanska J, Skurski P, Simons J. J. Phys. Chem. A. 2006;110:1261–1266. doi: 10.1021/jp052914j. [DOI] [PubMed] [Google Scholar]
- (29).Sobczyk M, Neff D, Simons J. Int. J. Mass Spectrom. 2008;269:149–164. [Google Scholar]
- (30).Turecek F, Syrstad EA. J. Am. Chem. Soc. 2003;125:3353–3369. doi: 10.1021/ja021162t. [DOI] [PubMed] [Google Scholar]
- (31).Turecek F. J. Am. Chem. Soc. 2003;125:5954–5963. doi: 10.1021/ja021323t. [DOI] [PubMed] [Google Scholar]; Turecek F, Syrstad EA, Seymour JL, Chen XH, Yao CX. J. Mass Spectrom. 2003;38:1093–1104. doi: 10.1002/jms.527. [DOI] [PubMed] [Google Scholar]
- (32).Syrstad EA, Turecek F. J. Am. Soc. Mass Spectrom. 2005;16:208–224. doi: 10.1016/j.jasms.2004.11.001. [DOI] [PubMed] [Google Scholar]
- (33).Yao CX, Turecek F. Phys. Chem. Chem. Phys. 2005;7:912–920. doi: 10.1039/b414764b. [DOI] [PubMed] [Google Scholar]; Yao CX, Syrstad EA, Turecek F. J. Phys. Chem. A. 2007;111:4167–4180. doi: 10.1021/jp0705020. [DOI] [PubMed] [Google Scholar]; Hayakawa S, Hashimoto M, Matsubara H, Turecek F. J. Am. Chem. Soc. 2007;129:7936–7949. doi: 10.1021/ja0712571. [DOI] [PubMed] [Google Scholar]; Turecek F, Jones JW, Towle T, Panja S, Nielsen SB, Hvelplund P, Paizs B. J. Am. Chem. Soc. 2008;130:14584–14596. doi: 10.1021/ja8036367. [DOI] [PubMed] [Google Scholar]
- (34).Jones JW, Sasaki T, Goodlett DR, Turecek F. J. Am. Soc. Mass Spectrom. 2007;18:432–444. doi: 10.1016/j.jasms.2006.10.005. [DOI] [PubMed] [Google Scholar]
- (35).Turecek F, Chen XH, Hao CT. J. Am. Chem. Soc. 2008;130:8818–8833. doi: 10.1021/ja8019005. [DOI] [PubMed] [Google Scholar]
- (36).Vertical electron affinity of the amide π* orbital is ca. -2.5 eV. The Coulomb stabilization energy varies with distance R (Å) as 14.4 eV Å/R (Å). See Ref. 27 and Seydou M, Modelli A, Lucas B, Konate K, Desfrancois C, Schermann JP. Eur. Phys. J. D. 2005;35:199–205.; Zhang H, Lu JF, Zhang SF, Tang K, Zhou ZY. Chin. J. Struct. Chem. 2007;26:1373–1379.
- (37).Laskin J, Futrell JH, Chu IK. J. Am. Chem. Soc. 2007;129:9598. doi: 10.1021/ja073748r. [DOI] [PubMed] [Google Scholar]; Frison G, van der Rest G, Turecek F, Besson T, Lemaire J, Maitre P, Chamot-Rooke J. J. Am. Chem. Soc. 2008;130:14916–14917. doi: 10.1021/ja805257v. [DOI] [PubMed] [Google Scholar]
- (38).Prell JS, O'Brien JT, Holm AIS, Leib RD, Donald WA, Willialms ER. J. Am. Chem. Soc. 2008;130:12680–12689. doi: 10.1021/ja8022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Al-Khalili A, Thomas R, Ehlerding A, Hellberg F, Geppert WD, Zhaunerchyk V, af Ugglas M, Larsson M, Uggerud E, Vedde J, Adlhart C, Semaniak J, Kaminska M, Zubarev RA, Kjeldsen F, Andersson PU, Osterdahl F, Bednarska VA, Paal A. J. Chem. Phys. 2004;121:5700–5708. doi: 10.1063/1.1782772. [DOI] [PubMed] [Google Scholar]; Uggerud E. Int. J. Mass Spectrom. 2004;234:45–50. [Google Scholar]; Bakken V, Helgaker T, Uggerud E. Eur. J. Mass Spectrom. 2004;10:625–638. doi: 10.1255/ejms.665. [DOI] [PubMed] [Google Scholar]
- (40).Pouthier V, Tsybin YO. J. Chem. Phys. 2008;129 doi: 10.1063/1.2965525. [DOI] [PubMed] [Google Scholar]
- (41).Li X, Cournoyer JJ, Lin C, O'Connor PB. J. Am. Soc. Mass Spectrom. 2008;19:1514–1526. doi: 10.1016/j.jasms.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Gunawardena HP, Gorenstein L, Erickson DE, Xia Y, McLuckey SA. Int. J. Mass Spectrom. 2007;265:130–138. [Google Scholar]
- (43).Chamot-Rooke J, van der Rest G, Dalleu A, Bay S, Lemoine J. J. Am. Soc. Mass Spectrom. 2007;18:1405–1413. doi: 10.1016/j.jasms.2007.04.008. [DOI] [PubMed] [Google Scholar]
- (44).Belyayev MA, Cournoyer JJ, Lin C, O'Connor PB. J. Am. Soc. Mass Spectrom. 2006;17:1428–1436. doi: 10.1016/j.jasms.2006.06.008. [DOI] [PubMed] [Google Scholar]
- (45).Brown RS, Lennon JJ. Anal. Chem. 1995;67:3990–3999. doi: 10.1021/ac00117a027. [DOI] [PubMed] [Google Scholar]
- (46).Schopfer FJ, Baker PRS, Freeman BA. Trends Biochem.Sci. 2003;28:646–654. doi: 10.1016/j.tibs.2003.10.006. [DOI] [PubMed] [Google Scholar]; Pacher P, Beckman JS, Liaudet L. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Han CC, Balakumar R. Tetrahedron Lett. 2006;47:8255–8258. [Google Scholar]; Tewari N, Nizar H, Mane A, George V, Prasad M. Synth. Commun. 2006;36:1911–1914. [Google Scholar]
- (48).Adamczyk M, Gebler JC, Wu J. Rapid Commun. Mass Spectrom. 2001;15:1481–1488. doi: 10.1002/rcm.394. [DOI] [PubMed] [Google Scholar]; Knight ZA, Schilling B, Row RH, Kenski DM, Gibson BW, Shokat KM. Nat. Biotechnol. 2003;21:1047–1054. doi: 10.1038/nbt863. [DOI] [PubMed] [Google Scholar]; Jalili PR, Sharma D, Ball HL. J. Am. Soc. Mass Spectrom. 2007;18:1007–1017. doi: 10.1016/j.jasms.2007.02.010. [DOI] [PubMed] [Google Scholar]
- (49).Jordan KD, Michejda JA, Burrow PD. J. Am. Chem. Soc. 1976;98:7189–7191. [Google Scholar]; Wentworth WE, Kao LW, Becker RS. J. Phys. Chem. 1975;79:1161–1169. [Google Scholar]; Ziatkis A, Lee CK, Wentworth WE, Chen ECM. Anal. Chem. 1983;55:1596–1599. [Google Scholar]; Wentworth WE, Limero T, Chen ECM. J. Phys. Chem. 1987;91:241–245. [Google Scholar]; Dillow GW, Kebarle P. J. Am. Chem. Soc. 1989;111:5592–5596. [Google Scholar]; Chowdhury S, Kebarle P. J. Am. Chem. Soc. 1986;108:5453–5459. [Google Scholar]; Desfrancois C, Periquet V, Lyapustina SA, Lippa TP, Robinson DW, Bowen KH, Nonaka H, Compton RN. J. Chem. Phys. 1999;111:4569–4576. [Google Scholar]; Fukuda EK, McIver RT. J. Am. Chem. Soc. 1985;107:2291–2296. [Google Scholar]
- (50).Sanger F. Biochem. J. 1945;39:507–515. doi: 10.1042/bj0390507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chen XH, Anderson VE, Chen YH. J. Am. Soc. Mass Spectrom. 1999;10:448–452. doi: 10.1016/s1044-0305(99)00013-6. [DOI] [PubMed] [Google Scholar]
- (52).Xie YM, Zhang J, Yin S, Loo JA. J. Am. Chem. Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- (53).Betowski LD, Enlow M, Aue DH. Int. J. Mass Spectrom. 2006;255:123–129. [Google Scholar]
- (54).Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Anal. Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Beavis RC, Chait BT. Anal. Chem. 1990;62:1836–1840. doi: 10.1021/ac00216a020. [DOI] [PubMed] [Google Scholar]
- (56). http://prospector.ucsf.edu.
- (57).Granovsky AA. PC GAMESS version 7.1. http://classic.chem.msu.su/gran/gamess/index.html.
- (58).Polasek M, Turecek F. J. Am. Chem. Soc. 2000;122:9511–9524. [Google Scholar]
- (59).Becke AD. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- (60).Lee CT, Yang WT, Parr RG. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- (61).Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, Dupuis M, Montgomery JA. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- (62).Zhao Y, Truhlar DG. Theor. Chem. Account. 2008;120:215–241. [Google Scholar]
- (63).Bode BM, Gordon MS. J. Mol. Graph. 1998;16:133–138. doi: 10.1016/s1093-3263(99)00002-9. [DOI] [PubMed] [Google Scholar]
- (64).Savitski MM, Kjeldsen F, Nielsen ML, Zubarev RA. Angew. Chem. Int. Edit. 2006;45:5301–5303. doi: 10.1002/anie.200601240. [DOI] [PubMed] [Google Scholar]
- (65).Savitski MM, Kjeldsen F, Nielsen ML, Zubarev RA. J. Am. Soc. Mass Spectrom. 2007;18:113–120. doi: 10.1016/j.jasms.2006.09.008. [DOI] [PubMed] [Google Scholar]
- (66).Chalkley RJ, Brinkworth CS, Burlingame AL. J. Am. Soc. Mass Spectrom. 2006;17:1271–1274. doi: 10.1016/j.jasms.2006.05.017. [DOI] [PubMed] [Google Scholar]
- (67).Shaffer SA, Sadilek M, Turecek F. J. Org. Chem. 1996;61:5234–5245. [Google Scholar]
- (68).Laskin J, Yang ZB, Lam C, Chu IK. Anal. Chem. 2007;79:6607–6614. doi: 10.1021/ac070777b. [DOI] [PubMed] [Google Scholar]
- (69).Yu W, Vath JE, Huberty MC, Martin SA. Anal. Chem. 1993;65:3015–3023. doi: 10.1021/ac00069a014. [DOI] [PubMed] [Google Scholar]; Qin J, Chait BT. J. Am. Chem. Soc. 1995;117:5411–5412. [Google Scholar]; Lee SW, Kim HS, Beauchamp JL. J. Am. Chem. Soc. 1998;120:3188–3195. [Google Scholar]
- (70).Sarver A, Scheffler NK, Shetlar MD, Gibson BW. J. Am. Soc. Mass Spectrom. 2001;12:439–448. doi: 10.1016/S1044-0305(01)00213-6. [DOI] [PubMed] [Google Scholar]; Lee SJ, Lee JR, Kim YH, Park YS, Park SI, Park HS, Kim KP. Rapid Commun. Mass Spectrom. 2007;21:2797–2804. doi: 10.1002/rcm.3145. [DOI] [PubMed] [Google Scholar]; Salzano AM, D'Ambrosio C, Scaloni A. Nitric Oxide, Part F: Oxidative and Nitrosative Stress in Redox Regulation of Cell Signaling. Vol. 440. Elsevier Academic Press Inc; San Diego: 2008. pp. 3–15. [Google Scholar]
- (71).Frankevich VE, Zhang J, Friess SD, Dashtiev M, Zenobi R. Anal. Chem. 2003;75:6063–6067. doi: 10.1021/ac034436j. [DOI] [PubMed] [Google Scholar]
- (72).Karas M, Gluckmann M, Schafer J. J. Mass Spectrom. 2000;35:1–12. doi: 10.1002/(SICI)1096-9888(200001)35:1<1::AID-JMS904>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]; Knochenmuss R, Zenobi R. Chem. Rev. 2003;103:441–452. doi: 10.1021/cr0103773. [DOI] [PubMed] [Google Scholar]; Knochenmuss R. Analyst. 2006;131:966–986. doi: 10.1039/b605646f. [DOI] [PubMed] [Google Scholar]
- (73).Hunter EPL, Lias SG. J. Phys. Chem. Ref. Data. 1998;27:413–656. [Google Scholar]
- (74).Freiser BS, Woodin RL, Beauchamp JL. J. Am. Chem. Soc. 1975;97:6893–6894. [Google Scholar]
- (75).Frazier JR, Christophorou LG, Carter JG, Schweinler HC. J. Chem. Phys. 1978;69:3807–3818. [Google Scholar]
- (76).The energetics and MOs of the excited states for the rest of species are available in Supporting Information.
- (77).Hodyss R, Cox HA, Beauchamp JL. J. Am. Chem. Soc. 2005;127:12436–12437. doi: 10.1021/ja052042z. [DOI] [PubMed] [Google Scholar]
- (78).Addario V, Guo YZ, Chu IK, Ling Y, Ruggerio G, Rodriquez CF, Hopkinson AC, Siu KWM. Int. J. Mass Spectrom. 2002;219:101–114. [Google Scholar]
- (79).Sohn CH, Loo JA, Beauchamp JL. unpublished.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.