Abstract
Reactive oxygen species can induce the formation of tandem DNA lesions. We recently showed that the treatment of calf thymus DNA with Cu2+/H2O2/ascorbate could result in the efficient formation of a tandem lesion where a 5,6-dihydroxy-5,6-dihydrothymidine (or thymidine glycol) is situated on the 5′ side of an 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG). In the present study, we assessed how the 5′-Tg-(8-oxodG)-3′ and 5′-(8-oxodG)-Tg-3′ tandem lesions are replicated by purified DNA polymerases and how they are recognized by base excision repair enzymes. Our results revealed that the tandem lesions blocked primer extension mediated by Klenow fragment and yeast polymerase η more readily than when the Tg or 8-oxodG was present alone. The mutagenic properties of Tg or 8-oxodG differed while they were present alone or in tandem. Moreover, the human 8-oxoguanine-DNA glycosylase (hOGG1)-mediated cleavage of 8-oxodG was compromised considerably by the presence of a neighboring 5′ Tg, whereas the presence of Tg as the adjacent 3′ nucleoside enhanced the 8-oxodG cleavage by hOGG1. The efficiency for the cleavage of Tg by endonuclease III was not affected by the presence of an adjoining 8-oxodG. These results supported the notion that the replication and repair of tandem single-nucleobase lesions depend on the types of lesions involved and their spatial arrangement.
Introduction
DNA is susceptible to damage by reactive oxygen species (ROS)1, which can be induced in human cells via normal aerobic metabolism and by exogenous processes including ionizing radiation and UV light, and the accumulation of ROS-induced DNA lesions is thought to be implicated in various human diseases (1). Other than single-nucleobase lesions, clustered DNA lesions, where two or more damaged nucleosides are located within 1-2 helical turns of DNA, can form upon interaction with ROS, particularly those formed upon exposure to ionizing radiation (2-4).
Clustered DNA lesions are also termed multiply damaged site (MDS). Owing to the intrinsic chemical and structural properties of different lesions within MDS and their close proximity, lesions at MDS are often more difficult to repair than when they are present alone (5-12). Among the clustered DNA lesions, tandem lesions, consisting of two neighboring damaged nucleotides on the same DNA strand, could be initiated from a single hydroxyl radical attack and, depending on the nature of the tandem lesions, molecular oxygen may or may not be involved in their formation (13-23).
Thymidine glycol (or 5,6-dihydroxy-5,6-dihydrothymidine, Tg) and 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) are major oxidatively induced lesions of thymidine and 2′-deoxyguanosine, respectively. We demonstrated recently that the tandem lesion with a Tg lying on the 5′ side of an 8-oxodG could form efficiently in calf thymus DNA upon treatment with Cu2+/H2O2/ascorbate (20).
Some lesions, when present in replicating DNA, can lead to replication fork stalling and/or give rise to mutations. Thymidine glycol blocks effectively DNA replication, but is not mutagenic under most conditions (24); on the other hand, 8-oxodG does not block appreciably the DNA replication and it can result in significant frequencies of G→T transversion mutation (25, 26). Tg could also arise from the deamination of 5-methylcytosine glycol (27), an oxidative lesion formed on 5-methylcytosine (28). In this respect, cytosine at a CpG dinucleotide site can be methylated, and approximately 5% cytosine residues are methylated in the human genome (29). The methylated CpGs are mutational hot spots in the human p53 tumor suppressor gene (30). Previously we found that oxidative intrastrand cross-link lesions could form at methylated CpG sites, which may account for the mCG→TT tandem double mutations induced by Fenton type reagents (31). The 5′-Tg-(8-oxodG)-3′ tandem lesion may also emanate from ROS attack at methylated CpG site thereby contributing to CpG mutagenesis. Along this line, a relatively high frequency of mCG→TT mutation was observed after the Cu2+/H2O2/ascorbate-treated pSP189 shuttle vector was propagated in nucleotide excision repair (NER)-deficient human XPA cells (32).
Cells have evolved with various strategies to minimize the deleterious effects of DNA lesions by an intricate DNA repair system and certain mechanisms to cope with unrepaired or highly repair-resistant DNA lesions (33). It was proposed that when a high-fidelity replication fork is arrested by DNA damage, translesion synthesis DNA polymerases can replace temporarily the replicative polymerases to bypass the damage site (34). Several studies have been carried out to assess the cytotoxic and mutagenic properties of clustered DNA lesions. Our previous in-vitro replication studies on two intrastrand cross-link lesions, G[8-5]C and G[8-5m]T, showed that they can either stall DNA replication performed by high-fidelity replicative polymerases or give rise to mutations by a translesion synthesis polymerase, yeast polymerase η (23, 35, 36). In these two intrastrand cross-link lesions, the C8 of guanine is covalently bonded with the C5 or methyl carbons of cytosine and thymine, respectively. In addition, altered mutagenic potential was found for 8-oxodG when it is present in a clustered DNA damage site (12, 37, 38).
The base excision repair (BER) pathway can allow for the efficient and accurate repair of ROS-induced single-nucleobase lesions (39). However, when these lesions are present as components of a MDS, the repair by the BER enzymes becomes difficult. A number of studies showed that the excision, by purified BER enzymes or by cell extracts, of clustered DNA lesions is indeed compromised, and the effects vary with the types of lesions involved and their spatial distribution (6-9, 12, 40).
Building upon our previous demonstration of the efficient formation of the 5′-Tg-(8-oxodG)-3′ tandem lesion in calf thymus DNA upon exposure to Cu2+/H2O2/ascorbate (20) and our successful synthesis of oligodeoxyribonucleotides (ODNs) containing both Tg and 8-oxodG (41), here we examined how the presence of the 5′-Tg-(8-oxodG)-3′ and 5′-(8-oxodG)-Tg-3′ tandem lesions in template DNA perturbs nucleotide incorporation by two DNA polymerases. One is a replicative DNA polymerase, the exonuclease-free Klenow fragment of E. coli DNA polymerase I, and the other is a member of the “Y” superfamily polymerases, Sacchramyces cerevisiae DNA polymerase η (pol η). Pol η is the gene product of Rad30 in budding yeast (42) and the variant form of xeroderma pigmentosum (XP-V) in humans (43), which was revealed to bypass efficiently many DNA lesions, including 8-oxodG and Tg (44, 45). We also assessed how these two types of tandem lesions are recognized by human 8-oxoguanine-DNA glycosylase (hOGG1) and E. coli endonuclease III.
Experimental Procedures
Materials
All unmodified ODNs used in this study were purchased from Integrated DNA Technologies (Coralville, IA). [γ-32P]ATP was obtained from Amersham Biosciences (Piscataway, NJ). The Klenow fragment (3′→5′ exo-) of E. coli DNA polymerase I and endonuclease III were from New England Biolabs (Ipswich, MA). One unit of Endonuclease III is defined as the amount of enzyme required to cleave 1 pmol of a 34-mer ODN duplex containing a single abasic site in a total reaction volume of 10 μl in 1 hour at 37°C in the Endonuclease III reaction buffer containing 10 pmol of fluorescently labeled ODN duplex. Human AP endonuclease 1 (APE1) was purchased from Enzymax (Lexington, KY). Yeast pol η and hOGG1 were expressed and purified following previously published procedures (46, 47).
Preparation of Substrates for In-vitro Replication and Repair Studies
The ODNs containing the cis-(5R,6S) diastereomer of Tg, an 8-oxodG, or both were synthesized previously (sequences shown in Table 1) (41). The dodecameric lesion-bearing substrate, e.g., d(ATGGCTgG*GCTAT) (“G*” represents 8-oxodG), was ligated with the 5′-phosphorylated d(GATCCTAG) in the presence of a template ODN, d(CCGCTCCCTAGGATCATAGCCAGCCAT), following previously described procedures (35). The desired lesion-containing 20-mer ODN was purified by using 20% denaturing polyacrylamide gel electrophoresis (PAGE) and desalted by ethanol precipitation. The purity of the product was further confirmed by PAGE analysis.
Table 1.
The sequences of ODNs used for enzymatic ligation (“G*” represents an 8-oxodG)
| ODNs | Sequences |
|---|---|
| 1, 5′-Tg-(8-oxodG)-3′ | 5′-ATG GCTg G*GC TAT-3′ |
| 2, 5′-(8-oxodG)-Tg-3′ | 5′-ATG GCG* TgGC TAT-3′ |
| 3, 5′-(8-oxodG)-dT-3′ | 5′-ATG GCG* TGC TAT-3′ |
| 4, 5′-Tg-dG-3′ | 5′-ATG GCG TgGC TAT-3′ |
| 5, control | 5′-ATG GCG TGC TAT-3′ |
Primer Extension Assays
The 20-mer lesion-containing ODNs or the unmodified template (20 nM) were annealed with a 5′ 32P-labeled 14- or 15-mer primer (10 nM). To the duplex mixture were added all four dNTPs at a concentration of 200 μM each and a DNA polymerase. The reaction was carried out in a buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol (DTT) at 37 °C for 60 min. The amounts of the polymerases are indicated in the figures. The reaction was terminated by adding a 2 volume excess of formamide gel-loading buffer [80% formamide, 10 mM EDTA (pH 8.0), 1 mg/mL xylene cyanol, and 1 mg/mL bromophenol blue]. The products were resolved on 20% (29:1) cross-linked polyacrylamide gels containing 8 M urea. Gel band intensities for the substrates and products were quantified by using a Typhoon 9410 variable-mode imager (Amersham Biosciences Co.).
Steady-state Kinetic Measurements
The steady-state kinetic analyses were performed as described previously (48). In this measurement, the primer-template complex (10 nM) was incubated with either Klenow fragment (5 ng) or yeast pol η (5 ng) in the presence of an individual dNTP at various concentrations as indicated in the figures. The reaction was carried out at room temperature with the same reaction buffer as described for the primer extension assays. The dNTP concentration was optimized for different insertion reactions to allow for less than 20% primer extension. The observed rate of nucleotide incorporation (Vobs) was plotted as a function of dNTP concentration, and the apparent Km and Vmax steady-state kinetic parameters for the incorporation of both the correct and incorrect nucleotides were determined by fitting the rate data with the Michaelis-Menten equation:
The kcat values were then calculated by dividing the Vmax values with the concentration of the polymerase used. The efficiency of nucleotide incorporation was determined by the ratio of kcat/Km, and the fidelity of nucleotide incorporation was calculated by the frequency of misincorporation (finc) with the following equation:
BER Assays
The 20mer 5′ 32P-labeled lesion-containing ODNs or a control substrate were annealed with their respective complementary strands by heating the mixture to 90 °C and cooling slowly to room temperature in a solution containing 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 1 mM EDTA. The duplex (10 nM) was incubated with either hOGG1 or endonuclease III in a 10-μL buffer solution at 37 °C for 30 min. The amounts of enzyme are shown in the figures. A buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2 and 1 mM DTT, along with 1 ng of APE1, was used for the hOGG1 cleavage assays, and a buffer bearing 20 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM DTT was employed for endonuclease III reactions. The reaction products were mixed with formamide gel-loading buffer, heated at 90 °C for 20 min to cleave the apurinic/apyrimidinic sites (AP sites), and the resulting products were resolved by 20% denaturing polyacrylamide gels. The level of the BER enzyme-induced cleavage was quantified based on the gel band intensities for the substrates and products by phosphorimaging analysis.
Results
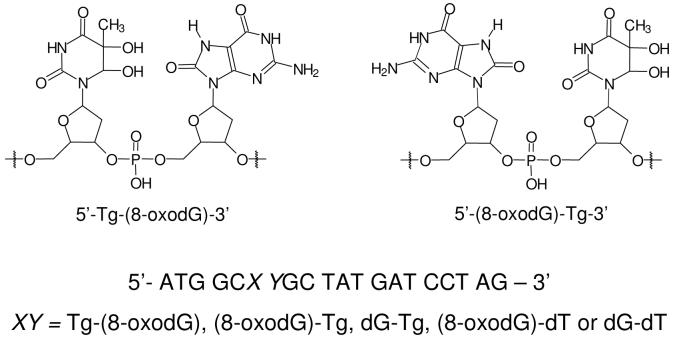
To assess how the tandem 5′-Tg-(8-oxodG)-3′ and 5′-(8-oxodG)-Tg-3′ lesions perturb DNA replication and how they are recognized by BER enzymes, we first constructed 12-mer ODN substrates carrying the cis-(5R,6S) diastereomer of Tg, an 8-oxodG, or both (Table 1) (41). These lesion-containing ODNs were further ligated with a 5′-phosphorylated 8-mer ODN to afford 20-mer lesion-containing substrates for in-vitro replication and repair studies (Figure 1).
Figure 1.
The structures of the 5′-Tg-(8-oxodG)-3′ and 5′-(8-oxodG)-Tg-3′ tandem lesions, and the sequences of the 20-mer lesion-containing substrates used in the present in-vitro replication and repair studies.
Increased Blocking Effects Induced by Tandem Lesions during DNA Replication in vitro
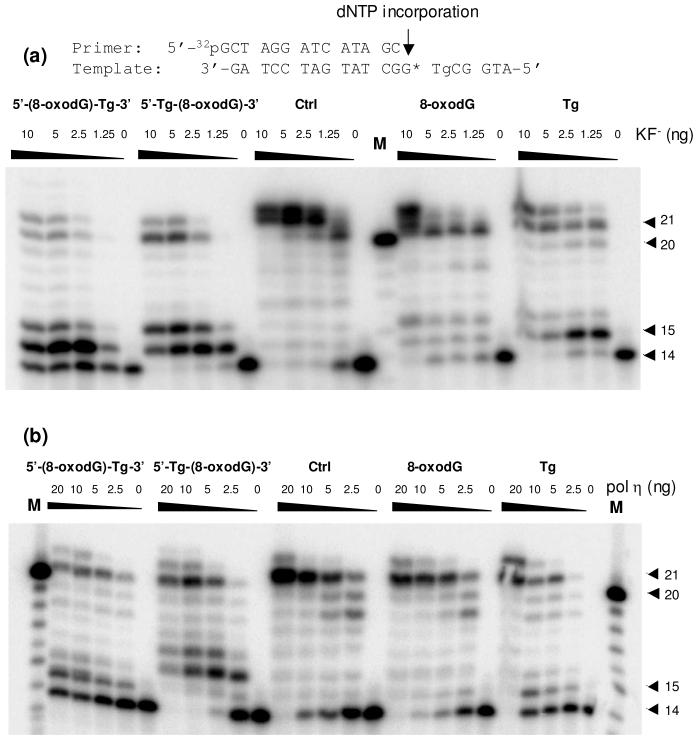
First, we performed primer extension assays on the four lesion-bearing substrates and an undamaged control substrate with Klenow fragment and yeast pol η. The results with the Klenow fragment showed that, in the presence of all four dNTPs, the synthesis catalyzed by the Klenow fragment stopped mostly after incorporating one or two nucleotides opposite the tandem lesions (Figure 2a). In addition, the Tg moiety of the tandem lesions could lead to a greater blocking effect than 8-oxodG on the Klenow fragment-mediated bypass of the tandem lesions. For instance, when the polymerase encounters the Tg first, i.e, for the 5′-(8-oxodG)-Tg-3′ tandem lesion, some of the primers remain unextended; only a trace amount of full-length product was detected (Figure 2a). This observation is in keeping with the fact that Tg is a replication-blocking lesion (24), whereas 8-oxodG does not block appreciably the DNA replication (25, 26). Together, the two tandem lesions block the Klenow fragment-mediated primer extension more readily than the two composing single-nucleobase lesions when present alone.
Figure 2.
Primer extension assays for nucleotide incorporation opposite tandem lesions, i.e., 5′-(8-oxodG)-Tg-3′ and 5′-Tg-(8-oxodG)-3′, a single 8-oxodG or Tg, and the undamaged control, with exo- Klenow fragment (a) and yeast pol η (b). 5′-[32P]-labeled d(GCTAGGATCATAGC) was used as the primer. Klenow fragment or yeast pol η at the indicated units/concentrations was incubated with 10 nM substrate and 200 μM dNTPs at 37 °C for 60 min. The products were subsequently resolved by using 20% denaturing polyacrylamide gels. The 21-mer was observed due to the presence of a 1-base overhang in the primer, and the 22-mer primer extension products were originated from the terminal transferase activity of the polymerase.
The primer extension assay with yeast pol η showed that this polymerase could bypass both tandem and isolated single-nucleobase lesions, and generate full-length replication products in the presence of all four dNTPs (Figure 2b). However, similar to what we found for the Klenow fragment, both tandem lesions were somewhat more difficult for yeast pol η to bypass than either single-nucleobase lesion.
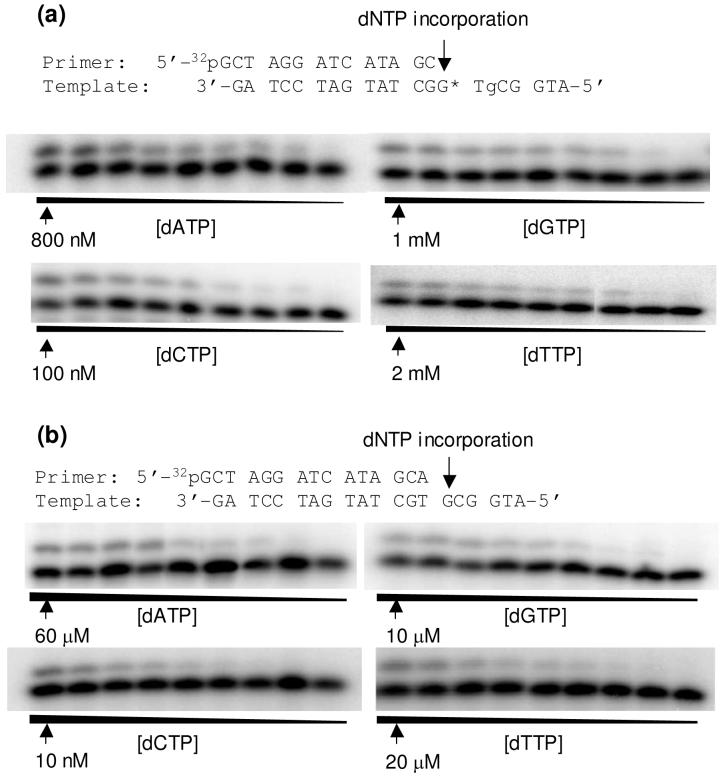
We next determined the steady-state kinetic parameters for nucleotide incorporation opposite both moieties of the tandem lesions, opposite an isolated 8-oxodG or Tg, or across an unmodified dG or dT by Klenow fragment and yeast pol η (Figure 3, and the steady-state kinetic parameters for nucleotide incorporation are summarized in Tables 2-4). It turned out that the Klenow fragment incorporated preferentially the correct nucleotide opposite the 3′ modified nucleoside in both tandem lesions, namely, dAMP and dCMP were the most favored nucleotides inserted opposite the Tg in the 5′-(8-oxodG)-Tg-3′ tandem lesion and the 8-oxodG in the 5′-Tg-(8-oxodG)-3′ tandem lesion, respectively (Tables 2 & 4). The presence of a 5′ neighboring 8-oxodG does not affect the efficiency of dAMP incorporation opposite Tg (Tables 2 & 4). Likewise, an adjacent 5′ Tg does not confer compromised efficiency in nucleotide incorporation opposite 8-oxodG (Tables 2 & 4). The nucleotide insertion opposite the 5′ component of the tandem lesion by Klenow fragment, however, became much more difficult; only little incorporation of dAMP opposite the Tg was observed when it lies on the 5′ side of the 8-oxodG.
Figure 3.
Example gel images for the steady-state kinetic measurements for the nucleotide incorporation opposite the 8-oxodG portion of the 5′-Tg-(8-oxodG)-3′ tandem lesion (a) or the corresponding dG site for the undamaged substrate. (b). Klenow fragment (5 ng) was incubated with 10 nM DNA substrate at room temperature for 10 min. The highest dNTP concentration is shown in the figure, and the ratio of dNTP concentrations between adjacent lanes was 0.5-0.6.
Table 2.
Steady-state kinetic parameters for Exo- Klenow fragment-mediated nucleotide incorporation opposite Tg and 8-oxodG in tandem lesion-containing substrates and opposite undamaged dG or dT in the control substratea
| Substrates | dNTP | kcat (min-1) | km (nM) | kcat / Km (nM-1 min-1) | finc |
|---|---|---|---|---|---|
| 5′-Tg-(8-oxodG)-3′ | 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | ||||
| dATP | 0.10 ± 0.01 | (2.9 ± 0.6) × 102 | 3.4 × 10-4 | 5.2 × 10-2 | |
| dGTP | 0.057 ± 0.003 | (1.4 ± 0.2) × 105 | 3.8 × 10-7 | 5.8 × 10-5 | |
| dCTP | 0.073 ± 0.008 | 11 ± 1 | 6.6 × 10-3 | 1.0 | |
| dTTP | 0.090 ± 0.008 | (1.5 ± 0.2) × 106 | 6.0 × 10-8 | 9.1 × 10-6 | |
| 15 mer Primer: 5′ - GCTAGGATCATAGCC - 3′ | |||||
| dATP | 0.098 ± 0.001 | (3.1 ± 0.3) × 104 | 3.2 × 10-6 | 1.0 | |
| 5′-(8-oxodG)-Tg-3′ | 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | ||||
| dATP | 0.21 ± 0.01 | 16 ± 0.75 | 1.3 × 10-3 | 1.0 | |
| dGTP | 0.065 ± 0.001 | (2.4 ± 0.1) × 104 | 2.7 × 10-6 | 2.1 × 10-3 | |
| dCTP | 0.049 ± 0.02 | (2.7 ± 1.2) × 105 | 1.8 × 10-7 | 1.4 × 10-4 | |
| dTTP | 0.030 ± 0.0003 | (2.2 ± 0.1) × 105 | 1.4 × 10-7 | 1.1 × 10-4 | |
| Undamaged 5′-dG-dT-3′ | 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | ||||
| dATP | 0.060 ± 0.001 | 0.71 ± 0.02 | 0.085 | 1.0 | |
| dGTP | 0.079 ± 0.005 | (6.0 ± 1.1) × 102 | 1.3 × 10-4 | 1.5 × 10-3 | |
| dCTP | 0.076 ± 0.003 | (4.0 ± 0.4) × 104 | 1.9 × 10-6 | 2.2 × 10-5 | |
| dTTP | 0.063 ± 0.001 | (1.1 ± 0.1) × 105 | 5.7 × 10-6 | 6.7 × 10-5 | |
| 15 mer Primer: 5′ - GCTAGGATCATAGCA - 3′ | |||||
| dATP | 0.057 ± 0.001 | (2.2 ± 0.2) × 104 | 2.6 × 10-6 | 2.8 × 10-4 | |
| dGTP | 0.076 ± 0.003 | (4.6 ± 0.4) × 103 | 1.7 × 10-5 | 1.8 × 10-3 | |
| dCTP | 0.079 ± 0.008 | 8.5 ± 3.4 | 9.3 × 10-3 | 1.0 | |
| dTTP | 0.11 ± 0.01 | (2.1 ± 0.3) × 104 | 5.2 × 10-6 | 5.6 × 10-4 | |
kcat and Km are average values based on three independent measurements
Table 4.
Steady-state kinetic parameters for nucleotide incorporation by Exo- Klenow fragment and yeast polymerase η on 8-oxodG- and Tg-containing substratesa
| Substrates | dNTP | kcat (min-1) | km (nM) | kcat / Km (nM-1 min-1) | finc |
|---|---|---|---|---|---|
| 5′-(8-oxodG)-dT-3′ | By Exo-Klenow fragment with 15 mer Primer: 5′ - GCTAGGATCATAGCA - 3′ | ||||
| dATP | 0.082 ± 0.005 | (9.7 ± 0.2) × 102 | 8.5 × 10-5 | 5.7 × 10-2 | |
| dGTP | 0.082 ± 0.001 | (3.7 ± 0.2) × 104 | 2.2 × 10-7 | 1.5 × 10-4 | |
| dCTP | 0.038 ± 0.003 | 26 ± 7 | 1.5 × 10-3 | 1.0 | |
| dTTP | 0.016 ± 0.001 | (4.8 ± 0.7) × 105 | 3.3 × 10-8 | 2.2 × 10-5 | |
| By yeast polymerase η with 15 mer Primer: 5′ - GCTAGGATCATAGCA - 3′ | |||||
| dATP | 0.13 ± 0.01 | (6.9 ± 1.3) × 102 | 1.9 × 10-4 | 5.1 × 10-2 | |
| dGTP | 0.065 ± 0.010 | (4.0 ± 0.9) × 103 | 1.6 × 10-5 | 4.3 × 10-3 | |
| dCTP | 0.063 ± 0.005 | 17 ± 4 | 3.7 × 10-3 | 1.0 | |
| dTTP | 0.041 ± 0.005 | (1.3 ± 0.04) × 103 | 3.2 × 10-5 | 8.6 × 10-3 | |
| 5′-dG-Tg-3′ | By Exo- Klenow fragment with 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | ||||
| dATP | 0.057 ± 0.005 | 36 ± 3 | 1.6 × 10-3 | 1.0 | |
| dGTP | 0.087 ± 0.014 | (2.1 ± 0.4) × 104 | 4.1 × 10-6 | 2.6 × 10-3 | |
| dCTP | 0.052 ± 0.005 | (1.1 ± 0.2) × 105 | 4.7 × 10-7 | 2.9 × 10-4 | |
| dTTP | 0.030 ± 0.003 | (2.0 ± 0.2) × 105 | 1.5 × 10-6 | 9.4 × 10-4 | |
| By yeast polymerase η with 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | |||||
| dATP | 0.053 ± 0.001 | 71.1 ± 0.2 | 7.5 × 10-4 | 1.0 | |
| dGTP | 0.070 ± 0.005 | (6.0 ± 1.0) × 103 | 1.2 × 10-6 | 1.6 × 10-3 | |
| dCTP | 0.067 ± 0.002 | (4.0 ± 0.4) × 105 | 1.7 × 10-7 | 2.3 × 10-4 | |
| dTTP | 0.058 ± 0.001 | (1.1 ± 0.1) × 105 | 5.3 × 10-7 | 7.1 × 10-4 | |
kcat and Km are average values based on three independent measurements
Unlike the nucleotide incorporation with the Klenow fragment, yeast pol η could insert nucleotides opposite both moieties of the tandem lesions. The efficiencies for the incorporation of the most favorable nucleotide, dCMP, opposite the 8-oxodG that is isolated, in 5′-Tg-(8-oxodG)-3′, or in 5′-(8-oxodG)-Tg-3′ are 3.7 × 10-3, 2.9 × 10-3 and 1.1 × 10-4 nM-1min-1, respectively (Tables 3 & 4). Thus, the presence of Tg as the 5′ neighboring nucleotide only resulted in marginal decrease (i.e., 22%) in efficiency of nucleotide incorporation; the existence of Tg as the 3′ adjoining nucleotide, however, led to a pronounced drop in efficiency for dCMP insertion, i.e., by ∼34 fold. This large drop is mainly due to the increase in Km, i.e., by ∼24 fold, for the 5′-(8-oxodG)-Tg-3′ lesion relative to the isolated 8-oxodG lesion (Tables 3 & 4). The latter compromised efficiency for dCMP incorporation reflects the difficulty experienced by the polymerase in extending the Tg:A base pair at the primer-template junction.
Table 3.
Steady-state kinetic parameters for nucleotide incorporation mediated by yeast pol η opposite Tg and 8-oxodG in tandem lesion-containing substrates and opposite undamaged dG and dT in the control substratea
| Substrates | dNTP | kcat (min-1) | km (nM) | kcat / Km (nM-1 min-1) | finc |
|---|---|---|---|---|---|
| 5′-Tg-(8-oxodG)-3′ | 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | ||||
| dATP | 0.29 ± 0.02 | (2.3 ± 0.3) × 103 | 1.3 × 10-4 | 4.5 × 10-2 | |
| dGTP | 0.060 ± 0.005 | (4.9 ± 0.9) × 103 | 1.2 × 10-5 | 4.1 × 10-3 | |
| dCTP | 0.099 ± 0.029 | 34 ± 16 | 2.9 × 10-3 | 1.0 | |
| dTTP | 0.060 ± 0.005 | (5.8 ± 0.7) × 103 | 1.0 × 10-5 | 3.4 × 10-3 | |
| 15 mer Primer: 5′ - GCTAGGATCATAGCC - 3′ | |||||
| dATP | 0.063 ± 0.001 | (2.9 ± 0.7) × 102 | 2.2 × 10-4 | 1.0 | |
| dGTP | 0.16 ± 0.01 | (6.3 ± 0.4) × 104 | 2.5 × 10-6 | 1.1 × 10-2 | |
| dCTP | 0.11 ± 0.002 | (6.3 ± 0.5) × 105 | 1.7 × 10-7 | 7.7 × 10-4 | |
| dTTP | 0.13 ± 0.01 | (2.7 ± 0.7) × 105 | 4.8 × 10-6 | 2.2 × 10-3 | |
| 5′-(8-oxodG)-Tg-3′ | 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | ||||
| dATP | 0.12 ± 0.002 | (3.3 ± 0.1) × 102 | 3.6 × 10-4 | 1.0 | |
| dGTP | 0.048 ± 0.002 | (1.4 ± 0.1) × 104 | 3.4 × 10-6 | 9.4 × 10-3 | |
| dCTP | 0.063 ± 0.005 | (2.5 ± 0.4) × 103 | 2.5 × 10-5 | 6.9 × 10-2 | |
| dTTP | 0.080 ± 0.002 | (5.3 ± 0.5) × 104 | 1.5 × 10-6 | 4.2 × 10-3 | |
| 15 mer Primer: 5′ - GCTAGGATCATAGCA - 3′ | |||||
| dATP | 0.084 ± 0.002 | (1.1 ± 0.1) × 105 | 7.6 × 10-7 | 6.9 × 10-3 | |
| dGTP | 0.15 ± 0.01 | (9.6 ± 0.5) × 105 | 1.6 × 10-7 | 1.5 × 10-3 | |
| dCTP | 0.046 ± 0.005 | (4.1 ± 0.6) × 102 | 1.1 × 10-4 | 1.0 | |
| dTTP | 0.17 ± 0.02 | (6.4 ± 0.5) × 105 | 2.7 × 10-7 | 2.5 × 10-3 | |
| 5′-dG-dT-3′ | 14 mer Primer: 5′ - GCTAGGATCATAGC - 3′ | ||||
| dATP | 0.055 ± 0.005 | 36 ± 1 | 1.5 × 10-3 | 1.0 | |
| dGTP | 0.080 ± 0.005 | (5.9 ± 0.5) × 104 | 1.4 × 10-6 | 9.3 × 10-4 | |
| dCTP | 0.077 ± 0.010 | (4.1 ± 0.1) × 103 | 1.9 × 10-5 | 1.3 × 10-2 | |
| dTTP | 0.13 ± 0.003 | (1.1 ± 0.9) × 105 | 1.2 × 10-6 | 8.0 × 10-4 | |
| 15 mer Primer: 5′ - GCTAGGATCATAGCA - 3′ | |||||
| dATP | 0.23 ± 0.02 | (5.2 ± 0.3) × 105 | 4.4 × 10-7 | 4.6 × 10-5 | |
| dGTP | 0.38 ± 0.01 | (5.8 ± 0.07) × 105 | 6.7 × 10-7 | 7.1 × 10-5 | |
| dCTP | 0.087 ± 0.010 | 9.2 ± 2.5 | 9.5 × 10-3 | 1.0 | |
| dTTP | 0.087 ± 0.010 | (1.8 ± 0.3) × 104 | 4.8 × 10-6 | 5.1 × 10-4 | |
kcat and Km are average values based on three independent measurements
The efficiencies for the insertion of the most favorable nucleotide, i.e., dAMP, opposite the Tg that is alone, in 5′-Tg-(8-oxodG)-3′, or in 5′-(8-oxodG)-Tg-3′ were 7.5 × 10-4, 2.2 × 10-4, and 3.6 × 10-4 nM-1min-1, respectively (Tables 3 & 4). The 5′ and 3′ adjacent 8-oxodG, therefore, led to the decreases in nucleotide incorporation efficiency by ∼2.1 and ∼3.4 fold, respectively, and the decrease arises again from the increase in Km (Tables 3 & 4). This result revealed that pol η encounters greater difficulty in extending the 8-oxoG:C base pair than a G:C base pair. The extent of decrease, however, is much less drastic than what we found for the 8-oxodG in the tandem lesions where Tg is the neighboring lesion (vide supra).
The Presence of Single-nucleobase Lesions in Tandem Affects their Mutagenic Potential
The steady-state kinetic parameters for yeast pol η-mediated nucleotide incorporation also revealed some notable differences in mutagenicity for the two single-nucleobase lesions while they are present alone or in tandem. In addition, the fidelity for nucleotide insertion is different for the tandem lesions with the Tg and 8-oxodG being in the opposite orientation. In this regard, the frequency of misincorporation of dAMP opposite 8-oxodG in the 5′-Tg-(8-oxodG)-3′ (4.5%) was similar as that opposite an isolated 8-oxodG (5.1%). However, the frequency for the misinsertion of dAMP opposite the 8-oxodG in 5′-(8-oxodG)-Tg-3′ was only 0.69% (Tables 3 & 4). The Tg component, however, exhibits greater mutagenic potential for both orientations of the tandem lesions than when it is present alone; the misinsertion of dGMP occurred at frequencies of 1.1% and 0.16% for the 5′-Tg-(8-oxodG)-3′ and isolated Tg, respectively (Tables 3 & 4). By contrast, dCMP was inserted opposite the Tg moiety of the 5′-(8-oxodG)-Tg-3′ tandem lesion at a relatively high frequency, i.e., 6.7% (Tables 3 & 4).
Very limited differences were found for the fidelity of Klenow fragment-mediated nucleotide incorporation opposite the Tg and 8-oxodG while they are isolated or neighboring to each other. The frequencies for the misincorporation of dAMP opposite 8-oxodG by Klenow fragment were similar, namely, 5.7% and 5.2% for isolated 8-oxodG and 5′-Tg-(8-oxodG)-3′ tandem lesion, respectively (Tables 2 & 4). In addition, the frequencies for the misinsertion of dGMP opposite the Tg were comparable, i.e., 0.21% and 0.26% for substrates containing 5′-(8-oxodG)-Tg-3′ tandem lesion and isolated Tg, respectively (Tables 2 & 4).
The Recognition of Tandem Lesions by BER Enzymes
We next investigated how efficiently the two tandem lesions can be recognized by two BER enzymes, i.e., hOGG1 and E. coli endonuclease III. Although hOGG1 is a bifunctional glycosylase harboring both glycosylase and AP lyase activities (49), previous kinetic studies showed that the hOGG1-mediated strand cleavage at 8-oxodG site is not very efficient, and the kcat values indicated that the purified protein may take more than 20 min to perform a single repair event in vitro (50). Human AP endonuclease 1 (APE1), however, can stimulate the DNA glycosylase activity of hOGG1 by cleaving the AP site produced by the latter (51). Thus, we employed APE1 to induce cleavage at the hOGG1-produced AP sites.
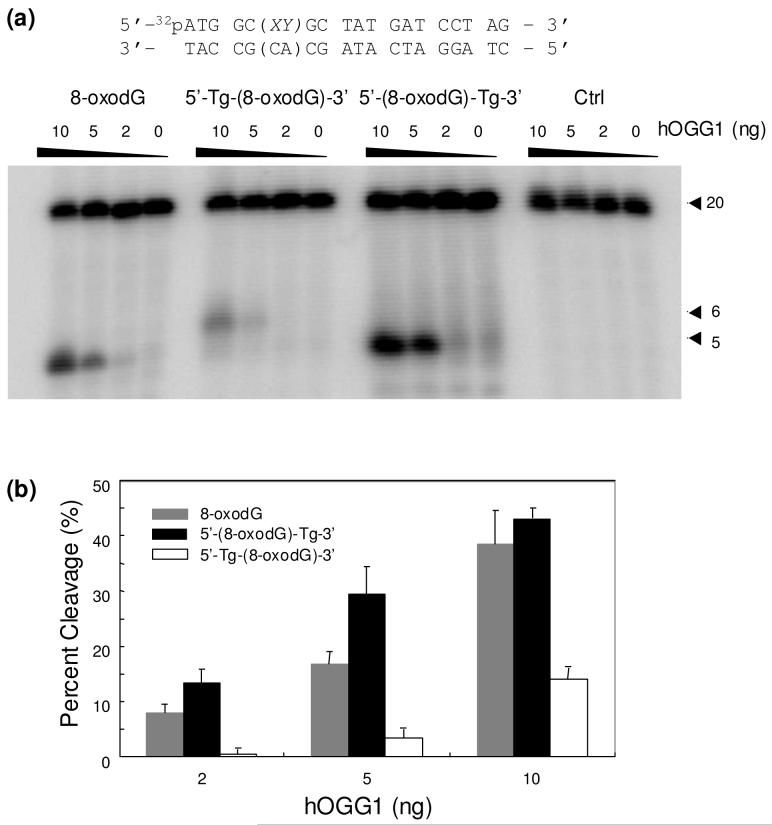
It turned out that the 8-oxodG in the two tandem lesions could be cleaved by hOGG1 (Figure 4); the efficiencies for the cleavage of 8-oxodG in the two tandem lesions are, however, considerably different from each other and from the hOGG1-mediated cleavage of an isolated 8-oxodG. In this respect, hOGG1 could remove 8-oxoguanine from the 5′-(8-oxodG)-Tg-3′ tandem lesion-bearing substrate more efficiently than from the substrate housing an isolated 8-oxodG (Figure 4). By contrast, the cleavage of 8-oxoguanine from the 5′-Tg-(8-oxodG)-3′ tandem lesion-carrying substrate is much less efficient than that from the substrate containing 8-oxodG alone; the cleavage of the former substrate was almost completely abolished at the lowest level of the enzyme used (Figure 4b). This result revealed that the presence of a vicinal Tg can perturb significantly the hOGG1-mediated cleavage of 8-oxodG, and this perturbation is dependent on the spatial arrangement of the two single-nucleobase lesions.
Figure 4.
(a) PAGE analysis of the hOGG1-mediated cleavage products of the substrates containing an 8-oxodG, the two tandem lesions, and unmodified GT. “XY” represents different lesions, and the 20 mer 5′-(CA)-3′- and 5′-(AC)-3′-containing complementary strands were used for the repair studies of the 5′-Tg-(8-oxodG)-3′- and 5′-(8-oxodG)-Tg-3′-containing substrates, respectively. (b) A summary of the quantification results of the percent cleavage products for different substrates. The values represent the mean ± standard deviation from three independent treatments and quantification experiments.
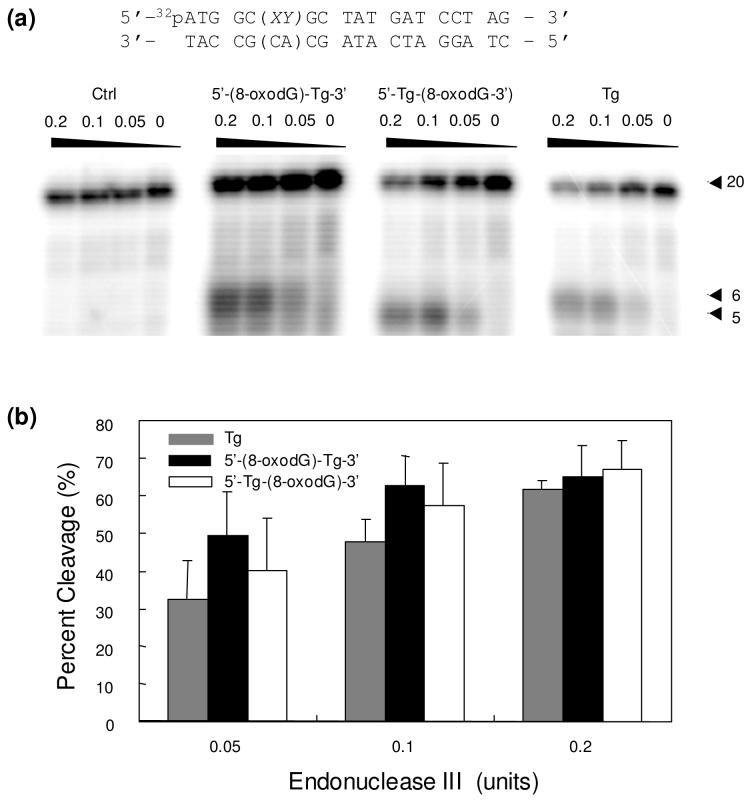
We next examined whether the presence of an adjoining 8-oxodG can affect the endonuclease III-mediated cleavage of Tg. Since endonuclease III has both glycosylase activity and a relatively robust AP lyase activity (52), no AP endonuclease was added for the endonuclease III-mediated cleavage reactions. In contrast to what we observed for hOGG1, the Tg in both tandem lesions could be cleaved by endonuclease III at comparable efficiencies as an isolated Tg (Figure 5).
Figure 5.
(a) PAGE analysis of the products arising from the endonuclease III-mediated cleavage of substrates housing Tg, 5′-Tg-(8-oxodG)-3′, 5′-(8-oxodG)-Tg-3′ and unmodified GT. (b) The quantification results of percent cleavage products for different substrates. The values represent the mean ± standard deviation from three independent treatments and quantification experiments.
Discussion
We demonstrated recently that the tandem lesion with a Tg being followed by an 8-oxodG could be induced in calf thymus DNA upon treatment with Fenton reagents under aerobic conditions (20). Although the formation of this type of lesion in cells remains to be assessed, the relatively high frequency of its formation in vitro suggests that it can be induced in cells. Here, we employed the ODN substrates containing Tg and 8-oxodG, either alone or neighboring each other, and examined how these lesions perturb DNA replication by using purified DNA polymerases and how efficiently they are recognized by two BER enzymes, hOGG1 and E. coli endonuclease III.
Our primer extension assay results revealed that the tandem lesions blocked DNA replication more effectively than the two isolated single-nucleobase lesions. In addition, the miscoding potentials of Tg and 8-oxodG, as revealed by steady-state kinetic measurements, are different while these lesions are present alone or in tandem. Our observation is consistent with previous findings that the tandem lesions, where an 8-oxodG is vicinal to an abasic site or a formylamine, can perturb differently the fidelity and efficiency of nucleotide incorporation opposite the lesion site from the situations where the composing lesions are present alone (37, 38). The alteration in the fidelity of nucleotide incorporation might be attributed to the local structure change imposed by the neighboring lesion.
BER assay results showed that Endonuclease III recognizes and cleaves the thymine glycol in the two tandem lesion-containing substrates at a similar efficiency as the substrate housing an isolated thymine glycol. On the other hand, the 8-oxodG in the two tandem lesions could be recognized differently by hOGG1; whereas the lesion with an adjacent 5′ Tg could be cleaved much less efficiently than an isolated 8-oxodG, the lesion with a neighboring 3′ Tg could be cleaved more readily than when the 8-oxodG was present alone.
Thermodynamic measurements revealed that the 5′-Tg-(8-oxodG)-3′ and 5′-(8-oxodG)-Tg-3′ tandem lesions destabilized duplex DNA to a similar extent, which is represented by a 5.1 kcal/mol increase in Gibbs free energy for duplex formation at 37 °C (41). Thus, the difference in recognition of the two tandem lesions by hOGG1 is not due to difference in overall destabilization to duplex DNA induced by the two tandem lesions. However, the above differential cleavage efficiencies for the three substrates can be rationalized from the nature of hOGG1-DNA interaction based on the X-ray structure of the hOGG1-DNA complex (53) and the structural perturbation to duplex DNA introduced by thymidine glycol (54, 55).
The X-ray co-crystal structure of hOGG1 and 8-oxodG-bearing duplex DNA revealed a marked structure alteration of the lesion-containing DNA (53). The modified nucleobase, 8-oxo-7,8-dihydroguanine (8-oxoGua) is extruded from the helix and is inserted deeply into an extrahelical active-site pocket of the enzyme. In addition, the X-ray structure showed the hydrogen bonding interaction between the nucleobase on the 5′ side of 8-oxoGua and Asn151 in the protein (53). The lack of aromaticity of Tg may compromise this interaction thereby resulting in decreased binding of the 5′-Tg-(8-oxodG)-3′-bearing substrate toward hOGG1. Furthermore, the structure showed that the formation of a catalytically competent protein-DNA complex necessitates significant bond rotation of the flanking 5′ phosphate so that its non-bridging oxygen atoms point inward towards the helix axis. The presence of Tg as the 5′ neighboring nucleoside may also perturb this bond rotation thereby decreasing the catalytic proficiency of the enzyme. The above factors together may contribute to the poorer hOGG1-mediated cleavage of 5′-Tg-(8-oxodG)-3′ than an isolated 8-oxodG.
The X-ray structure showed no direct contact between the protein and the 3′ flanking nucleobase and no significant bond rotation in the phosphate group on the 3′ side of 8-oxodG (53). On the other hand, previous molecular modeling studies showed that the methyl group in the cis-(5R,6S) isomer of Tg favors an axial conformation, which results in a steric clash between the methyl group and the 5′ neighboring nucleobase thereby destabilizing the 5′ base pair (54, 55). Therefore, the destabilization of the 8-oxoGua:Cyt (“Cyt” represents cytosine) base pair induced by the 3′ vicinal Tg may result in the facile extrusion of 8-oxoGua from the helix thereby enhancing the cleavage efficiency of the enzyme toward the 5′-(8-oxodG)-Tg-3′ substrate.
The relatively poor cleavage of 8-oxodG from the 5′-Tg-(8-oxodG)-3′-tandem lesion-containing substrate may render this lesion a substrate for the NER pathway. In this regard, both Tg and 8-oxodG can be recognized by mammalian NER machinery (56). The formation of this tandem lesion at methylated CpG site, therefore, may account for the occurrence of high frequency of mCG→TT mutation while CpG-methylated pSP189 shuttle vector was replicated in NER-deficient XPA cells (32).
Together, the above results revealed that when the two commonly observed ROS-induced lesions, i.e., Tg and 8-oxodG, are neighboring to each other, they impose greater challenges to DNA replication apparatus and confer different mutagenic properties than when these lesions are present alone. Moreover, the 5′-Tg-(8-oxodG)-3′ tandem lesion can be recognized by hOGG1 much less efficiently than an isolated 8-oxodG. Therefore, the efficient formation of the 5′-Tg-(8-oxodG)-3′ tandem lesion, coupled with the elevated difficulty in the hOGG1-mediated repair of its 8-oxodG component, underscores the biological significance of this tandem lesion. To our knowledge, this represents the first replication and repair study on a tandem single-nucleobase lesion [i.e., 5′-Tg-(8-oxodG)-3′] whose efficient formation in isolated DNA has been demonstrated.
Acknowledgment
This work was supported by the National Institutes of Health (R01 CA101864) and Yong Jiang was supported in part by the University of California Toxic Substances Research and Teaching Program. The authors also want to thank Professors Olga S. Fedorova and John-Stephen A. Taylor for providing the expression vectors for hOGG1 and yeast pol η.
Footnotes
- ROS
- Reactive oxygen species
- MDS
- multiply damaged site
- Tg
- 5,6-dihydroxy-5,6-dihydrothymidine
- 8-oxodG
- 8-oxo-7,8-dihydro-2′-deoxyguanosine
- ODN
- oligodeoxyribonucleotide
- hOGG1
- human 8-oxoguanine-DNA glycosylase
- BER
- base excision repair
- NER
- nucleotide excision repair
- AP site
- apurinic/apyrimidinic site
- pol η
- polymerase η
- XP
- xeroderma pigmentosum
- PAGE
- polyacrylamide gel electrophoresis
References
- (1).Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- (2).Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Nikjoo H, O’Neill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- (4).Sutherland BM, Bennett PV, Sutherland JC, Laval J. Clustered DNA damages induced by x rays in human cells. Radiat. Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- (5).Budworth H, Dianov GL. Mode of inhibition of short-patch base excision repair by thymine glycol within clustered DNA lesions. J. Biol. Chem. 2003;278:9378–9381. doi: 10.1074/jbc.M212068200. [DOI] [PubMed] [Google Scholar]
- (6).Budworth H, Dianova II, Podust VN, Dianov GL. Repair of clustered DNA lesions - Sequence-specific inhibition of long-patch base excision repair by 8-oxoguanine. J. Biol. Chem. 2002;277:21300–21305. doi: 10.1074/jbc.M201918200. [DOI] [PubMed] [Google Scholar]
- (7).Chaudhry MA, Weinfeld M. The action of Escherichia coli endonuclease-III on multiply damaged sites in DNA. J. Mol. Biol. 1995;249:914–922. doi: 10.1006/jmbi.1995.0348. [DOI] [PubMed] [Google Scholar]
- (8).David-Cordonnier MH, Cunniffe SMT, Hickson ID, O’Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- (9).David-Cordonnier MH, Laval J, O’Neill P. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem. 2000;275:11865–11873. doi: 10.1074/jbc.275.16.11865. [DOI] [PubMed] [Google Scholar]
- (10).Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- (11).Lomax ME, Cunniffe S, O’Neill P. 8-oxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- (12).Pearson CG, Shikazono N, Thacker J, O’Neill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bellon S, Ravanat JL, Gasparutto D, Cadet J. Cross-linked thymine-purine base tandem lesions: Synthesis, characterization, and measurement in gamma-irradiated isolated DNA. Chem. Res. Toxicol. 2002;15:598–606. doi: 10.1021/tx015594d. [DOI] [PubMed] [Google Scholar]
- (14).Box HC, Budzinski EE, Dawidzik JB, Gobey JS, Freund HG. Free radical-induced tandem base damage in DNA oligomers. Free Radic. Biol. Med. 1997;23:1021–1030. doi: 10.1016/s0891-5849(97)00166-4. [DOI] [PubMed] [Google Scholar]
- (15).Box HC, Budzinski EE, Dawidzik JB, Wallace JC, Iijima H. Tandem lesions and other products in X-irradiated DNA oligomers. Radiat. Res. 1998;149:433–439. [PubMed] [Google Scholar]
- (16).Hong H, Cao H, Wang Y, Wang Y. Identification and quantification of a guanine-thymine intrastrand cross-link lesion induced by Cu(II)/H2O2/ascorbate. Chem. Res. Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hong IS, Carter KN, Sato K, Greenberg MM. Characterization and mechanism of formation of tandem lesions in DNA by a nucleobase peroxyl radical. J. Am. Chem. Soc. 2007;129:4089–4098. doi: 10.1021/ja0692276. [DOI] [PubMed] [Google Scholar]
- (18).Zhang Q, Wang Y. Independent generation of 5-(2 ′-deoxycytidinyl)methyl radical and the formation of a novel cross-link lesion between 5-methylcytosine and guanine. J. Am. Chem. Soc. 2003;125:12795–12802. doi: 10.1021/ja034866r. [DOI] [PubMed] [Google Scholar]
- (19).Zhang Q, Wang Y. Generation of 5-(2 ′-deoxycytidyl)methyl radical and the formation of intrastrand cross-link lesions in oligodeoxyribonucleotides. Nucleic Acids Res. 2005;33:1593–1603. doi: 10.1093/nar/gki301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Jiang Y, Wang Y, Wang Y. Efficient induction of tandem thymine glycol/8-oxoguanine lesion in DNA by Cu(II)/H2O2/ascorbate. J. Am. Chem. Soc. 2008 Revision pending. [Google Scholar]
- (21).Wang Y. Bulky DNA lesions induced by reactive oxygen species. Chem. Res. Toxicol. 2008;21:276–281. doi: 10.1021/tx700411g. [DOI] [PubMed] [Google Scholar]
- (22).Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007;35:7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Jiang Y, Hong H, Cao H, Wang Y. In vivo formation and in vitro replication of a guanine-thymine intrastrand cross-link lesion. Biochemistry. 2007;46:12757–12763. doi: 10.1021/bi7012195. [DOI] [PubMed] [Google Scholar]
- (24).Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. USA. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- (26).Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G->T and A->C substitutions. J. Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- (27).Bienvenu C, Cadet J. Synthesis and kinetic study of the deamination of the cis diastereomers of 5,6-dihydroxy-5,6-dihydro-5-methyl-2′-deoxycytidine. J. Org. Chem. 1996;61:2632–2637. doi: 10.1021/jo951900e. [DOI] [PubMed] [Google Scholar]
- (28).Zuo S, Boorstein RJ, Teebor GW. Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res. 1995;23:3239–3243. doi: 10.1093/nar/23.16.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and Distribution of 5-Methylcytosine in Human DNA from Different Types of Tissues or Cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pfeifer GP. p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res. 2000;450:155–166. doi: 10.1016/s0027-5107(00)00022-1. [DOI] [PubMed] [Google Scholar]
- (31).Cao H, Wang Y. Quantification of oxidative single-base and intrastrand cross-link lesions in unmethylated and CpG-methylated DNA induced by Fenton-type reagents. Nucleic Acids Res. 2007;35:4833–4844. doi: 10.1093/nar/gkm497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lee DH, O’Connor TR, Pfeifer GP. Oxidative DNA damage induced by copper and hydrogen peroxide promotes CG -> TT tandem mutations at methylated CpG dinucleotides in nucleotide excision repair-deficient cells. Nucleic Acids Res. 2002;30:3566–3573. doi: 10.1093/nar/gkf478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- (34).Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- (35).Gu C, Wang Y. LC-MS/MS identification and yeast polymerase η bypass of a novel γ-irradiation-induced intrastrand cross-link lesion G[8-5]C. Biochemistry. 2004;43:6745–6750. doi: 10.1021/bi0497749. [DOI] [PubMed] [Google Scholar]
- (36).Gu C, Wang Y. Thermodynamic and in vitro replication studies of an intrastrand G[8-5]C cross-link lesion. Biochemistry. 2005;44:8883–8889. doi: 10.1021/bi050036+. [DOI] [PubMed] [Google Scholar]
- (37).Gentil A, Le Page F, Cadet J, Sarasin A. Mutation spectra induced by replication of two vicinal oxidative DNA lesions in mammalian cells. Mutat. Res. 2000;452:51–56. doi: 10.1016/s0027-5107(00)00034-8. [DOI] [PubMed] [Google Scholar]
- (38).Kalam MA, Basu AK. Mutagenesis of 8-oxoguanine adjacent to an abasic site in simian kidney cells: tandem mutations and enhancement of G-->T transversions. Chem. Res. Toxicol. 2005;18:1187–1192. doi: 10.1021/tx050119r. [DOI] [PubMed] [Google Scholar]
- (39).David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry. 2008;47:4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wang Y, Wang Y. Synthesis and thermodynamic studies of oligodeoxyribonucleotides containing tandem lesions of thymidine glycol and 8-oxo-2 ′-deoxyguanosine. Chem. Res. Toxicol. 2006;19:837–843. doi: 10.1021/tx060032l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Johnson RE, Prakash S, Prakash L. Efficient bypass of a thyminethymine dimer by yeast DNA polymerase, Pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- (43).Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- (44).Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- (45).Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Cannistraro VJ, Taylor JS. DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase eta. J. Biol. Chem. 2004;279:18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- (47).Kuznetsov NA, Koval VV, Zharkov DO, Nevinsky GA, Douglas KT, Fedorova OS. Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 2005;33:3919–3931. doi: 10.1093/nar/gki694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- (49).Shinmura K, Kasai H, Sasaki A, Sugimura H, Yokota J. 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) DNA glycosylase and AP lyase activities of hOGG1 protein and their substrate specificity. Mutat. Res. 1997;385:75–82. doi: 10.1016/s0921-8777(97)00041-4. [DOI] [PubMed] [Google Scholar]
- (50).Asagoshi K, Yamada T, Terato H, Ohyama Y, Monden Y, Arai T, Nishimura S, Aburatani H, Lindahl T, Ide H. Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J Biol Chem. 2000;275:4956–4964. doi: 10.1074/jbc.275.7.4956. [DOI] [PubMed] [Google Scholar]
- (51).Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Warner HR, Demple BF, Deutsch WA, Kane CM, Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc. Natl. Acad. Sci. USA. 1980;77:4602–4606. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- (54).Clark JM, Pattabiraman N, Jarvis W, Beardsley GP. Modeling and molecular mechanical studies of the cis-thymine glycol radiation damage lesion in DNA. Biochemistry. 1987;26:5404–5409. doi: 10.1021/bi00391a028. [DOI] [PubMed] [Google Scholar]
- (55).Brown KL, Adams T, Jasti VP, Basu AK, Stone MP. Interconversion of the cis-5R,6S- and trans-5R,6R-thymine glycol lesions in duplex DNA. J. Am. Chem. Soc. 2008;130:11701–11710. doi: 10.1021/ja8016544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]