Abstract
The conserved membrane-proximal external region (MPER) of the HIV-1 gp41 envelope protein is the established target for very rare but broadly neutralizing monoclonal antibodies (NAbs) elicited during natural human infection. Nevertheless attempts to generate an HIV-1 neutralizing antibody response with immunogens bearing MPER epitopes have met with limited success. Here we show that the MPER peptide (residues 662–683) forms a labile α-helical trimer in aqueous solution and report the crystal structure of this autonomous folding subdomain stabilized by addition of a C-terminal isoleucine-zipper motif. The structure reveals a parallel triple-stranded coiled coil in which the neutralization epitope residues are buried within the interface between the associating MPER helices. Accordingly, both the 2F5 and 4E10 NAbs recognize the isolated MPER peptide but fail to bind the trimeric MPER subdomain. We propose that the trimeric MPER structure represents the prefusion conformation of gp41, preceding the putative prehairpin intermediate and the postfusion trimer-of-hairpins structure. As such, the MPER trimer should inform the design of new HIV-1 immunogens to elicit broadly neutralizing antibodies.
The envelope glycoprotein (Env) of HIV-1 mediates virus attachment and entry into target cells and offers a primary target for vaccine development (1). The native, prefusion form of the Env complex is thought to be a trimer comprising three gpl20 surface glycoproteins associated non-covalently with three membrane-anchored gp41 subunits (2–4). This structure is unstable, and its binding to cell-surface receptors (CD4 and a coreceptor, usually CCR5 or CXCR4) triggers large-scale conformational changes in gp41 that ultimately promote formation of a highly stable trimer-of-hairpins structure and thereby fusion of the viral and cellular membranes (5, 6). Much of what is known about the structure-function relationships of gp41 has emerged from biophysical and structural studies, showing that the core of the postfusion trimer-of-hairpins is a bundle of six α-helices formed by antiparallel association of two conserved heptad-repeat (HR) regions in the gp41 ectodomain (7–11). The first repeat (HRN) is adjacent to the N-terminal fusion peptide, which is exposed and inserted into the target cell membrane in the fusion process, while the second (HRC) immediately precedes the transmembrane segment (Figure 1a). Hence, formation of the six-helix bundle allows the two membrane attachment points to come together in the postfusion conformation (9). The accumulated evidence suggests that activation of gp41-induced membrane fusion proceeds via a regulated sequence of structural transitions with one or more on-pathway intermediate(s) (5, 6). The structure of gp41 in its native and intermediate states remains unknown.
FIGURE 1.
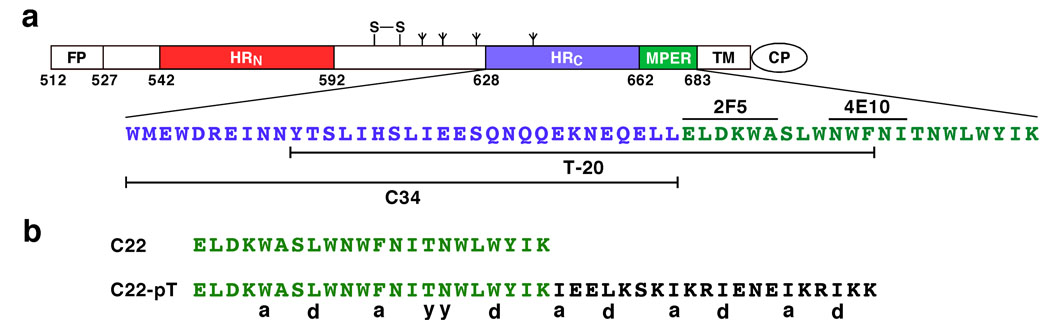
Membrane-proximal external region in HIV-1 gp41. (a) Schematic diagram of gp41. The positions of the fusion peptide (FP), the two 4–3 heptad repeats of hydrophobic residues (HRN and HRC), the disulfide bond (S–S), the potential N-linked glycosylation sites (tree-like symbols), the membrane-proximal external region (MPER), the transmembrane segment (TM) and the cytoplasmic region (CP) are shown. The amino acid sequence of the C-terminal ectodomain segment is shown. The locations of C34, T-20 (enfuvirtide) and the core epitopes for two broadly neutralizing human mAbs, 2F5 and 4E10, are indicated. Residues are numbered according to their position in HIV-1 HXB2 gp160. (b) Constructs for experimental characterization of trimerizing MPER. The amino acid sequences of C22 and C22-pT are shown. The C22-pT peptide contains a C-terminal 20-residue isoleucine-zipper trimer sequence. The observed core residue positions (a, d or y) for the C22-pT trimer are indicated (see also Figure 3 legend).
The design of HIV-1 immunogens to elicit broadly neutralizing antibodies remains one of the most important challenges confronting biomedical research today. The difficulty lies with lack of information on natural immunity to the virus, the extraordinary sequence diversity of Env and continued viral genetic variation (1). While there have been many disappointments in the creation of a NAb-based vaccine, a few human monoclonal antibodies (mAbs) derived from B cells of HIV-1-infected individuals do have significant breadth of neutralizing activities that cross virus isolates and phylogenetic clades (1). These rare mAbs include the two most broadly reactive NAbs, 2F5 and 4E10/zl3, that recognize adjacent but distinct epitopes on the membrane-proximal external region (MPER) of the gp41 ectodomain (12). However, all attempts to elicit comparable anti-gp41 antibodies with immunogens bearing these neutralization epitope sequences have thus far failed. It has been postulated that the induction of 2F5- and 4E10-like NAbs responses requires presentation of the MPER in the prefusion form of the trimeric Env complex (1, 13, 14). Great strides have been made in characterizing the molecular architecture of the native Env spike on virions, but no atomic structure of the gpl20/gp41 complex is available (2–4). Recent electron tomographic studies of Env spikes are interpreted to reveal a native trimer held together in part by close contacts at the gp41 base (2, 4) or anchored to the plasma membrane in a tripod4ike structure (3). Missing from the analysis to date is a structural description of the gp41-gp41 interactions in the native configuration. Here we describe the identification and crystal structure of trimeric MPER solved to 2.0 Å resolution and discuss the implications of this autonomous folding subdomain for native Env structure, gp41 fusion activation and antibody-mediated neutralization.
MATERIALS AND METHODS
Protein Expression and Purification
The pC22 and pC22-pT constructs (Figure 1b) were appended to the TrpLE’ leader sequence (15) and cloned into the pET24a vector (Novagen) by using standard molecular biology techniques. All mutants were created by site-directed DNA mutagenesis; the generated sequences were confirmed by DNA sequencing. The C22 and C22-pT peptides were expressed in the E. coli strain BL21(DE3)/pLysS (Novagan), purified from inclusion bodies and cleaved from the TrpLE' leader sequence with cyanogen bromide as described (15). All peptides were purified to homogeneity by reverse-phase HPLC (Waters, Inc) on a Vydac C18 preparative column (Hesperia, CA) using a water–acetonitrile gradient in the presence of 0.1% trifluoroacetic acid and lyophilized. Peptide identities were confirmed by electrospray mass spectrometry (PerSeptive Biosystems Voyager Elite, Cambridge, MA). Protein concentrations were determined by using the method of Edelhoch (16).
Biophysical Experiments
Circular dichroism (CD) spectra were measured on a 62A/DS CD spectrometer (Aviv Associates, Lakewood, NJ) in TBS (50 mM Tris-HCl, pH 8.0, 150 mM NaCl) supplemented with 7 mM SDS at 10°C. A [θ]222 value of −33 000 deg cm2/dmol was taken to correspond to 100% helix. Sedimentation equilibrium experiments were carried out on an XL-A analytical ultracentrifuge (Beckman Coulter) equipped with an An-60 Ti rotor (Beckman Coulter). Peptide samples were dialyzed overnight against TBS (pH 8.0) and 7 mM SDS, loaded at initial concentrations of 2.5, 5, 10, 30 and 100 µM and analyzed at rotor speeds of 30, 33 and 37 krpm for C22, and 21 and 24 krpm for C22-pT at 10 and 25°C. Data sets were fitted to a single-species model.
Crystallization and Structure Determination
Crystals of C22-pT were grown at room temperature by the hanging-drop vapor-diffusion method by mixing equal volumes of peptide solution (8 mg/mL in 5% (v/v) acetic acid) and precipitant (0.1 M sodium HEPES, pH 7.6, 0.1 M ammonium dihydrogenphosphate, 50% (v/v) MPD). Crystals belong to space group P21 and contain six monomers in the asymmetric unit (Table 1). The crystals were harvested in the precipitant buffer and frozen in liquid nitrogen. Diffraction data were collected on beamline X4A at the National Synchrotron Light Source (Brookhaven, NY). The images were indexed and integrated by using DENZO (17). The intensities were scaled in P21 symmetry with SCALEPACK (17); the systematic absence of intensities indicates a 2-fold screw axis. Initial phases were determined by molecular replacement with MOLREP (18) using the structure of the GCN4-pII trimer (PDB entry 1GCM) as a search model. Two trimers were oriented and placed in the asymmetric unit with a correlation coefficient of 0.391 and an R factor of 0.577. In order to remove model bias, this model and the dataset for C22-pT were directly fed to Arp/Warp (19), which allowed ~83% of the final model to be automatically traced. The resulting electron density map enabled most of the side chains to be docked. Density interpretation and manual model building were carried out with O (20). The C22-pT structure wasrefined by using Refmac (21), resulting in the R free of 31.6% and the R work of 25.6% between 19.6 and 2.00 Å resolution. At this stage, phosphate ions, MPDs and water molecules were modeled in the electron density. Crystallographic refinement was concluded with Refmac using TLS groups assigned for each C22-pT monomer (22). The final structure consists of residues 1–42 (monomer A), 3–42 (monomer B), 3–42 (monomer C), 2–42 (monomer D), 3–42 (monomer E) and 2–42 (monomer F) in the asymmetric unit, 5 phosphate ions, 8 MPDs and 78 water molecules. All protein residues but one (Lys-C41) are in the most favored regions of the Ramachandran plot. Lys-C41 lies in an additionally allowed region of the Ramachandran space and is the second residue from the C terminus of monomer C. Coiled-coil parameters were calculated by using TWISTER (23). The rms deviations were calculated with LSQKAB in CCP4i program suite (24). Buried surface areas were calculated from the difference of the accessible side chain surface areas of the trimer structure and of the individual helical monomers by using CNS 1.0 (25).
Table 1.
Crystallographic Data Collection and Refinement Statistics
| resolution (Å) | 19.6–2.00 (2.07–2.00) |
| space group | P21 |
| unit cell parameter | a = 42.53 Å, b = 48.16 Å, c = 81.78 Å, β = 95.4° |
| no. of unique reflections | 22 010(2 171) |
| no. of reflections >2σI | 172 46 |
| multiplicity | 3.7(3.6) |
| Rmerge (%) | 4.9 (52.8) |
| I/σ(I) | 14.1 (2.3) |
| completeness (%) | 98.0 (97.5) |
| no. of molecules in the asymmetric unit | 6 |
| solvent content (%) | 54 |
| no. of protein atoms in refinement | 2 241 |
| no. of water molecules | 78 |
| no. of PO43− ions | 5 |
| no. of MPD molecules | 8 |
| Rcryst/Rfree, % | 21.3/26.9 |
| average B factors (Å2) | 57.4 |
| rmsd (bond lengths) (Å) | 0.026 |
| rmsd (bond angles) (deg) | 2.0 |
| rmsd (torsion angles) (deg) | 4.8 |
Values for the highest resolution shell are given in brackets.
SPR Measurements
Biacore experiments were carried out on a Biacore 3000 (Biacore, Inc.) with the CM5 sensor chip composed of carboxymethylated dextran covalently attached to a gold surface (Biacore) at 25°C. The running buffer was 10 mM HEPES (pH 7.4) containing 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) surfactant P20 (HBS-EP) supplemented with 7 mM SDS. Approximately 1000 response units (RU) of 2F5 or 4E10 antibody at a flow rate of 5 µl/min were immobilized to the sensor chip via amino coupling in two experimental flow-cells. A mock-treated flow-cell was used as a control. All flow-cells were then blocked with 1 M ethanolamine-HCl (pH 8.0) and washed using 10 mM glycine-HCl (pH 2.5). For kinetic measurements, 2.5 µM of peptide antigens were injected at a flow rate of 5 µl/min, and sensorgrams acquired with a 60-s association phase and a >120-s dissociation phase. The sensor surfaces were regenerated between each experiment with two 10 µl injections of 10 mM glycine-HCl (pH 3.0). Identical injections over blank surfaces were subtracted from the response data for analysis of binding interactions. Antibody binding to peptide antigens was evaluated by using BiaEvaluation software (Biacore).
RESULTS
The MPER Peptide Self-Associates to Form an α-Helical Trimer
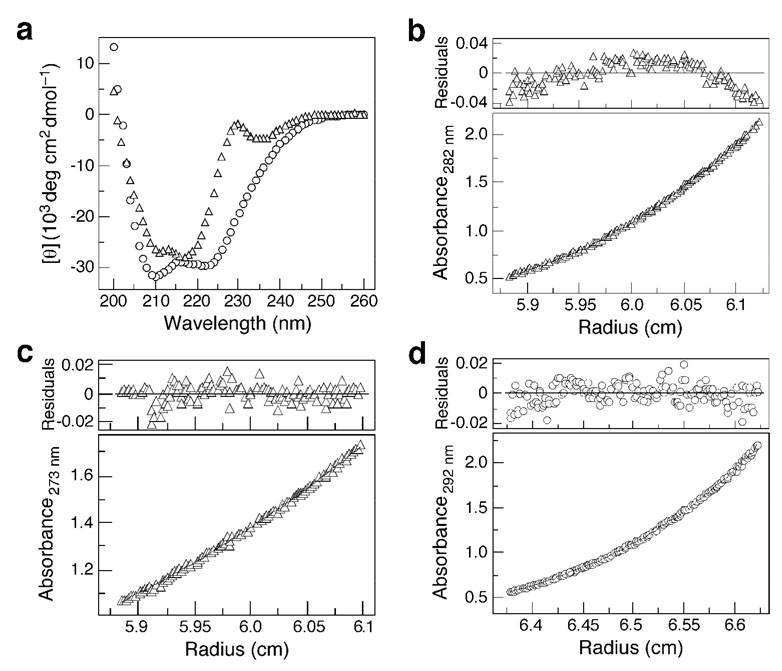
The HIV-1 MPER segment (amino acids 662–683; denoted C22) is among the most highly conserved within the HIV-1 Env and is unusually rich in the amino acid tryptophan (Figure 1a). The C22 peptide is only marginally soluble in aqueous solutions but attains a solubility of >500 µM in TBS buffer (pH 8.0) supplemented with a submicella concentration of SDS (i.e., 7 mM). This behavior is consistent with binding of SDS to the hydrophobic side chains of the peptide. The CD spectrum of C22 has double minima at 211 nm and 217 nm as well as a minor positive band around 230 nm (Figure 2a). This CD signal differs from the characteristic signature of an α-helical conformation (with minima at 208 nm and 222 nm), which can be attributed to Trp residues whose indole chromophore is known to contribute significant ellipticity in the far-UV CD spectrum of helical peptide proteins (26). While not a quantitative measure, the mean residue ellipticity at either 208 nm or 222 nm would suggest ~70% helical structure at 10°C. Sedimentation equilibrium experiments indicate that C22 sediments as a trimer at 10°C: the apparent molecular mass is 9.5 kDa in the concentration range of 10 µM to 100 µM (the expected molecular mass is 8.8 kDa) (Figure 2b). A very slight deviation of residuals from an ideal trimer may indicate the presence of <5% monomer in the sample. The monomelic form of C22 at 5 µM peptide concentration and 25°C is demonstrated by the sedimentation equilibrium data (Figure 2c). These results indicate that the C22 peptide associates intermolecularly to form a folded α-helical trimer structure under native conditions (low temperatures).
FIGURE 2.
Solution properties of C22 and C22-pT. (a) CD spectra of 10 µM solution of C22 (triangles) and C22-pT (circles) in TBS (pH 8.0) and 7 mM SDS at 10°C. (b) Equilibrium sedimentation data (30 krpm) of C22 (10 µM) in TBS (pH 8.0) and 7 mM SDS at 10°C. The data fit closely to a trimeric complex. The deviation in the data from the linear fit for a trimeric model is plotted (upper), (c) Equilibrium sedimentation data (37 krpm) of C22 (5 µM) in TBS (pH 8.0) and 7 mM SDS at 25°C. The deviation in the data from the linear fit for a monomelic model is plotted (upper), (d) Equilibrium sedimentation data (21 krpm) of C22-pT (30 µM) in TBS (pH 8.0) and 7 mM SDS at 10°C. The data fit closely to a trimeric complex. The deviation in the data from the linear fit for a trimeric model is plotted (upper).
Design of the C22-pT Chimera
Previous studies have suggested that favorable hydrophobic packing interactions between the predicted transmembrane helices of gp41 can contribute to the trimerization of the protein (27, 28). Based on the trimeric helical structure of C22 reported here, we posit that the trimerization specificity of native gp41 may reside in the MPER segment abutting the transmembrane anchor. In attempting to engineer a stabilized MPER peptidic molecule for crystallographic studies, we appended a soluble isoleucine-zipper motif (GCN4-pT) to C22 at the C terminus in place of the transmembrane anchor (Figure 1b). In this 20-residue GCN4-pT sequence of the classical GCN4-pII coiled coil (29), four hydrophobic surface residues were substituted with charged Lys and Arg side chains, which are expected to increase solubility. The resulting C22-pT peptide exhibits a characteristic α-helical CD spectrum with negative minima at ~222 nm and ~210 nm in TBS (pH 8.0) and 7 mM SDS (Figure 2a). The mean residue ellipticity of the folded peptide at 222 nm is −29,700 deg cm2/dmol, consistent with a >90% helical structure. Sedimentation equilibrium studies demonstrate that C22-pT forms a discrete trimer, with an apparent molecular mass of 17.3 kDa over a 40-fold range of peptide concentration (Figure 2d). Thus, the C-terminally appended GCN4-pT sequence appears to effectively replace the transmembrane anchor and thus stabilize the homotrimeric conformation of the parent C22 molecule. A similar approach was successfully used to create soluble trimers of viral fusion protein ectodomains without distortion of their triple-stranded coiled-coil structures (9, 30).
Structure of C22-pT
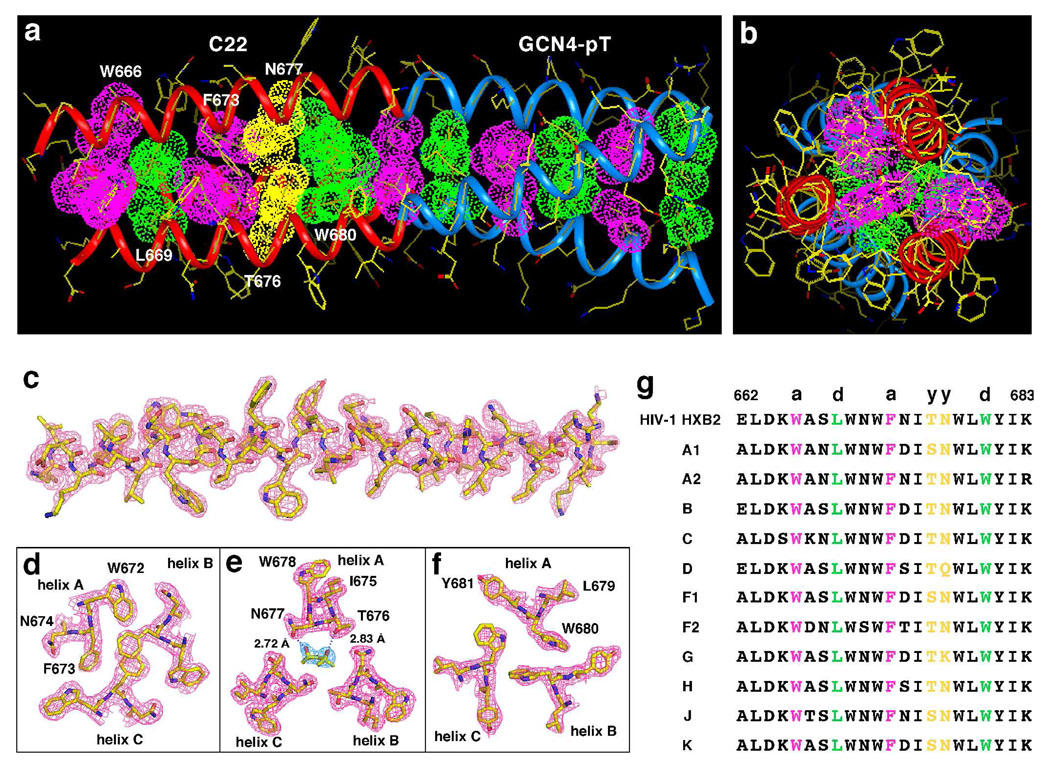
To investigate the basis for trimerization interaction specificity of the MPER, the X-ray crystal structure of the C22-pT peptide was determined at 2.0 Å resolution by molecular replacement (Table 1). Crystals of C22-pT contain two trimers in the asymmetric unit, giving rise to six independently refined monomers. The final experimental electron density map is of excellent quality and reveals the positions of all of the amino acid residues except for two at the N-terminal end. Glu-662 and Leu-663 are disordered to a different degree among different chains. Although it is possible that this variation is due to the differences in local lattice contact environments, these two N-terminal residues are more likely to be intrinsically flexible. This structural disorder is consistent with the measured molar ellipticity of C22-pT in solution (Figure 2a). The quality of the structure was verified by PROCHECK, with all residues in the most favored α-helical region of the Ramachandran plot. A representative portion of the 2F o – F c electron density map is shown in Figure 3c. Despite asymmetric crystal contacts, the six individual chains in the C22-pT structure have essentially the same conformation and degree of order. The average rms difference in Cα atom positions between any pair of chains is 0.48 Å, and the average B-factors vary from 50.3 to 56.5 Å2.
FIGURE 3.
The C22-pT peptide forms a parallel three-stranded coiled coil, (a) Lateral view of the trimer. The Cα backbones of the C22 and GCN4-pT portions of C22-pT are showed in red and blue, respectively. Pink van der Waals surfaces identify residues at the a positions, green surfaces identify residues at the d positions and yellow surfaces identify residues at the y positions, (b) Axial view of the trimer. The view is from the N terminus looking down the superhelical axis, (c) The 2Fo – Fc electron density map (contoured at 1.0σ) superposed onto the final model of the A helix, (d) Cross-section of the trimer in the Phe-673(a) layer. The 2Fo – Fc electron density map at 1.0σ contour is shown with the refined molecular model. The aromatic ring planes of helices A and C incline by ~45° toward the C terminus of each helix with respect to the plane perpendicular to the superhelical axis, while that of helix B is tilted by ~45° toward the N terminus. (e) Cross-section of the trimer in the Thr-676–Asn-677(y) layer (60). A single MPD molecule is located in the trimeric coiled-coil core. Hydrogen bonds are denoted by blue dotted lines, (f) Cross-section of the trimer in the Trp-680(d) layer. (g) Sequence conservation of MPER in HIV-1 Envs. The consensus sequences of the MPER segment are shown for subtypes of the HIV-1 M group (61). The observed a, d and y core positions are indicated above the HXB2 MPER sequence.
As anticipated, the C22-pT trimer consists of three continuous, parallel α helices wrapped in a slight left-handed superhelical twist (Figure 3a). This coiled coil has a straight supercoil axis and forms a cylinder that is ~60 Å in length and ~24 Å in diameter. An approximate 3-fold axis of symmetry coincides with the superhelical axis. C22-pT displays 4–3 hydrophobic heptad continuity at the junction of the chimera (i.e., the Trp-580(d) layer of the MPER is followed by the isoleucine a layer of GCN4-pT) (Figure 1b). Alteration of the phasing of this heptad register by addition of either one or two alanine residues at the N terminus of the GCN4-pT sequence causes the resulting molecule to form an insoluble aggregate in solution. In addition, the GCN4-pT fraction adopts the same three-stranded coiled-coil conformation as in the original GCN4-pII structure (rmsd for equivalent Cα atoms of 0.49 Å) (29). It is possible but unlikely that addition of the GCN4-pT sequence imposes the complete structure of the C22 trimer. Therefore, we assume that C22-pT retains the intrinsic structural features of three-stranded coiled-coil interactions in the MPER.
Trimerizing MPER Interactions
The MPER trimer interface of the C22-pT coiled coil consists of five cross-sectional layers of interhelical interactions: Trp-666(a), Leu-669(d), Phe-673(a), Thr-576-Asn-577(y) and Trp-680(d) (Figure 3a,g). The four nonpolar a and d residues make side-by-side contacts in a “knobs-into-holes” pattern (29) and thus provide a stabilizing hydrophobic core. The side chains of the d residues, for example, pack into holes formed between the side chains of residues in the d and e positions of an adjacent helix (Figure 3f). The angles between the Cα–Cα and Cα–Cβ vectors at these a and d layers are 120° and 150°, respectively. This type of acute packing arrangement in the interior of the coiled coil is characteristic of parallel trimers (29). All of these hydrophobic a and d residues adopt their well-populated rotamer conformations in α-helices. We note that the Phe-673 side chain of helix B is oriented differently from those of helices A and C, thus making stacking interactions and van der Waals contacts with that of the neighboring C helix (Figure 3d). This non-ideal geometry for knobs-into-holes packing results from avoidance of steric clashing between the Phe-673 side chain and Trp-672 at the g position of helix A, because the indole ring of the latter residue points back into the interior of the coiled coil (Figure 3d). In general, the interfacial residues at the buried core positions of the MPER are either absolutely conserved or highly restricted among HIV-1 sequences from M groups, whereas exposed polar residues display great sequence variability (Figure 3g). This pattern of sequence conservation presumably reflects selective pressure on the MPER coiled-coil interactions to nucleate trimer formation, and to regulate activation of the metastable native state, as discussed below.
The adjacent Thr-576 and Asn-577 side chains project obliquely to the trimer interface in a distinguishing hexagonal arrangement, thus giving rise to a specific non-close-packed core with weak interhelical contacts (Figure 3e). An electron density peak is located in a prominent cavity in the middle of this so-called y layer and was identified as a MPD molecule (a precipitant that is present in the crystallization buffer) (Figure 3e). The two hydroxyl groups of the bound MPD are well positioned to make hydrogen bonds with both the Thr-576 and Asn-577 side chains of helix A, and its carbon atoms form optimal van der Waals contacts (3.0–3.6 Å) with the same side chains of the other two helices. Variability at the Thr-576 and Asn-577 positions among HIV-1 isolates is limited to serine and polar amino acids, respectively (Figure 3g).
The presence of the Thr-576 and Asn-577 residues in the hydrophobic core of the MPER trimer markedly alters its overall coiled-coil conformation and geometry. In contrast to the C-terminal GCN4-pT fraction that has a superhelical pitch of 139 Å, the pitch of the N-terminal MPER segment is 492 Å. This unusually large pitch allows the Thr-576 and Asn-577 side chains to face toward the axis of supercoil rotation and mesh in the non-close-packed cross-section, as recognition sites for intermolecular interactions must be positioned in relation to the exact pitch of the helix. As a consequence, the backbones of the three MPER helices are less highly curved, resulting in an underwound superhelix. Thus the MPER trimer is significantly less tightly packed than the canonical GCN4-pT coiled coil. Approximately 1940 Å2 of solvent-accessible surface area (30% of total accessible surface area of the three α-helical chains) is buried in the MPER trimeric coiled coil. Relative to the side chains of isolated helices, residues at the a, d and y positions of the MPER trimer are substantially buried (>57%), those at the e and g positions are partly buried (~30% and 42%, respectively) and the b, c and f positions remain completely exposed. Together these results suggest that the non-close-packed core may function to achieve knobs-into-holes close packing of the apolar residues while minimizing the deformations of the supercoiled MPER helices. Such recurring heptad-repeat anomalies may serve a general role in facilitating polymorphic structural transitions in viral fusion proteins (31, 32).
2F5 and 4E10 Do Not Bind Trimeric MPER
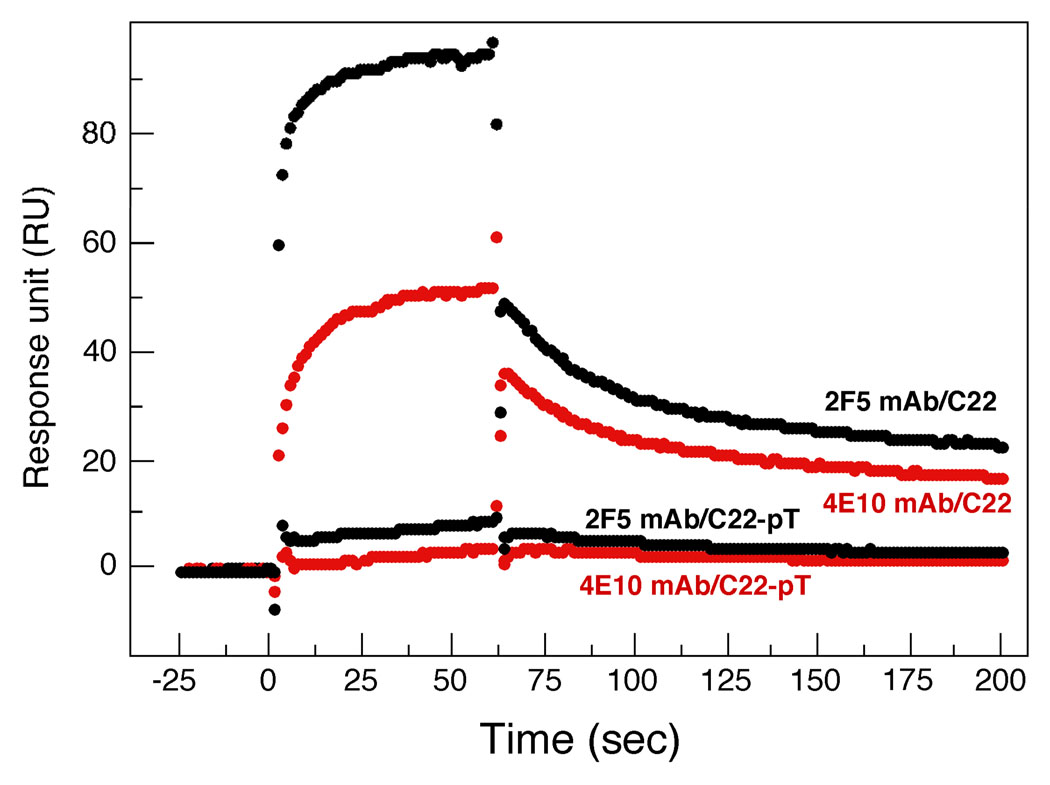
The neighboring linear MPER epitopes targeted by the 2F5 and 4E10 NAbs (Figure 1a) appear to be poorly exposed on the surface of both HIV-1-infected cells and virions (1, 12). The exposure of these neutralizing epitopes is enhanced or triggered by receptor binding but is lost by formation of the stable six-helix bundle core, indicating that changes in accessibility of the MPER occur during the fusion process (33, 34). Our structural study indicates that four hydrophobic residues (Trp-666, Trp-672, Phe-673 and Trp-680) critical for the neutralization activity of 2F5 and 4E10 (13) are buried within the MPER trimer interface (Figure 3). We performed SPR experiments to compare the binding of 2F5 and 4E10 to monomeric vs. trimeric MPER by using the C22 and C22-pT peptides. Experimental conditions for these Biacore analyses were chosen (e.g., 2.5 µM peptide concentration) so that C22 and C22-pT are predominantly a monomer and trimer in solution, respectively (as judged from sedimentation equilibrium data; see Figure 2c). As expected, C22 interacts specifically with both immobilized 2F5 and 4E10 (Figure 4). The reactivity of C22 to 2F5 is greater than that to 4E10, in accord with results of previous studies (12, 35). In contrast, little or no antibody binding to C22-pT is observed under the same conditions. These results support our hypothesis that the trimeric coiled-coil conformation of the MPER is a significant structural factor that regulates the conversion of the native state of Env to the extended intermediate, thus explaining why the latter conformation is a prime target of the 2F5 and 4E10 antibodies.
FIGURE 4.
Binding of mAbs 2F5 and 4E10 to the MPER peptide assessed by surface plasmon resonance. Representative sensograms for binding monomeric C22 at 2.5 µM and trimeric C22-pT at 2.5 µM to immobilized 2F5 (black) and 4E10 (red) at 25°C are shown. Specific interactions of 2F5 and 4E10 with C22 but not C22-pT are observed. Data represent three independent binding experiments, each of which was carried out in duplicate.
DISCUSSION
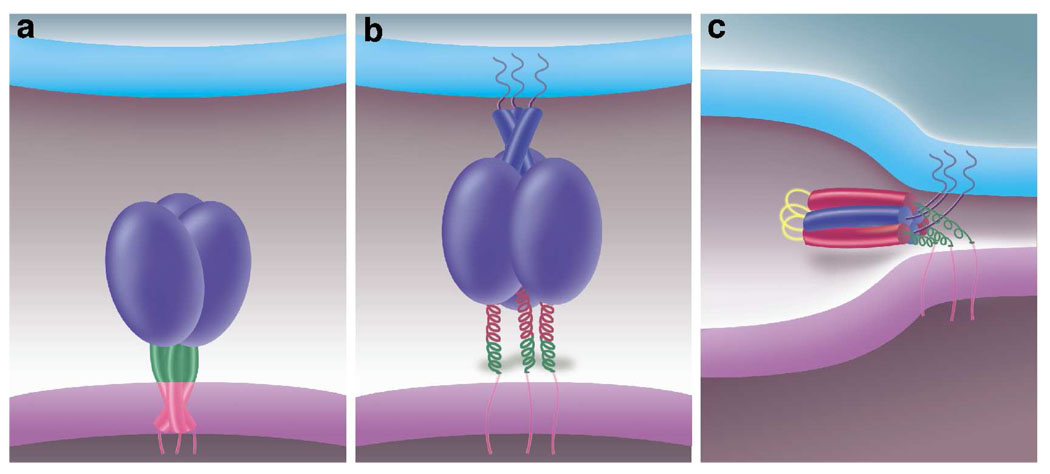
The unexpected identification and crystal structure of the MPER subdomain of gp41 described here, together with the results of immunological reactivity (33, 34) and the cryoelectron tomographic structure of the native Env spike (2, 4), lead us to propose that the labile three-stranded coiled coil represents the native, prefusion conformation of gp41. Three C-terminal MPER segments, one from each gp41 subunit, interact laterally with each other to create a superhelical substructure at the bottom of the ectodomains, whereas the fusion peptides and HRN regions are sequestered in the interior of the native Env complex as a result of interactions with the gpl20 chains (Figure 5a). They may also contribute to the prefusion Env trimer structure primarily from the three V1/V2 loop regions (4, 36). According to present theories, binding of gpl20 to CD4 and a chemokine receptor alters gpl20/gp41 intersubunit interactions in the native state so as to initiate cooperative folding of a HRN coiled-coil trimer that presents the fusion peptides to the target membrane (5, 6). As a result, the native state of gp41 converts into an extended homotrimeric prehairpin intermediate (Figure 5b). We can speculate that dissociation of the MPER trimer as the three helices splay apart may act as a trigger for the conformational change at the C-terminal part of the gp41 ectodomain. The MPER helix displays substantial affinity for lipid bilayers (14, 35, 37), a phenomenon postulated to facilitate this structural transition. The two key features of this gp41 refolding seem to be a coiled coil-to-lipid-embedded helix transition in MPER and reorientation of the C-terminal ectodomain segment, so that the HRC regions can fold-back onto the newly formed HRN trimer surface (Figure 5c). Thus, temporal collapse of the native MPER subdomain might serve as a possible mechanism for achieving controlled activation of membrane fusion.
FIGURE 5.
A potential sequence of conformational changes for HIV-1 gp41. (a) A schematic of the native Env trimer on the surface of the viral membrane (at the base of the diagram). A three-chain coiled-coil structure (formed by the MPER of each gp41 subunit) located just N-terminal to the transmembrane anchors is shown in green as a stem of native gp41. The gp120 chain is depicted as a purple ball. (b) Following interaction of gp120 to cell-surface receptors, the native state of gp41 converts to a transient prehairpin intermediate. Now the N-terminal HRN coiled coil (blue) is formed, relocating the fusion peptides to allow them to interact with the target bilayer (at the top of the diagram), while the C-terminal segments of the ectodomains open up. (c) Postfusion, the HRC regions (red) are shown as jackknifed and packed against the HRN coiled coil to form the trimer-of-hairpins structure. The fusion peptides and the transmembrane anchors are now adjacent in the fused membrane. Fusion ensues as the initial lipid stalk progresses through local hemifusion and then opening and enlargement of a fusion pore.
Polymorphic Interactions of the MPER
Entry of HIV-1 into its target cell requires receptor binding-activated structural rearrangements in gp41 that are coupled to the apposition and merger of the virus and cell membranes (5, 6). Current thinking proposes that prefusion gp41 is trapped in a metastable native state that precludes formation of the thermodynamically preferred trimer-of-hairpins structure. The three-chain coiled-coil structure of the MPER subdomain presented here (Figure 4), combined with previous knowledge, provides an opportunity to elucidate the molecular events involved in the control of HIV-1 membrane fusion. First, our proposal that formation of the MPER trimer is important for the native, prefusion conformation of gp41 is consistent with a large body of mutagenesis data on the structure-fusion activity relationships of Env. Deletion of the MPER sequence (residues 666–682) abolishes viral infectivity and membrane fusion, even though the mutant Env protein exhibits normal cell-surface expression, gp160 precursor processing, gp120/gp41 association and CD4 binding (38). Alanine substitutions that have a deleterious effect on infectivity map to the Trp-666, Leu-669, Trp-672, Phe-673, Ile-675 and Leu-679 residues (39, 40) that appear to play a critical role in stabilizing the trimeric MPER structure determined here (Figure 3g). In keeping with this notion, biochemical studies have shown that the N-terminal Δ660–671 deletion affects formation of the oligomeric structure of native gp41 (40). Thus, it seems reasonable to postulate that mutations which destabilize the MPER subdomain of gp41 prematurely induce fusion-associated conformational changes in the absence of target membranes and cause viral inactivation.
Second, the MPER segment of gp41 undergoes structural transitions in the course of the fusion reaction, as inferred from changes in antibody binding and sensitivity to limited proteolysis (33, 34, 39, 40). Both the 2F5 and 4E10 NAbs likely target the extended prehairpin conformation rather than the native gp41 structure as discussed above, and they also show enhanced reactivity to MPER peptides with different lengths in the presence of lipids (14, 35, 40). Chemical cross-linking and fluorescence-quenching experiments show that the MPER peptides self-associate to form homo-oligomers in solution and membranes (41, 42). On the other hand, NMR studies in detergent micelles suggest that the MPER segment forms a straight or bent amphipathic helix lying in the outer leaflet of the lipid bilayer (14, 37). We therefore propose that in the native state of gp41, the MPER folds as a trimeric coiled coil, and is converted to a membrane-bound helix in the prehairpin intermediate (Figure 5). Alterations in the bulk hydrophobic side chains from the coiled-coil trimer interface to the lipid-embedded state could influence the net free energy change of this structural transition. Moreover, release of constraints imposed by the MPER trimer contacts in the prehairpin intermediate may permit the refolding of each gp41 chain independently of the other two in order to attain the still more stable trimer-of-hairpins state. Indeed, mutations in the HRN region of gp41 that impair fusion kinetics by destabilizing the six-helix bundle enhance viral sensitivity to 2F5 mAb (43). A coiled coil-to-lipid-embedded helix transition is likely to be a common theme in viral fusion proteins since many protein ectodomains contain characteristic hydrophobic and often relatively Trp-rich membrane-proximal sequences (44).
Third, the MPER of gp41 is thought to play a role in a final conformational step of protein refolding in the fusion pathway, although no structural details have been available (39). In the completed postfusion six-helix bundle core, the outer-layer HRC regions have zipped up alongside the HRN trimer surface, which brings MPER into proximity with residues connecting the fusion peptide and the HRN coiled coil (Figure 5c). Recent studies suggest that highly favorable interactions between these two membrane-proximal elements of postfusion gp41 can facilitate the transition from a “hemifusion” state to complete membrane fusion (11, 39, 42). A comparable step has been defined in the extensively studied system of influenza virus hemagglutinin (45). It has also been proposed that the MPER of gp41 can interact with the fusion peptide to promote membrane perturbations that ensure fusion pore opening (46, 47). It is possible that the C-terminal ectodomain segments of gp41 participate in the assembly of postfusion trimers into an oligomeric, lipidic fusion pore (42). Alternatively, the lipid-embedded helical state of MPER might directly couple to the process of membrane deformation and fusion (14, 46, 47). Further studies are required to determine the precise mechanism by which MPER contributes to fusion pore formation and thus merger of the two bilayers.
Inhibitors of HIV-1 Entry
Viral entry is an important part of the HIV-1 life cycle that can be successfully inhibited. Two such antiviral drugs are the fusion inhibitor enfuvirtide (T-20) and the CCR5 antagonist maraviroc, which are used as salvage therapy for a growing number of AIDS patients who no longer respond to antiretroviral therapy (48). Peptides derived from the C-terminal ectodomain of gp41 are potent inhibitors of HIV-1 infection (49, 50). These inhibitors, such as C34 and T-20 (see Figure 1a), are thought to work by binding to the trimeric coiled coil of the postfusion gp41 inner core (6, 48). Nonetheless, T-20 contains an additional MPER sequence (at positions 670–673) whose binding site extends beyond this HRN coiled-coil region yet provides the key determinant of inhibitory activity in T-20 (Figure 1a) (50). A second-generation fusion inhibitor, T-1249, designed to overcome viral resistance to T-20, includes the additional HRN pocket-binding sequence and demonstrates enhanced potency and a different resistance profile (51). Moreover, C34 is fully active in vitro against T-20-resistant viral strains (52), reinforcing the suggestion that the target for the antiviral effect of T-20 may be distinct from that of C34 (42). T-20 may retard or block the MPER coiled coil-to-helix transition postulated here to be essential for gp41 activation, thereby disrupting trimer-of-hairpins formation and blocking viral entry. Consistent with this mode of action, there is substantial evidence to indicate that T-20 acts on the transiently exposed prehairpin state of gp41 rather than its native conformation or a post-lipid mixing stage (11). The trimeric MPER crystal structure may serve as a staring point for elucidating the mechanism of action of T-20 and how its antiviral effect is abrogated by the evolution of resistant viruses in vivo.
Development of orally bioavailable therapeutic agents for inhibition of HIV-1 membrane fusion is the subject of intense pharmaceutical efforts. Discovery of small-molecule inhibitors that target the gp41 HRN coiled-coil pocket represent an attractive approach in this endeavor. Indeed, a set of structurally related chemical compounds have been identified that inhibit formation of the six-helix bundle with antiviral activity in the low micromolar range (53). Based on the crystal structure of C22-pT, native gp41 is expected to contain a prominent central cavity in the MPER coiled-coil trimer (Figure 3e). The size of this cavity (approximately 440 Å3) makes it ideal for binding by a small molecule of roughly 500 Da, raising the possibility that it could serve as a target for drug development. Interestingly, an MPD molecule is observed to be anchored to the cavity in the C22-pT crystal structure, with favorable hydrophobic and hydrogen-bonding interactions between the bound MPD and the participating amino acid residues (Figure 3e). The availability of the atomic structure of C22-pT alone or in combination with a specific cavity-binding molecule (e.g., MPD) would enable rational drug development approaches. For example, chemical libraries can be screened with C22-pT to identify novel cavity-binding compounds.
Implications for Vaccine Design
Intensive efforts have been directed toward creation of a soluble recombinant Env (gp140) protein that forms the native trimeric structure as found on the infectious virion for vaccine and/or structural studies over the past decade. There are many hurdles to the production of such a trimeric gp140 that is truncated immediately before the transmembrane anchor (i.e., just C-terminal to the MPER). A major issue is that the native state of Env trimer is labile, because the inter-subunit interactions (gp120-gp41 and gp41-gp41) are non-covalent and weak; this inherent instability is essential for unleashing the postfusion trimer-of-hairpins conformati54. Hence several protein engineering strategies have been adopted to stabilize gp140 trimers for use as immunogens (54–59). Our studies suggest that trimerization of native gp41 is initiated at the C terminus of its ectodomain by formation of a labile trimeric α-helical coiled coil. Our structural data of the stable C22-pT subdomain should guide future attempts to design and engineer the native gpl40 trimer for structural analysis and for assessment of its potential as a vaccine immunogen.
ACKNOWLEDGMENTS
We thank D. Eliezer, N. Kallenbach and N. Lue and for comments on manuscript; and John Schwanof for assistance at beamline X4A at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY).
ABBREVIATIONS
- HIV-1
human immunodeficiency virus type 1
- NAbs
neutralizing antibodies
- Env
envelope glycoprotein
- HR
heptad repeat
- mAbs
monoclonal antibodies
- HPLC
high-performance liquid chromatography
- CNBr
cyanogen bromide
- CD
circular dichroism
- TBS
Tris-buffered saline
- GuHCl
guanidine hydrochloride
- [θ]222
molar ellipticity at 222 nm
- rms
root mean square
- SPR
surface plasmon resonance.
Footnotes
This work was supported by National Institutes of Health Grants R01 AI42382 (to M.L.) and R01 AI45463 and R37 AI36082 (to J.P.M.).
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (entry 3G9R).
REFERENCES
- 1.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 2.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathogens. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gpl20 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 7.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 8.Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 9.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 10.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 2005;79:1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun ZY, Oh KJ, Kim M, Yu J, Brusic V, Song L, Qiao Z, Wang JH, Wagner G, Reinherz EL. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Shu W, Ji H, Lu M. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry. 1999;38:5378–5385. doi: 10.1021/bi990199w. [DOI] [PubMed] [Google Scholar]
- 16.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 19.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D. 1993;49:129–149. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 20.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likehood method. Acta Crystallog.r D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Schomaker V, Trueblood KN. Correlation of internal torsional motion with overall molecular motion in crystals. Acta Crystallog.r B. 1998;54:507–514. [Google Scholar]
- 23.Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- 24.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallog. D Biol. Crystallog.r. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallog.r. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Yong W, Deng Y, Kallenbach NR, Lu M. Atomic structure of a tryptophan-zipper pentamer. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16156–16161. doi: 10.1073/pnas.0405319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenz O, Dittmar MT, Wagner A, Ferko B, Vorauer-Uhl K, Stiegler G, Weissenhorn W. Trimeric membrane-anchored gp41 inhibits HIV membrane fusion. J. Biol. Chem. 2005;280:4095–4101. doi: 10.1074/jbc.M411088200. [DOI] [PubMed] [Google Scholar]
- 28.Shang L, Yue L, Hunter E. Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection. J. Virol. 2008;82:5417–5428. doi: 10.1128/JVI.02666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbury PB, Kim PS, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 30.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 32.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 33.de Rosny E, Vassell R, Jiang S, Kunert R, Weiss CD. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J. Virol. 2004;78:2627–2631. doi: 10.1128/JVI.78.5.2627-2631.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry. 2007;46:1398–1401. doi: 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- 35.Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, Haynes BF. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gpl20 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schibli DJ, Montelaro RC, Vogel HJ. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40:9570–9578. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 38.Salzwedel K, West JT, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellamy-McIntyre AK, Lay CS, Baar S, Maerz AL, Talbo GH, Drummer HE, Poumbourios P. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J. Biol. Chem. 2007;282:23104–23116. doi: 10.1074/jbc.M703485200. [DOI] [PubMed] [Google Scholar]
- 40.Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saez-Cirion A, Arrondo JL, Gomara MJ, Lorizate M, Iloro I, Melikyan G, Nieva JL. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 2003;85:3769–3780. doi: 10.1016/S0006-3495(03)74792-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kliger Y, Gallo SA, Peisajovich SG, Munoz-Barroso I, Avkin S, Blumenthal R, Shai Y. Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J. Biol. Chem. 2001;276:1391–1397. doi: 10.1074/jbc.M004113200. [DOI] [PubMed] [Google Scholar]
- 43.Follis KE, Larson SJ, Lu M, Nunberg JH. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 2002;76:7356–7362. doi: 10.1128/JVI.76.14.7356-7362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallaher WR, Ball JM, Garry RF, Griffin MC, Montelaro RC. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res. Human Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shnaper S, Sackett K, Gallo SA, Blumenthal R, Shai Y. The C- and the N-terminal regions of glycoprotein 41 ectodomain fuse membranes enriched and not enriched with cholesterol, respectively. J. Biol. Chem. 2004;279:18526–18534. doi: 10.1074/jbc.M304950200. [DOI] [PubMed] [Google Scholar]
- 47.Suarez T, Gallaher WR, Agirre A, Goni FM, Nieva JL. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 2000;74:8038–8047. doi: 10.1128/jvi.74.17.8038-8047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug. Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 49.Jiang S, Lin K, Strick N, Neurath AR. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 50.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eron JJ, Gulick RM, Bartlett JA, Merigan T, Arduino R, Kilby JM, Yangco B, Diers A, Drobnes C, DeMasi R, Greenberg M, Melby T, Raskino C, Rusnak P, Zhang Y, Spence R, Miralles GD. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J. Infect. Dis. 2004;189:1075–1083. doi: 10.1086/381707. [DOI] [PubMed] [Google Scholar]
- 52.Rimsky LT, Shugars DC, Matthews TJ. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13938–13943. doi: 10.1073/pnas.0601036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 2000;74:4746–4754. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatatos L, Lim M, Cheng-Mayer C. Generation and structural analysis of soluble oligomeric gpl40 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res. Human Retroviruses. 2000;16:981–994. doi: 10.1089/08892220050058407. [DOI] [PubMed] [Google Scholar]
- 57.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gpl20 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dey AK, David KB, Klasse PJ, Moore JP. Specific amino acids in the N-terminus of the gp41 ectodomain contribute to the stabilization of a soluble, cleaved gpl40 envelope glycoprotein from human immunodeficiency virus type 1. Virology. 2007;360:199–208. doi: 10.1016/j.virol.2006.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng Y, Liu J, Zheng Q, Yong W, Lu M. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure. 2006;14:889–899. doi: 10.1016/j.str.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuikem C, Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Wolinsky S, Korber B. HIV Sequence Compendium 2008. Los Alamos, New Mexico.: Los Alamos National Laboratory, Thoretical Biology and Biophysics; 2008. [Google Scholar]